Sample Preparation for In Situ Cryotomography of Mammalian Cells
Summary
This method provides an accessible and flexible protocol for the preparation of electron microscopy (EM) grids for in situ cellular cryotomography and correlative light and electron microscopy (CLEM).
Abstract
In situ cellular cryotomography is a powerful technique for studying complex objects in their native frozen-hydrated cellular context, making it highly relevant to cellular biology and virology. The potential of combining cryotomography with other microscopy modalities makes it a perfect technique for integrative and correlative imaging. However, sample preparation for in situ cellular tomography is not straightforward, as cells do not readily attach and stretch over the electron microscopy grid. Additionally, the grids themselves are fragile and can break if handled too forcefully, resulting in the loss of imageable areas. The geometry of tissue culture dishes can also pose a challenge when manipulating the grids with tweezers. Here, we describe the tips and tricks to overcome these (and other) challenges and prepare good-quality samples for in situ cellular cryotomography and correlative imaging of adherent mammalian cells. With continued advances in cryomicroscopy technology, this technique holds enormous promise for advancing our understanding of complex biological systems.
Introduction
In situ cellular cryotomography is a powerful technique allowing for the study of biologically-relevant structures in cells without chemical fixation. By attaching cells to EM grids and plunge-freezing the grids in a cryogen, objects of interest are frozen in their natural cellular contexts without the formation of crystalline ice from intracellular water1,2. Both chemical fixation and crystalline ice formation can disrupt the structures of relevant molecules, such as proteins and lipids, reducing the biological accuracy of images obtained using these techniques3,4. In tomography, grids are imaged at incremental angles using electron microscopy, and these images are then used to construct three-dimensional representations of the target region imaged5. In situ cryotomography can be used alongside other microscopy techniques for integrative and correlative imaging, such as cryofluorescence imaging, soft x-ray tomography, and cryoFIB/SEM (cryogenic Focused Ion Beam/Scanning Electron Microscopy)6,7,8,9,10,11. Integration of multiple techniques allows for more information to be obtained about a structure or process than any single microscopy technique could achieve.
Despite all of the benefits of in situ cellular cryotomography, sample preparation can prove to be challenging for a variety of reasons. Due to their fragility, forceful manipulation of electron microscopy grids can lead to damage, with the thin carbon layer in particular being delicate and prone to tearing, reducing the imageable area of the grids. Electron microscopy grids are also difficult to manipulate due to their small size and are prone to becoming detached from the surface of the wells or microslide used to grow cells. Manipulation of the grids within the wells or microslides can prove difficult due to the geometry of these. Improper preparation of the grids (e.g., allowing them to float) can lead to low cell density and reducing the number of potential imaging areas, especially when cells are not prone to attach to the grids themselves. For direct cellular cryotomography, cells must spread very thin, which can be disrupted for many reasons, including improper temperatures or rough handling of the grids.
Through a variety of optimizations, the techniques presented in this article are meant to handle these most common pitfalls which arise during the preparation of electron microscopy grids for cryotomography. The use of 5/15 angled tweezers allows for the manipulation of grids within well plates or microslides. A fibronectin solution applied to both sides of the grids prior to plating makes floating grids less likely, which is beneficial in ensuring that grids have adequate cell density and that the grids are less likely to be damaged due to manipulation. By keeping the grids incubated at 37°C until just before plunge freezing, we also ensure that the cells are kept in a comfortable environment to prevent the cells from retracting their thin edges. Blotting the grids from the back side also prevents damage to the cells from mechanical force. Altogether, these measures increase the success rate of sample preparation for in situ cellular cryotomography studies, increasing the accessibility of this imaging approach.
Protocol
1. Grid preparation
NOTE: In the experimental design, plan for a maximum of 8-12 grids total per plunge-freezing session and 4-5 grids per well. More than that will lead to a very long plunge-freezing session, which may cause increased cell stress, ice contamination, and user errors.
- Glow discharge
- Load an appropriate number of grids with a perforated carbon support film to be glow discharged at 15 mA for 45 s at 0.39 mBar atmosphere in a glow discharge device.
- Make sure the carbon side of the grid is facing up. When finished, transfer the grids to a clean container lined with filter paper (Figure 1A1).
NOTE: Other grids may be used as long as the material is not toxic to cells (which excludes any copper grid). Finder grids are useful for correlative studies. Grids made of other materials are also possible. Many labs have had good results growing cells on top of SiO2 support grids8,9.
- Adherent treatment
- Transfer the grids, fibronectin, PBS, plates, and tweezers to a biosafety cabinet. Treat both sides of the grids with 20 µg/mL bovine fibronectin by micropipetting two generously sized dots of adherent solution on a sterile surface, such as the lid of a 6 well plate. Ensure that the dots are large enough to envelop the entire grid (around 100 µL is recommended).
NOTE: Prior to treatment with the selected adherent solution, there is the option of photo-micropatterning the grid here to optimize the attachment of cells on the center of grid squares. Reducing the ability of cells to attach to the bars will increase the number of imageable cells. This is ideal for experiments involving cryoFIB milling. - Use 5/15 tweezers to manipulate the grids. Be sure to treat both sides of the grid in solution.
NOTE: Other adhesive solutions may be used (fibrinogen, collagen, other extracellular matrix components, etc.). Fibronectin is used here, as it gives good results with a wide variety of cell lines. (U2OS, HeLa, Vero, Calu-3, Tzm-bl). The 5/15 tweezer is useful as its geometry allows improved maneuverability around the 6-well plates, 35 mm dishes, and micro slides. This results in less mechanical stress on the grids and a more intact carbon film. Make sure all required materials are placed in the biosafety cabinet prior to beginning, as this reduces the possibility for grid contamination.
- Transfer the grids, fibronectin, PBS, plates, and tweezers to a biosafety cabinet. Treat both sides of the grids with 20 µg/mL bovine fibronectin by micropipetting two generously sized dots of adherent solution on a sterile surface, such as the lid of a 6 well plate. Ensure that the dots are large enough to envelop the entire grid (around 100 µL is recommended).
- Attaching grids
- Once treated with bovine fibronectin, the grid will become sticky. Using 5/15 tweezers, gently touch the non-carbon surface of the grid to the bottom of an empty dish/plate and open the tweezers. The grid should easily attach to the new surface (Figure 1A2).
NOTE: Try to avoid placing grids in the direct center or the edges of the dish/plate. When adding cells, there is a tendency for cell density to accumulate in the center of the plate. This can result in grids with excessive cellular density. Alternatively, 3D-printed grid holders may be used here instead of attaching cells directly to the surface of the dish/plate12.
- Once treated with bovine fibronectin, the grid will become sticky. Using 5/15 tweezers, gently touch the non-carbon surface of the grid to the bottom of an empty dish/plate and open the tweezers. The grid should easily attach to the new surface (Figure 1A2).
- Incubation
- Incubate the grids for 30 min in the fibronectin adherent solution (20 µg/mL). To ensure that the grids do not become dry during this process, micropipette 1-2 drops of more adherent solution directly on top of grids every 15 min (Figure 1A3).
NOTE: The incubation period will vary depending on the adherent solution used. For bovine fibronectin, the recommended time is 30 min.
- Incubate the grids for 30 min in the fibronectin adherent solution (20 µg/mL). To ensure that the grids do not become dry during this process, micropipette 1-2 drops of more adherent solution directly on top of grids every 15 min (Figure 1A3).
- Washing
- Following the incubation period, remove excess adherent solution via micropipetting. Wash the grids by micropipetting drops of PBS directly on top of the grids. Repeat this 2-3 times. (Figure 1A4).
- Sterilization
- Use UV light to sterilize the grids in the biosafety cabinet for 1 h. Place the grids as close to the UV source as possible to maximize sterilization (Figure 1A5). To ensure that the grids do not become dry during this process, micropipette 1-2 drops of PBS directly on top of grids every 10 min.
NOTE : Steps 1.4-1.6 can be performed concurrently.
- Use UV light to sterilize the grids in the biosafety cabinet for 1 h. Place the grids as close to the UV source as possible to maximize sterilization (Figure 1A5). To ensure that the grids do not become dry during this process, micropipette 1-2 drops of PBS directly on top of grids every 10 min.
2. Seeding grids
- Count cells:
- Detach and count the cells using mechanical or enzymatic methods. Determine the optimum number of cells to use for seeding prior to counting. For U2OS seeded in a 6-well plate, use anywhere between 6 x 104 to 1.6 x 105 cells per well (Figure 1B1).
NOTE: The number of cells seeded will vary according to the cell type, the surface area of the well, dish or microslide used, the time between seeding and plunge freezing, and the experimental design. Test a range of cell numbers to identify conditions that will result in 0.25-1 cells per grid square at the time of plunge freezing. It is important to note that high cell density reduces the speed of heat transfer and may impact the vitrification process. To combat this, some labs have found success using cell strainers, which act to prevent cell clumps from forming13. Finally, this protocol results in the random attachment of cells to the carbon layer. If a targeted attachment is necessary, photo-micropatterning may be used (see step 1.3)14.
- Detach and count the cells using mechanical or enzymatic methods. Determine the optimum number of cells to use for seeding prior to counting. For U2OS seeded in a 6-well plate, use anywhere between 6 x 104 to 1.6 x 105 cells per well (Figure 1B1).
- Adding cells and growth medium : Add cells via micropipette to the wells, avoiding bubbles (Figure 1B2). In a 6-well plate, a total well volume of 1.5-2.0 mL is recommended.
NOTE: It is recommended to wet around the grids carefully before adding the entire volume. This will help prevent grids from detaching and floating in the solution. Gently move the plate side-to-side to facilitate homogeneous cell distribution on the well. Do not swirl the plate in a circular motion, as this will result in excessive cell density allocating in the center of the well. - Incubation: Incubate the slides at 37 °C for an appropriate time according to the experimental design, which will depend on downstream applications. For this example (transfection of HIV molecular clones), incubate between 16-24 h (Figure 1B3).
NOTE: Longer incubation times will result in more cell division cycles. Therefore, adjust the starting number of cells for seeding accordingly.
3. Transfection
- Preparing transfection mixture:
- Prepare an appropriate cationic liposome reagent for plasmid transfection into the selected cell line.
- In this transfection example, use 1 µg of total plasmid DNA per well (in a 6 well plate) to be transfected at a 1:3 ratio of HIViΔEnv plasmid to psPAX2 plasmid. For each well, dilute 1 µg of DNA into 50 µL of serum-free DMEM. Homogenize the mixture via repeatedly micropipetting or gentle swirling of the plate.
- For each well, dilute 3 µL of transfection reagent in serum-free DMEM, then homogenize the mixture again by micropipetting or gentle swirling. Add the entire diluted transfection reagent mixture to the diluted DNA mixture (step 3.1.2) and incubate at room temperature (RT) for 15 min (Figure 1C1).
NOTE: While this mixture is incubating, replace the media in the wells with 1.5 mL of fresh DMEM. Be careful not to dislodge the grids from the bottom of the well.
- To apply the transfection mixture, pipette 100 µL of the mixture from step 3.1.3 dropwise to each well, concentrating the drops over the grids to ensure contact (Figure 1C2). Change the growth medium 16-24 h after transfection (Figure 1C3).
4. Plunge freezing
NOTE: Keep cells at 37 °C until needed for blotting. Additionally, be sure to make a note of the carbon side of the grids throughout the process.
- Cell density
- Check the grids in an inverted optical microscope and note cell densities on each grid.
NOTE: Grids with less than 0.25 cells per grid square (or one cell every 4 grid squares), are considered 'empty'. Grids with 0.25-1 cells per grid square are considered 'normal'. Grids with more than 1 cell per grid square are considered 'full'. - Take note of the cell density on each grid to allow the dialing of blotting times later during plunge-freezing. Add 2-3 mL of PBS to an empty plate/dish and warm to 37 °C.
- Check the grids in an inverted optical microscope and note cell densities on each grid.
- Preparing fiducials
- To prepare gold fiducials for downstream cryotomography applications, pipette 50 µL of 10 nm colloidal gold bead solution into a clean 1.5 mL tube. Add 2 µL of 1 mg/mL BSA to the tube and homogenize by micropipetting repeatedly.
- Centrifuge the tube at 15,000-20,000 x g for 15 min. Micropipette to remove as much of the supernatant as possible without disturbing the pellet.
NOTE: The pellet will be very loose. - Add 50 µL of PBS to the pellet and resuspend. Centrifuge the tube again at 15,000-20,000 x g for 15 min.
- Using a 10 µL tip, directly aspirate 4 µL of the pellet, and transfer it to a clean 1.5 mL tube (Figure 1D1). Use 1 µL of the washed and concentrated gold per grid.
NOTE: The type of fiducials used depends on downstream applications. For TEM tomography, use 10 nm gold beads. For soft X-ray tomography, use 200 nm gold beads. For correlation microscopy, use fluorescent latex beads. Fiducials are normally not necessary for experiments involving cryoFIB milling, as the milling process will destroy them.
- Blotting
- Set up the environmental chamber to 95% humidity and 30°C (Figure 1D1). Select a grid using the 5/15 tweezers.
- Wash the grids with PBS and transfer them to plunging tweezers (Figure 1D2). Once secure, remove the 5/15 tweezers and slide the clamp on the plunging tweezers.
- Insert the grid into the plunge freezer, keeping the clamp secure (Figure 1D3). Add 1 µL of gold fiducials to the back side of the grid (Figure 1D4). Add 2-3 µL of PBS to the carbon side of the grid. Use automated blotting to blot the back side of the grid and plunge freeze into liquid ethane (Figure 1D5).
NOTE: It is recommended to have between 3-4 µL of volume per grid for optimal blotting. Grids may retain a variable amount of PBS after washing. Adjust the added volume of PBS to account for pre-existing liquid retention on the grid. It is recommended that the gold fiducials be added to the back of the grid. Addition to the front can result in pools of gold fiducials near the point of insertion, whereas addition from the back results in a more homogenized solution. Blotting duration depends on the cell dentistry of the grid. 2 s for empty, 3 s for normal, and 4 s for full grids, respectively. The type of plunge freezer available may determine how blotting is approached: The Leica GP2 plunge freezer is the one used in this protocol and is capable of automated single-sided blotting by default. Thermo Fisher Vitrobot does automated double-sided blotting, though this often damages the cells attached to the carbon side. Manual blotting in the Vitrobot is also possible, as well as manual gravity plungers.
Representative Results
Following the cotransfection of HIViGFPΔEnv and psPAX2, all of the grids had minimal tearing in the carbon layer. Grids were imaged using phase light microscopy and fluorescent light microscopy 24 h after incubation with the transfection reagent (Figure 2). Cells on both the mock grids and the co-transfected grids contained viable cells in multiple grid squares.
psPAX2 codes for all structural and enzymatic proteins of HIV-1 without any fluorescence tagging. HIViGFPΔEnv is similar to psPAX2 but with codes for a GFP-tagged HIV Gag protein. Both plasmids are ΔEnvelope. The cotransfection results in native-like assembly and budding of fluorescent HIV-1 particles, making this a great system for CLEM studies of HIV in Biosafety Level 1 condition. The co-transfected grids showed a subset of cells exhibiting green fluorescence, indicating successful cotransfection. No cells on the mock grid exhibited fluorescence, further validating the cotransfection using HIViGFPΔEnv and psPAX2. After viewing the grids using light-based microscopy, grids were plunge frozen and moved to long-term storage in liquid nitrogen.
Figure 3 depicts results from grids produced using the same experimental method but utilizing slightly different plasmid constructs. U2OS-containing grids were co-transfected using different HIV clones (HIVmCherryΔEnv, and NL4-3ΔEnvGFP at a 1:6 ratio). Since a higher mass of fluorescently tagged plasmids were used, these grids enabled the observation of a larger number of transfected cells, providing an advantage while capturing images using cryoCLEM and cryoET. Using cryoCLEM, full grid atlases were generated for each grid using cryofluorescence microscopy to record the locations of all co-transfected cells. With the locations of cells known, cryoET was performed. A full low-magnification grid atlas was collected and overlaid with the fluorescent atlas collected at cryofluorescence (Figure 3A). Cryotomograms were collected at cellular sites capturing intricate details of the viral life cycle, including the assembly and budding of HIV from cells (Figure 3B).
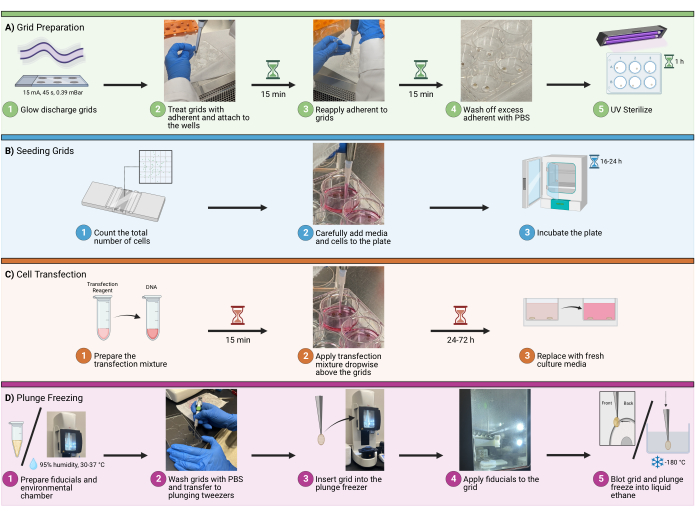
Figure 1: Cell seeding on grids workflow. A schematic depicting the overall procedure to seed cells on cryoEM grids. The process is divided into four major steps, including (A) preparing grids in the wells for seeding, (B) adding the appropriate amount of cells to each well, (C) the optional transfection of cells for fluorescent imaging, and (D) the plunge freezing of grids to allow for vitrification of the sample. Please click here to view a larger version of this figure.
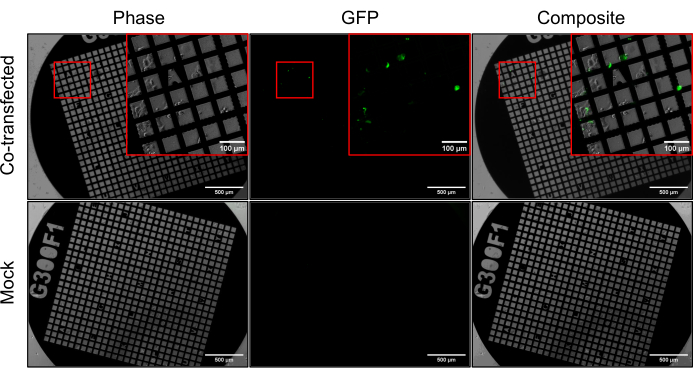
Figure 2: Cotransfection of U2OS using HIViGFPΔEnv and psPAX2. U2OS cells were co-transfected with GFP-containing HIViGFPΔEnv and psPAX2 in a 1:3 ratio. Grids were imaged by phase contrast and fluorescence microscopy. Cells that are shown to have GFP expression indicate successful cotransfection. Scale bar: 500 µm. Scale bar (insets): 100 µm. Please click here to view a larger version of this figure.
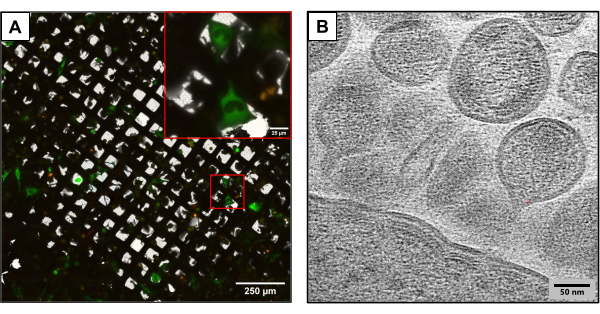
Figure 3: Potential downstream cryo-based methods. (A) A cryoCLEM image with co-transfected U2OS cells. Cells in green represent HIV-producing cells and are used to measure cotransfection success. Red puncta represent mCherry tagged HIV-1 Gag. Scale bar: 250 µm. Scale bar (inset): 25 µm (B) A cryoET image of multiple HIV particles budding from the plasma membrane of U2OS cells. Scale bar: 50 nm Please click here to view a larger version of this figure.
Discussion
Here, we have provided an accessible, flexible, and reproducible protocol to seed cells on electron microscopy grids for in situ cryoelectron tomography applications. This method can be easily adapted to fit the needs of downstream applications and/or experimental requirements. In addition to great flexibility, we have described a workflow that optimizes and reduces common pitfalls in grid seeding, notably extensive damage to the carbon layer, low cell density, and poor structural integrity of thin cell projections.
Although the protocol described here does provide several alternatives, there are some critical steps that should be followed to optimize general outcomes. One of the biggest issues with grid cell seeding is the detachment and floating of grids from the well or microslide. Therefore, it is important to fully wet the grid with an adherent solution on both sides and prevent it from drying during the incubation period. If using 3D-printed grid holders, be aware that multiple changes of media to these holders have the potential to produce floating grids since the air trapped under the grid can force it out of the holder.
Our choice of tweezers also improves grid quality in the way of providing a geometrically favorable way of manipulating the grids without extensive grid bending that would damage the carbon layer. Keeping the cells at 37 °C for as long as possible before plunging reduces cell suffering and improves the number of thin imageable cells on the grid. Finally, blotting from the gold side will protect the cells from harsh mechanical forces that could lead to damage to fragile cellular structures.
While not included in this protocol, grid photo-micropatterning has been shown to increase the number of imageable cells by optimizing their attachment to the center of grid squares14. Finally, 3D-printed grid holders have recently been used to reduce grid damage by limiting direct grid manipulation12.
It may be important to note that this protocol is optimized for imaging thin edges and protrusions from cells for the application of cryotomography. We suggest troubleshooting a variety of conditions from our recommendations in the protocol to find the best outcome for the downstream applications of choice. Overall, this protocol provides a reliable yet versatile method of seeding cells onto grids that can be tweaked for specific needs.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We would like to thank the Mansky lab for access to plunge-freezing equipment. Parts of this work were carried out in the characterization facility of the University of Minnesota, which receives partial support from the National Science Foundation (NSF) through the Materials Research Science and Engineering Center (MRSEC; Award Number DMR-2011401) and the National Neuroscience Curriculum Initiative (NNCI; Award Number ECCS-2025124) programs. We would like to thank funding from the Behavior of HIV in Viral Environments center (B-HIVE; 1U54AI170855-01) and the Duke Center for HIV Structural Biology (DCHSB; U54AI170752) center.
Materials
| 10 nm colloidal gold bead solution | Sigma-Aldrich | 741957 | |
| 6 well multidish, 100/CS | Fisher Scientific | FB012927 | |
| Allegra V-15R Benchtop Centrifuge, IVD 120 V 60 Hz | Beckman-Coulter | C63125 | |
| Au G300F1 with R2/2 Quantifoil carbon | Quantifoil | TEM-G300F1-AU | |
| Bovine serum albumin | MilliporeSigma | A9647 | |
| BRAND counting chamber BLAUBRAND Neubauer improved | Sigma-Aldrich | BR717805-1EA | |
| DMi1 Inverted Microscope | Leica | 22A00G119 | |
| Dulbecco's modified eagle's medium – high glucose, no glutamine | Gibco | 11-960-044 | |
| Dumont 5/15 tweezer | Electron Microscopy Sciences | 0103-5/15-PO | |
| EM GP2 | Leica | 587085 | Automated plunge freezer |
| Fetal Bovine Serum | Gibco | A5209 | |
| Fibronectin from bovine plasma, cell culture grade | MilliporeSigma | F1141 | |
| GenJet version II in vitro DNA transfection reagent | SignaGen Laboratories | SL100489 | |
| GlutaMAX I 100x | Fisher Scientific | 35050061 | Media supplement |
| Neslab EX-211 Heating Circulator | Neslab | Out of production | Water bath for media warming |
| Original Portable Pipet-Aid Pipette Controller | Drummond Scientific | 4-000-100 | |
| PBS, pH 7.4 | Gibco | 10010023 | |
| Pelco easyGlow device | Pelco | 91000S | Glow discharge device |
| Penicillin-Streptomycin | Sigma-Aldrich | P0781 | Media supplement |
| Pipetman P1000, 100–1000 µL, Metal Ejector | Gilson | F144059M | |
| Pipetman P2, 0.2–2 µL, Metal Ejector | Gilson | F144054M | |
| Pipetman P20, 2–20 µL, Metal Ejector | Gilson | F144056M | |
| Whatman number 2 filter paper, 55 mm | Whatman | 28455-041 | Blotting paper |
References
- Zhang, P., Mendonça, L. Analysis of Viruses in the Cellular Context by Electron Tomography. Encyclopedia of Virology. 1, 242-247 (2021).
- Fäßler, F., Dimchev, G., Hodirnau, V. -. V., Wan, W., Schur, F. K. M. Cryo-electron tomography structure of Arp2/3 complex in cells reveals new insights into the branch junction. Nature Communications. 11, 6437 (2020).
- McDowall, A. W., et al. Electron microscopy of frozen hydrated sections of vitreous ice and vitrified biological samples. Journal of Microscopy. 131 (Pt 1), 1-9 (1983).
- Stewart, M., Vigers, G. Electron microscopy of frozen-hydrated biological material. Nature. 319 (6055), 631-636 (1986).
- Gan, L., Jensen, G. J. Electron tomography of cells. Quarterly Reviews of Biophysics. 45 (1), 27-56 (2012).
- Yang, J. E., Larson, M. R., Sibert, B. S., Shrum, S., Wright, E. R. CorRelator: Interactive software for real-time high precision cryo-correlative light and electron microscopy. Journal of Structural Biology. 213 (2), 107709 (2021).
- Mendonça, L., et al. Correlative multi-scale cryo-imaging unveils SARS-CoV-2 assembly and egress. Nature Communications. 12 (1), 4629 (2021).
- Klumpe, S., et al. A modular platform for automated cryo-FIB workflows. eLife. 10, e70506 (2021).
- Wagner, F. R., et al. Preparing samples from whole cells using focused-ion-beam milling for cryo-electron tomography. Nature Protocols. 15 (6), 2041-2070 (2020).
- Klein, S., et al. IFITM3 blocks influenza virus entry by sorting lipids and stabilizing hemifusion. Cell Host & Microbe. 31 (4), 616.e20-633.e20 (2023).
- Shah, P. N. M., et al. Characterization of the rotavirus assembly pathway in situ using cryoelectron tomography. Cell Host & Microbe. 31 (4), 604.e4-615.e4 (2023).
- Fäßler, F., Zens, B., Hauschild, R., Schur, F. K. M. 3D printed cell culture grid holders for improved cellular specimen preparation in cryo-electron microscopy. Journal of Structural Biology. 212 (3), 107633 (2020).
- Hoffmann, P. C., et al. Electron cryo-tomography reveals the subcellular architecture of growing axons in human brain organoids. eLife. 10, e70269 (2021).
- Toro-Nahuelpan, M., et al. Tailoring cryo-electron microscopy grids by photo-micropatterning for in-cell structural studies. Nature Methods. 17 (1), 50-54 (2020).

