Analysis of DNA Double-strand Break (DSB) Repair in Mammalian Cells
Summary
This article describes GFP-based fluorescence in vivo assays that separately quantify homologous recombination and nonhomologous end joining in mammalian cells.
Abstract
DNA double-strand breaks are the most dangerous DNA lesions that may lead to massive loss of genetic information and cell death. Cells repair DSBs using two major pathways: nonhomologous end joining (NHEJ) and homologous recombination (HR). Perturbations of NHEJ and HR are often associated with premature aging and tumorigenesis, hence it is important to have a quantitative way of measuring each DSB repair pathway. Our laboratory has developed fluorescent reporter constructs that allow sensitive and quantitative measurement of NHEJ and HR. The constructs are based on an engineered GFP gene containing recognition sites for a rare-cutting I-SceI endonuclease for induction of DSBs. The starting constructs are GFP negative as the GFP gene is inactivated by an additional exon, or by mutations. Successful repair of the I-SceI-induced breaks by NHEJ or HR restores the functional GFP gene. The number of GFP positive cells counted by flow cytometry provides quantitative measure of NHEJ or HR efficiency.
Protocol
In this protocol we describe the method for analysis of DNA DSB repair with chromosomally integrated reporter constructs1,2 where DSBs are induced in vivo by transient expression a rare cutting endonuclease I-SceI3. The integrated assay provides the advantage of analyzing DSB repair within the chromosomal context. However, this protocol requires prolonged cell passaging, which may be problematic when working with primary cells.
An alternative approach is an extrachromosomal assay where reporter constructs are digested in vitro with I-SceI or HindIII endonucleases and then transfected into cells as linear DNA. The exctrachromosomal assay protocol4 is similar to the integrated assay described below with the following modifications. For the extrachromosomal assay omit the integration of reporter constructs and instead co-transfect the cells with 0.5 μg of NHEJ reporter plasmid linearized in vitro with I-SceI or HindIII and 0.1 μg of pDsRed2-N1 as transfection control. For the HR assay co-transfect 2 μg HR reporter plasmid linearized with I-SceI or HindIII and 0.1 μg pDsRed2-N1. Analyze the cells by FACS three days after transfection. The extrachromosomal assay allows for the analysis of DSB repair in primary isolates, avoiding extensive cell passaging, and facilitating the analysis of multiple cell lines.
1. Reporter Constructs for NHEJ and HR
The NHEJ reporter cassette5 contains a GFP gene with an engineered 3 kb intron from the Pem1 gene (GFP-Pem1). The Pem1 intron contains an adenoviral exon flanked by recognition sequences for HindIII and I-SceI endonucleases (Figure 1a) for induction of DSBs. The I-SceI sites are in inverted orientation. I-SceI has a nonpalindromic recognition sequence, hence two inverted sites generate incompatible DNA ends (Figure 1c). Incompatible DNA ends best mimic the naturally occurring DSBs. The intact NHEJ cassette is GFP negative as the adenoviral exon disrupts the GFP ORF. Upon induction of DSBs by I-SceI, the adenoviral exon is removed and NHEJ restores function of the GFP gene (Figure 2). The unique feature of this NHEJ reporter is that it can detect a wide spectrum of NHEJ events since the intron can tolerate deletions and insertions.
The HR reporter cassette1 (Figure 1b) is also based on the GFP-Pem1. In the HR cassette, the first exon of the GFP-Pem1 contains a 22 bp deletion combined with the insertion of three restriction sites, I-SceI/HindIII/I-SceI. The deletion ensures that GFP cannot be reconstituted by an NHEJ event. The two I-SceI sites are in inverted orientation, so that I-SceI digestion leaves incompatible ends (Figure 1c). The first copy of GFP-Pem1 is followed by a promoter-less/ATG-less first exon and intron of GFP-Pem1. The intact construct is GFP-negative. Upon induction of a DSB by I-SceI digestion the functional GFP gene is reconstituted by intramolecular or intermolecular gene conversion between the two mutated copies of the first GFP-Pem1 exon. Since the second copy of the GFP gene is lacking the first ATG codon and the second exon, crossing over or single-strand annealing will not restore the GFP activity. This design allows for the exclusive detection of gene conversion, which is the predominant HR pathway in mammalian cells (Figure 3).
2. Integration of the Reporter Constructs into the Genome
- Make a high quality preparation of NHEJ or HR reporter plasmid. For best results use Qiagen Endo Free Kit. Measure plasmid concentration and purity by absorbance at 260/280 nm.
- Linearize 10 μg of the reporter plasmid by digesting with 50 U of NheI in 50 μl (total volume of the reaction) for 6 hours in 37°C incubator (do not use the dry block, to prevent evaporation). Purify DNA from the digestion solution with Qiagen QiaexII Kit, elute DNA into 20 μl of 10 mM Tris-HCl pH 8.0. Measure DNA concentration and purity by absorbance at 260/280 nm. Usually, you will lose 20-30% of DNA during purification. Confirm the yield and digestion by running a small aliquot (2 μl) on a gel, along with an undigested reporter construct. The purified linearized reporter construct can be stored at 20°C.
- In preparation for transfection, bring the cells to the best growing condition. If starting from a frozen vial, split the cells twice before transfection. If starting from a plate of confluent cells, split them once.
- Grow the cells to 70-80% confluence (usually two days, if plated 5×105 cells/100 mm plate, for normal human fibroblasts). It is critical for the best transfection efficiency to bring the cells into logarithmic growth. Actively growing culture contains cells in M stage (seen as rounded cells attached to the surface).
- Transfect the cells using Amaxa Nucleofector following manufacturer’s recommendations. For fibroblasts use NHDF solution and program U20. For transfection, harvest the cells from two 100 mm plates (~2×106 cells). It is critical not to over trypsinize the cells. Stop trypsinization as soon as 80% of cells detached from the plate. Resuspend the cells in NHDF solution. Cells are sensitive to NHDF solution, therefore, minimize the incubation time and mix the cells gently. Transfect the cells with 0.5 μg of linearized reporter construct. These transfection conditions, when used with normal human fibroblasts result in the integration of a single copy of a reporter construct for the majority of integrants.
- Select the cells with chromosomally integrated reporter constructs by adding 1 mg/mL geneticin (G418) 24h after transfection. Continue selection for 7-10 days.
- Pick the G418 resistant colonies (you can pick individual colonies or pool the resistant clones). Expand the culture to approximately five 100 mm plates. Freeze three plates for future use and expand remaining two plates to the desired cell number (usually ten 100 mm plates are sufficient for five transfections).
- When the cells reach 70-80% confluence (two days after plating 5×105 cells per 100 mm plate for fibroblasts), the cells are ready to be transfected with I-SceI expressing plasmid.
3. Induction of DSB by Transient Expression of I-SceI Endonuclease
- Transfect ~2×106 cells (two plates) of logarithmically growing culture with a mixture of 5 μg I-SceI expressing plasmid and 0.1 μg pDsRed2-N1 (Clontech) as a transfection efficiency control using Amaxa Nucleofector (use NHDF solution and U20 program for fibroblasts).
- At the same time prepare three calibration controls for FACS by trasfecting ~2×106 cells with (i) 5 μg EGFP-expressing plasmid; (ii) 5 μg pDsRed2-N1; (iii) 5 μg of a control plasmid that does not express a fluorescent protein (we are using a plasmid encoding HPRT minigene).
- Grow the cells for four days to provide sufficient time for expression of GFP and DsRed proteins.
- (Optional) Three days after transfection verify the efficiency of transfection and expression of GFP and DsRed proteins by fluorescent inverted microscope with filters for detection of GFP and DsRed.
4. Analysis of the GFP+ and DsRed+ Cells by Flow Cytometry
- Four days after transfection harvest I-SceI transfected cells and the control cells. Resuspend the cells in 500 μL of PBS and transfer cells to the FACS tubes. At this stage it is critical to harvest all the cells and have cells of round shape. To achieve this, it is recommended to use a longer trypsinization time or a stronger trypsin. Confirm that 99% of cells are detached from the plate and have round shape by observing the cells under microscope. Keep the cells on ice protected from light (cover ice bucket with foil). It is possible to store the cells on ice for a few hours. For the best results analyze the cells immediately after harvesting.
- Calibrate FACS with GFP, DsRed, and the negative controls. Adjust voltage and color compensation in order to include all the fluorescent cells in the analysis (Figure 4). Keep in mind that the fluorescent intensity of the experimental samples will be lower than that of the controls.
- Analyze I-SceI transfected cells by FACS. We count at least 20,000 cells for each treatment. To prevent potential interference between GFP and DsRed fluorescence it is recommended to keep the percent of GFP+ or DsRed+ cells below 25%. If the percentage of DsRed+ or GFP+ cells is higher reduce the amount of pDsRed2-N1 or I-SceI plasmid. In our experiments we only needed to adjust the amount of pDsRed2-N1 plasmid, but not of the I-SceI plasmid.
- The percent of GFP+ cells corresponds to the efficiency of DNA DSB repair and the percent of DsRed+ cells indicates the efficiency of transfection. Calculate the relative efficiency of DNA DSB repair as the ratio of GFP+ cells to DsRed+ cells.
- The repaired end junctions may be sequenced to test the accuracy of repair. The reporter constructs contain E.coli origin of replication and kanamycin resistance genes (Figure 1). This enables rescuing the constructs by digesting genomic DNA with EcoRI enzyme, recircularization, and transformation into competent E.coli cells. The rescued plasmids are then analyzed by restriction digestion and sequencing.
5. Representative Results
Typical FACS results of the control transfections and of NHEJ and HR experiments are shown in Figures 4 and 5. Fluorescent intensity and the number of fluorescent cells will be lower in the I-SceI transfected cells containing integrated NHEJ and HR constructs (Figure 5) compared to the controls (Figure 4) due to the difference in the copy number of GFP and DsRed constructs in these cells. Each cell in the experiment contains only one copy of the integrated reporter construct, thus successful repair events will reconstitute the GFP gene in a fraction of the cells. Transfection with 0.1 μg pDsRed2-N1 in human fibroblasts delivers approximately 1 plasmid copy per cell.
The efficiency of NHEJ and HR is calculated as a ratio of GFP+/DsRed+ cells. NHEJ is a more efficient process than HR in human cells2. In human fibroblasts the NHEJ efficiency is typically 0.6-1.3, and HR efficiency is 0.05-0.3. Since DSB efficiency is measured as a ratio GFP+/DsRed+ cells variations in the mixture of I-SceI and DsRed2-N1 plasmids may affect the result. The plasmid quality may also affect the results by changing transfection efficiency. Therefore, to obtain consistent measurements, it is important to use the same plasmid mix within one experiment. Furthermore, the DSB repair efficiency is affected by cell type, cell cycle phase, and chromosomal location of the integrated constructs.
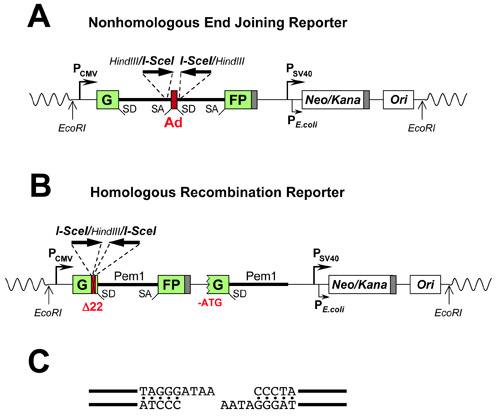
Figure 1. Reporter constructs for the analysis of DNA DSB repair by NHEJ (A) and HR (B). (C) Incompatible DNA ends generated by digestion of two inverted I-SceI sites. SD, splice donor; SA, splice acceptor; shaded squares, polyadenylation sites; Neo/Kana, single ORF controlled by two promoters: SV40 conferring neomycin resistance in mammalian cells; and β-lactamase conferring kanamicyn resistance in E. coli; Ori, E. Coli pUC origin of replication.
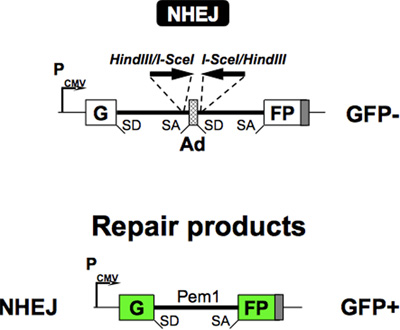
Figure 2. Reporter construct for the analysis of NHEJ. In this construct the GFP gene is inactive; however upon induction of a DSB and successful NHEJ the construct becomes GFP+. Shown below is a repair product of this reporter by NHEJ.
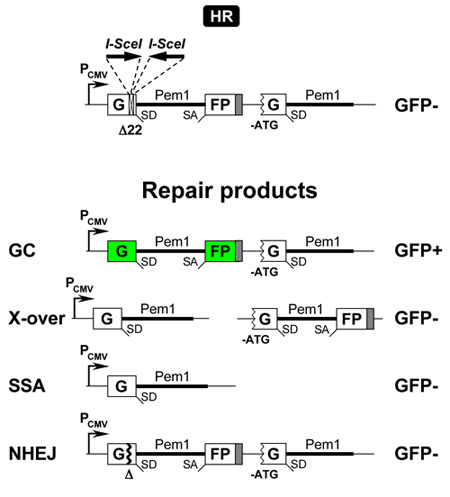
Figure 3. Reporter construct for the analysis of HR by gene conversion. Upon induction of the DSBs by I-SceI, HR by gene conversion (GC) reconstitutes an active GFP gene. Shown below are repair products of this reporter by the major DNA repair pathways: GC, crossing-over (X-
over), single strand annealing (SSA), and NHEJ. X-over repair product is shown for intramolecular recombination, exramolecular recombination will give the same product but located on one chromosome. Genes expressing active GFP protein (GFP+) are indicated by green color.
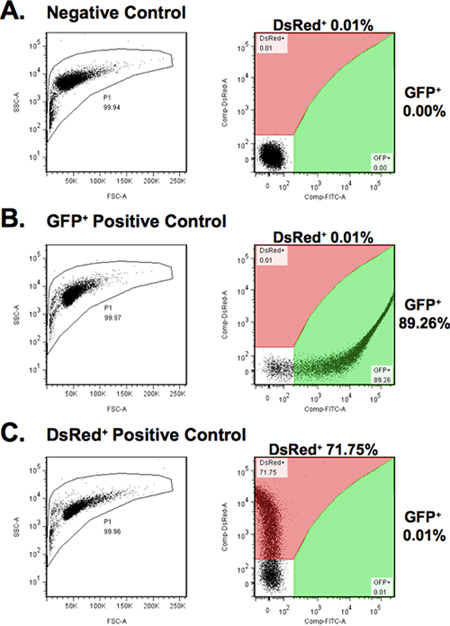
Figure 4. Calibration of the parameters for FACS analysis. (A) Fibroblasts transfected with the pHPRT plasmid, used as a negative control for excluding auto fluorescent cells. (B) Fibroblasts transfected with pEGFP plasmid, used for setting the gating for GFP+ cells (green area). (C) Fibroblasts transfected with pDsRed2-N1 plasmid, used for setting the gating for DsRed+ cells (red area).
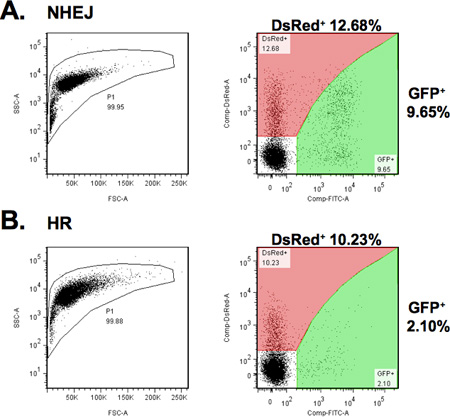
Figure 5. Typical results of the analysis of NHEJ (A) and HR (B) in normal human fibroblasts with chromosomally integrated reporter constructs.
Discussion
The fluorescent NHEJ and HR reporter assays provide a quantitative way for separately measuring each DSB repair pathway in vivo. The assays are very sensitive, as FACS can reliably detect 10 GFP+ cells in 20,000 cells. The assays may be adapted for measuring repair events in “real time” by detecting the appearance of GFP+ cells within minutes or hours after induction of DSBs2. Furthermore, the analysis of GFP+ cells does not rely on additional cell proliferation, allowing DSB repair to be analyzed at different cell cycle stages after treatment with various drugs that arrest cell division6. This provides an important advantage over reporter constructs based on reconstitution of a selectable marker.
The NHEJ process is intrinsically error-prone generating various small rearrangements at the ends junctions. The unique property of our NHEJ reporter is that repair events occur within an intron, which tolerates deletions and insertions, thus allowing detection of a wide spectrum of NHEJ events. The HR construct is based on the same modified GFP gene as the NHEJ construct allowing the comparative analysis of NHEJ and HR. The use of fluorescent NHEJ and HR assays will facilitate the studies of mammalian DSB repair.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The original GFP-Pem1 was a gift from Dr. Lei Li. This work was supported by grants from the NIH and the Ellison Medical Foundation to V.G. and A.S.
Materials
| Material Name | Type | Company | Catalogue Number | Comment |
|---|---|---|---|---|
| EndoFree Plasmid Maxi kit | Qiagen | 12362 | ||
| Qiaex II Gel Extraction Kit | Qiagen | 20021 | ||
| Amaxa Nucleofector | Lonza | AAD-1001 | ||
| Geneticin (G418) | Invitrogen | 11811-031 | ||
| pDsRed2-N1 | Clontech | 632406 | ||
| Round bottom tubes | BD Falcon | 352058 | FACS tubes |
References
- Mao, Z., Seluanov, A., Jiang, Y., Gorbunova, V. TRF2 is required for repair of nontelomeric DNA double-strand breaks by homologous recombination. Proc Natl Acad Sci U S A. 104, 13068-13073 (2007).
- Mao, Z., Bozzella, M., Seluanov, A., Gorbunova, V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst). 7, 1765-1771 (2008).
- Rouet, P., Smih, F., Jasin, M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci U S A. 91, 6064-6068 (1994).
- Mao, Z., Jiang, Y., Xiang, L., Seluanov, A., Gorbunova, V. DNA repair by homologous recombination, but not by nonhomologous end joining, is elevated in breast cancer cells. Neoplasia. 11, 683-691 (2009).
- Seluanov, A., Mittelman, D., Pereira-Smith, O. M., Wilson, J. H., Gorbunova, V. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc Natl Acad Sci U S A. 101, 7624-7629 (2004).
- Mao, Z., Bozzella, M., Seluanov, A., Gorbunova, V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle. 7, 2902-296 (2008).

