A Set of Screening Techniques for a Quick Overview of the Neutrophil Function
Summary
This protocol features a set of neutrophil functional assays to be used as a screening method to cover functions from different signaling pathways. The protocol includes an initial and simple evaluation of cell viability, purity, reactive oxygen species production, real-time migration, phagocytosis, and a preliminary suggestion of neutrophil extracellular traps.
Abstract
Neutrophils are known as one of the first lines of defense in the innate immune response and can perform many particular cellular functions, such as chemotaxis, reverse migration, phagocytosis, degranulation of cytotoxic enzymes and metabolites, and release of DNA as neutrophil extracellular traps (NETs). Neutrophils not only have tightly regulated signaling themselves, but also participate in the regulation of other components of the immune system. As fresh neutrophils are terminally differentiated, short-lived, and highly variable among individuals, it is important to make the most of the collected samples. Researchers often need to perform screening assays to assess an overview of the many neutrophil functions that may be affected by specific conditions under evaluation. A set of tests following a single isolation process of normal density neutrophils was developed to address this need, seeking a balance between speed, comprehensiveness, cost, and accuracy. The results can be used to reason and guide in-depth follow-up studies. This procedure can be carried out in an average time of 4 h and includes the evaluation of cell viability, reactive oxygen species (ROS) production, real-time migration, and phagocytosis of yeast on glass slides, leaving enough cells for more detailed approaches like omics studies. Moreover, the procedure includes a way to easily observe a preliminary suggestion of NETs after fast panoptic staining observed by light microscopy, with a lack of specific markers, albeit enough to indicate if further efforts in that way would be worthwhile. The diversity of functions tested combines common points among tests, reducing the analysis time and expenses. The procedure was named NeutroFun Screen, and although having limitations, it balances the aforementioned factors. Furthermore, the aim of this work is not a definite test set, but rather a guideline that can easily be adjusted to each lab's resources and demands.
Introduction
Neutrophils are the most abundant innate immune cells in human blood and are known to play a major part in infection and inflammation, being the first responders to arrive at the site of tissue damage1. In recent years, there has been a growing recognition of the crucial role that neutrophils play in a variety of diseases and in supporting homeostasis2. Neutrophils not only have tightly regulated signaling themselves, but also participate in the regulation of other components of the immune system3,4,5. Therefore, investigating neutrophils and their many unusual cellular functions, such as chemotaxis, reverse migration6, phagocytosis7, respiratory burst8, and the release of neutrophil extracellular traps (NETs)7, is imperative in numerous research contexts where it is necessary to assess the potential neutrophil functional, morphological, or molecular changes triggered by specific conditions under analysis.
Freshly isolated neutrophils are terminally differentiated, short-lived, highly dynamic, and easily activated9. However, an efficient storage method that does not affect the neutrophil responses has not yet been achieved, making it challenging to perform multiple assays that must be uninterrupted. Furthermore, previously described functional analyses10,11, based on assays that require cytometry and/or fluorescent staining, may not be a viable choice when a broad and initial evaluation of the neutrophil is needed.
To address these issues, this protocol describes a set of tests that can be carried out following a single isolation process, including the evaluation of cell viability, reactive oxygen species (ROS) production, real-time migration, and phagocytosis of Saccharomyces cerevisiae, whose results can be used to reason in-depth follow-up studies. This procedure, named NeutroFun Screen, was designed to encompass the leading effector activities, except degranulation, and can be completed in an average time of 4 h, including 1 h of activation. Additionally, the remaining cells can be used for more detailed approaches like omics studies. The advantage of this method lies in its balance between speed, comprehensiveness, cost, and accuracy.
Furthermore, there is a way to easily observe a preliminary suggestion of NETs, without specific markers, but enough to indicate if further efforts in that direction would be worthwhile. The diversity of functions tested aims to combine common points among tests, reducing the analysis time and expenses. The main goal of this method is to provide a balanced, functional analysis regarding speed, comprehensiveness, cost, and accuracy that allows for an overview of the neutrophil's response, making it a useful initial step in investigating the effects of novel stimuli on normal density neutrophils.
Protocol
All experiments strictly followed the ethical guidelines set by the institutional review board at the University of Brasilia (process 13364819.0.0000.5558), and samples were identified by codes to ensure donor anonymity. The cells were obtained from normal healthy male donors aged 18-35 years, who signed the informed consent and met the following eligibility criteria: non-smokers/vapers, no chronic health conditions, and no history of inflammatory conditions in the last 14 days.
1. Blood collection
- Aseptically place 0.3 mL of 5,000 IU/mL heparin (see Table of Materials) in a sterile 20 mL syringe to heparinize it.
- Apply a venous tourniquet about 4 in above the puncture site and identify the median cubital or cephalic vein for venipuncture.
NOTE: Ensure that the total tourniquet time does not exceed 1 min. - Clean the puncture site with 70% alcohol and perform the venipuncture.
- Gently invert the syringe three or four times after collecting the blood to mix the blood and heparin properly.
2. Neutrophil isolation
NOTE: Polymorphonuclear leukocytes (PMNs) are isolated through density gradient centrifugation followed by hypotonic lysis of the remaining red blood cells (RBC), as previously described11 with some changes. This method is not mandatory to perform the screening assays, and can be replaced as long as the chosen method results in a viability of >97%, priming or activation of <3% of the PMNs, and yields enough cells for all assays, replicates, and conditions. Performing these steps under aseptic conditions and using endotoxin-free solutions are mandatory to avoid cell activation.
- Make 12 mL dilutions of 60% and 70% separation media (commercially available; see Table of Materials) in 50 mL conical tubes.
- Prepare the gradient from bottom to top by adding 4 mL at a time of the 60% dilution over the 70% dilution, using a 5 mL pipette. Do this gently to prevent mixing the interface.
- Carefully layer 12 mL of heparinized blood on top of the density gradient. Centrifuge at 200 x g for 15 min at room temperature.
NOTE: From this step onward, until PMN activation, all the reagents and tubes used must be kept in a cooler filled with ice. - Discard the plasma/mononuclear cell layer, then gently transfer the layer above the erythrocyte pellet into two 15 mL conical tubes with approximately 7.5 mL in each. Make up the tube volume with Hank's balanced salt solution (HBSS; see Table of Materials).
- Centrifuge at 300 x g for 5 min at 19 °C.
- Wash the cell pellet with HBSS.
- Discard the supernatant by pouring the tube and gently resuspend the pellet in 7 mL of HBSS.
- Centrifuge at 300 x g for 5 min at 19 °C to remove all the separation media.
- Perform hypotonic lysis of the remaining RBCs.
- Discard the supernatant and combine the pellets in a single tube.
- Resuspend the RBC/PMN pellet in 3 mL of sterile H2O and add 3 mL of HBSS (2x) within 25 s to restore the osmolarity. Then, centrifuge at 300 x g for 5 min at 19 °C.
- Repeat steps 2.8.1 and 2.8.2 for a white, erythrocyte-free pellet.
NOTE: The supernatant must be removed as soon as possible to minimize the contact of neutrophils with RBC breakdown products. Alternatively, the second hypotonic lysis can be replaced by gently resuspending the residual RBC layer and removing all the supernatant, as the remaining RBCs will sediment above the PMN pellet.
- Discard the supernatant by pouring the tube, gently resuspend the PMNs in the remaining buffer, and transfer them to an ice-cold microtube.
NOTE: Ensure to annotate the volume while transferring the resuspended cells with a micropipette. - Transfer 3 x 1 µL of the cell suspension to a clean glass slide (three wells of 1 µL each) and stain with fast panoptic (see Table of Materials) for morphology and purity evaluation12.
- To stain with fast panoptic, immerse the slide five times in panoptic fixative n° 1, six times in eosin n° 2, and twice in hematoxylin n° 3, with each immersion lasting 1 s.
- Gently wash the glass slide with distilled water.
- Allow to drain and air-dry.
- Observe under a microscope and count 300 random cells in each well, thus differentiating the neutrophils from other granulocytes.
- Transfer 1 µL of the cell suspension to 49 µL of 0.2% trypan blue dye13 and count the cells using a Neubauer chamber, distinguishing between dead and viable cells.
- Adjust the cell concentration to 6,667 cells/µL using a solution of 50% autologous plasma and 50% HBSS supplemented with calcium and magnesium. Divide the 6,667 cells/µL suspension evenly among the microtubes corresponding to the conditions to be tested, including the negative control.
NOTE: Any cell concentration similar to the circulating neutrophils in the model organism can be used, but it is important to use the same cell concentration in all experiments for reproducibility.
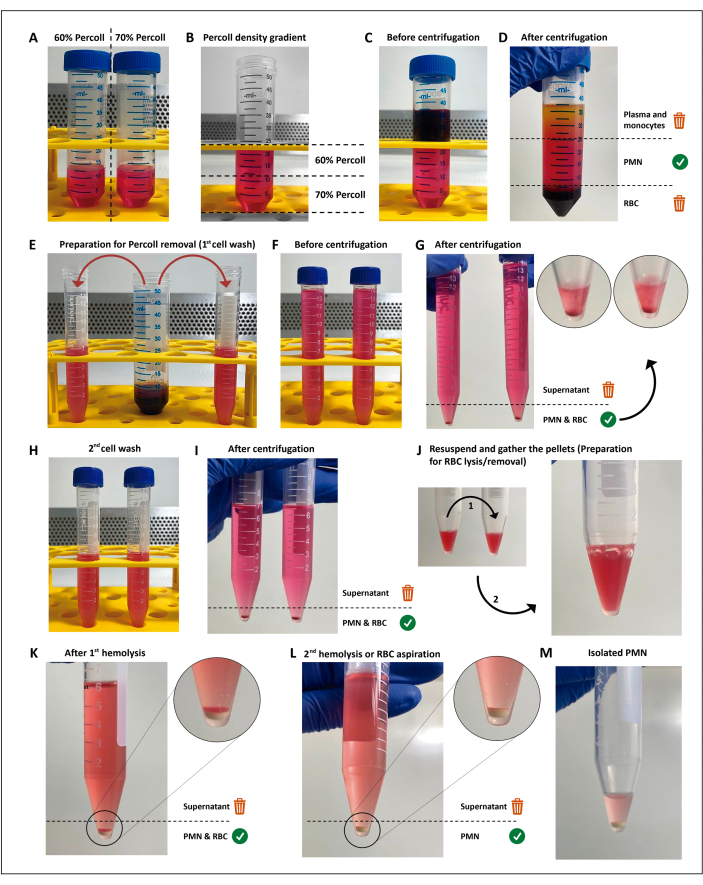
Figure 1: The neutrophil isolation protocol. Two concentrations of the separation media (percoll) (A) are stacked (B), then the blood is layered on top of the separation gradient (C). After centrifugation, the PMN is in the central layer (D), which is divided into two 15 mL tubes (E). The cell suspension is washed twice in HBSS and centrifuged (G–I) to remove the media, then the cells are resuspended, and residual RBCs are submitted to two rounds of hypotonic lysis (J–M). Please click here to view a larger version of this figure.
3. Preparation for neutrophil activation
- In 1.5 mL microtubes, prepare an activation system for each condition so that the final cell concentration is 6,600 cells/µL. For instance, to test the effects of 100 nM fMLP (N-formyl-methionyl-leucyl-phenylalanine; see Table of Materials), add 5 µL of 10 µM fMLP to 495 µL of the 6,667 cell/µL suspension. For the negative (unstimulated) control, add HBSS containing Ca2+ and Mg2+.
NOTE: To demonstrate this methodology, the following final concentrations of the stimuli were used: 100 nM fMLP, 16 µM of fallaxin, a naturally occurring antimicrobial peptide14, and 100 nM PMA (phorbol 12-myristate 13-acetate) (see Table of Materials). - Incubate at 37 °C without rotation.
NOTE: All aliquots for functional assays are taken from this cell suspension, hereafter referred to as the activation system.
4. Nitrotetrazolium blue chloride (NBT) assay for evaluating ROS production
- Preparation of NBT working solution: For each experimental condition, prepare an NBT (see Table of Materials) working solution of 6 mM using the following steps:
- Dissolve 0.0005 g of NBT in 10 µL of dimethyl sulfoxide (DMSO) and vortex for at least 15 min.
- Add 90 µL of HBSS Ca2+Mg2+ and vortex up to 2 min.
NOTE: All steps involving NBT must be performed in the dark.
- Perform NBT slide test.
- After 20 min of cell activation, gently mix the cell suspension and carefully transfer 2 µL of PMN to a clean glass slide. Incubate for 20 min in a humidified chamber at 37 °C.
NOTE: Do not spread the cell suspension too much over the slide; otherwise it may dry out before incubation. - Add 1 µL of the NBT working solution over the cells and further incubate for 20 min protected from light.
- Dry the slide with hot air and fix it with a drop of methanol in each well for 1 min. Stain with 0.03% safranin (see Table of Materials) for 1 min.
- Gently wash the glass slide with distilled water.
- Allow the slide to air-dry and observe under a microscope.
- Count 100 random cells in each well, differentiating neutrophils with and without formazan deposits.
- After 20 min of cell activation, gently mix the cell suspension and carefully transfer 2 µL of PMN to a clean glass slide. Incubate for 20 min in a humidified chamber at 37 °C.
- Perform NBT spectrophotometry assay.
- After 40 min of cell activation, gently mix the cell suspension and transfer 90 µL of PMNs from the activation system to a clean microtube. Then, carefully add 20 µL of the 6 mM NBT solution. Incubate in the dark for 20 min at 37 °C.
- Add 100 µL of 10% Sodium dodecyl sulfate (SDS; see Table of Materials) and vortex.
- Sonicate using a tip-sonicator at an amplitude of 60%, five cycles of 15 s each with 15 s intervals. Centrifuge at 12,000 x g for 5 min.
- Transfer 60 µL of the supernatant to a clear-bottom 96-well plate and measure the absorbance of the formazan product at 570 nm.
5. Phagocytosis assay
- Prepare a 33,000 yeasts/µL suspension for each condition, as described below:
- Add approximately 0.75 mg of dry yeast (Saccharomyces cerevisiae; see Table of Materials) to 200 µL of HBSS Ca2+Mg2+ and incubate in a thermomixer at 100 °C with 500 rpm for at least 15 min.
- Homogenize the mixture by vortexing and transfer 5 µL of yeast suspension to 45 µL of 0.2% trypan blue dye. Count the yeasts using a Neubauer chamber.
- Adjust the concentration of the initial suspension to 33,000 yeast cells/µL using HBSS Ca2+Mg2+. Keep the suspension on ice until use.
- After 20 min of cell activation, gently mix the cell suspension and transfer 5 µL of the activation system to 5 µL of the 33,000 yeast/µL Saccharomyces cerevisiae suspension in a new sterile microtube.
NOTE: The neutrophil to yeast ratio is 1:5 (PMN:yeast). - Immediately transfer 6 µL of the PMN/yeast suspension into three wells of a clean glass slide (2 µL each) and incubate the slide in a humidified chamber for 40 min.
NOTE: Do not spread the cell suspension too much over the slide; otherwise, it may dry out before incubation. - Dry the slide under hot air and stain with fast panoptic, as described in step 2.9 above.
NOTE: The third step of the fast panoptic staining is critical to the microscopy analysis of the slide. Staining the slide for ≥3 s at this step can render it unfit for analysis, since it will be difficult to differentiate yeast from neutrophil nuclear lobes. - Observe the slides under the microscope, counting 100 random neutrophils of each well and discriminating between PMNs positive and negative for phagocytosis.
NOTE: At least one yeast particle within or in direct contact with the PMN cellular membrane indicates a PMN positive for phagocytosis. If interested in the yeast/neutrophil ratio, count the number of yeast particles engulfed as well.
6. Real-time PMN chemotaxis assay
NOTE: The migration assay is performed similarly to the protocol described previoulsy15, with the following adaptations:
- Prepare the chemotactic gradient by adding 160 µL of chemoattractant (e.g., fMLP, IL-8, C5, or LTB4; see Table of Materials) to the lower chamber of an impedance-based real time cell analyzer (RTCA) plate. For negative controls and blanks, add 160 µL of HBSS Ca2+Mg2+.
- Attach the upper chamber and add 25 µL of HBSS Ca2+Mg2+. Incubate at room temperature for at least 1 h to form the chemotactic gradient.
- After 60 min of cell activation, gently mix the cell suspension and place 60 µL of cell suspension in the upper chamber. Add 60 µL of HBSS Ca2+Mg2+ to the blank.
- Place the RTCA plate and program the RTCA software to measure the cell index (CI) every 60 s for 2 h.
NOTE: The RTCA plates can be washed for reuse as previously described16. In summary, wash the RTCA chambers and electrodes with phosphate-buferred saline (PBS) three times, then with type I ultrapure water twice. Incubate the lower and upper chamber with 0.25% trypsin 0.53 mM ethylenediaminetetraacetic acid (EDTA) for 40 min. Wash with ultrapure water three times.
7. NET suggestive assay
- After 10 min of cell activation, gently mix the cell suspension and transfer 4 µL of the PMNs from each activation system under evaluation, divided into two wells of a clean glass slide. Incubate in a humidified chamber at 37 °C for 30 min.
- Add 1 µL of DNAse I to one of the wells and incubate for 20 min at 37 °C (in a wet chamber).
- Dry the slide and stain with fast panoptic, as previously described in step 2.9 of the "neutrophil isolation" section.
- Evaluate the slides under a microscope.
NOTE: Look for any indication of NET release, characterized by the presence of web-like structures. Once identified, confirm if the DNAse I treatment was capable of removing such structures. This assay is suggestive of NET formation, as additional tests are required to confirm their presence.
Representative Results
The density-based isolation method used in this study (Figure 1) met the criteria for the proposed experiments. Neutrophil parameters obtained from this method included viability ≥98%, purity ≥94%, and cell yield ≥1.5 x 107, with no activation detectable by the screening tests. Two relevant steps in the isolation of PMNs are anticoagulation and RBC removal. Keeping the anticoagulated blood tube or syringe at a gentle rocking before layering over the density gradient and choosing an RBC removal method to prevent both activation and contamination can influence the experiment yield and reproducibility.
To assess the functional overview of neutrophil function, a workflow was developed that allowed the proposed screening assays to be performed with at least two researchers in an average time of 4 h, with 1 h of activation and 2 h of real-time migration monitoring, as shown in Figure 2.
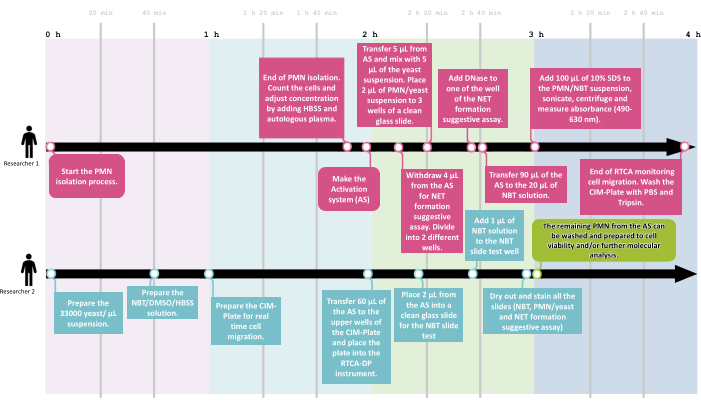
Figure 2: The NeutroFun Screen workflow. To assess the panel of functional responses triggered by specific conditions under evaluation, the NeutroFun Screen workflow includes the participation of at least two researchers. Researcher 1 (R1) starts the PMN isolation process, followed by cell concentration adjustment and activation system incubation, while in parallel, researcher 2 (R2) prepares the materials to carry out the next assays, which are divided between the two: R1 performs phagocytosis, NET, and spectrophotometric NBT assays; R2 performs real-time migration and NBT slide tests. Please click here to view a larger version of this figure.
The classic NBT assay17 was optimized for both slide and spectrophotometric assessment of the formazan formation resulting from the reaction of NBT with the ROS generated by PMNs. The spectrophotometric assay allowed for relative quantification among conditions, while the microscopy assay was useful for descriptive and semi-quantitative analysis, evaluating the regularity of crystal distribution, morphology, and number of cells containing formazan. The results of the spectrophotometric assay are shown in Figure 3A, indicating that 100 nM PMA induced elevated ROS production (in an average ratio of 3:2:1) compared to fMLP and control groups. The NBT slide test results (Figure 3B) corroborated the spectrophotometric results, also showing PMA as the most intense ROS production-inducing stimulus compared to fMLP (Figure 3C), as previously demonstrated by the direct measurement of ROS by cytometry18. Regarding cell counting, the number of neutrophils containing formazan crystals showed a ratio of 7:4:1 in the PMA:fMLP:control conditions, also revealing different formazan distribution patterns between fMLP and PMA-activated PMNs; fMLP treatment resulted in intensively activated individual cells, while PMA induced the formation of formazan crystals scattered throughout the cytoplasm in the majority of PMNs. Furthermore, a few morphological characteristics that differentiated the conditions were observed, such as expressive cell aggregation after PMA treatment. The absence of such typical activation characteristics and low levels of formazan on the control group indicated that the resting state was not significantly disturbed. Therefore, the NBT assay proved to be a simple, inexpensive, and reliable test for neutrophil ROS production that can be used to overview the capacity and intensity of a given stimulus to induce ROS production.
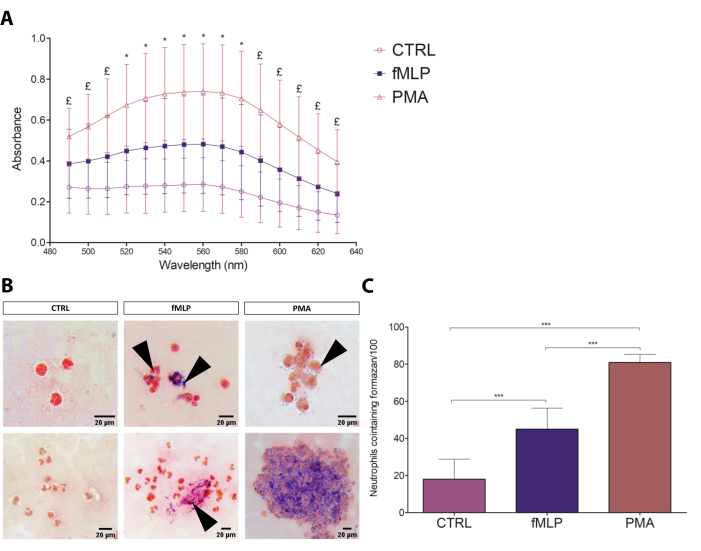
Figure 3: NBT test. (A) The spectrophotometric assessment of neutrophil respiratory burst to different conditions (negative control: 100 nM fMLP and 100 nM PMA) by formazan absorbance (490-630 nm). Data from four donors are presented as mean ± standard deviation (SD). * = all groups are different from each other (p≤ 0.05); £ = The comparisons CTRL versus PMA and fMLP versus PMA show significant differences (p≤ 0.05). (B) The slide test can be analyzed qualitatively, characterizing the intensity of formazan deposits, its location within the cell, and the cell aggregation. Black arrows point to formazan crystals. Scale bars: 20 µm. (C) Number of PMNs containing formazan, counted by optical microscopy. Data from eight donors presented as mean ± SD. *** p < 0.0001, Student's t-test. Please click here to view a larger version of this figure.
The phagocytosis slides were used to determine the phagocytosis rate as a percentage of neutrophils performing phagocytosis, as shown in Figure 4. Although both 100 nM fMLP and 16 µM antimicrobial peptide induced an apparently increased yeast engulfment, the difference was not statistically significant until the dual stimulation, which significantly enhanced the phagocytosis compared to the control group and might suggest a response upon dual stimulation.
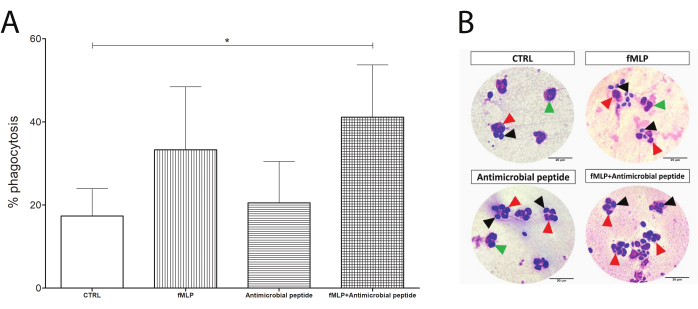
Figure 4: Phagocytosis test. Neutrophils were evaluated by their phagocytosis capacity (counting of neutrophils containing yeast). Although the exposure to 100 nM fMLP or a 16 µM antimicrobial peptide prior to yeast incubation enhanced the phagocytosis compared to the control group, (A) the increase was not statistically significant. Interestingly, an initial incubation with fMLP, followed by addition of the antimicrobial peptide, resulted in significantly enhanced phagocytosis in human neutrophils (asterisk between CTRL and the fMLP+ antimicrobial peptide). (B) Black arrows show yeast cells, green arrows show neutrophils alone, and red arrows point to phagocytosing neutrophils. Data from five donors presented as mean ± SD. Scale bars: 20 µm. Please click here to view a larger version of this figure.
An example of real-time cell migration results is shown in Figure 5, exhibiting the well-known chemotactic effect of 100 nM fMLP on neutrophils. The cell migration kinetics was adjusted to the Gompertz model, enabling the comparison of its parameters.
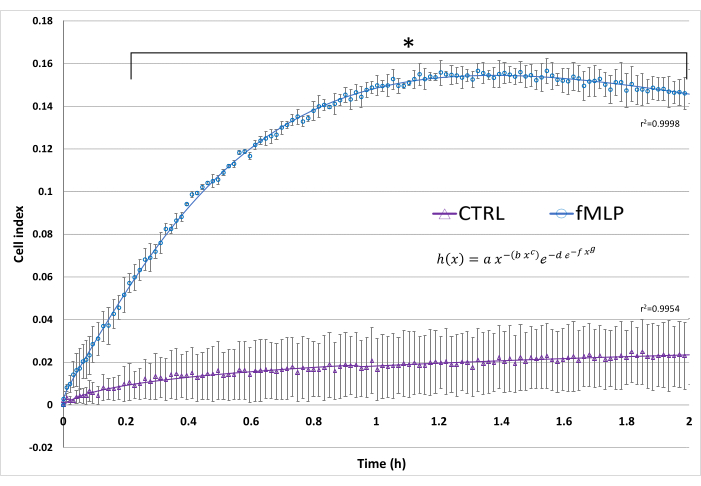
Figure 5: Real-time cell migration. The cell index reflects the electrical impedance resulting from cell migration. In this assay, fMLP was used as a positive control of neutrophil migration by adding 100 nM fMLP to the lower chamber. A negative control (CTRL) contained only HBSS in the lower chamber. The superimposed curves represent the curve fitting to the Gompertz model, modified to best fit ascending and descending slopes. Data from three donors, presented as mean ± SD.* p < 0.05, Student's t-test. Please click here to view a larger version of this figure.
Finally, a simple optical microscopy test suggestive of the presence of NET is presented, in which the PMNs are stained with fast panoptic. It is possible to observe that for some specific NET-inducing stimuli, such as PMA or some antimicrobial peptides, even after a short activation time (1 h) it is possible to observe filamentous web-like structures near the cells, while this is not possible in other conditions like the negative control and fMLP (Figure 6). To better support this claim, DNase I was added after 40 min of cell activation with 100 nm PMA or an antimicrobial peptide, removing the NET-like structures (Figure 6). Although this method is not suitable for NET characterization and can be influenced by artifacts, it is a cheap, simple, and interesting starting point to justify further investments in analyzing NETs, especially in contexts wherein the effects of the stimuli are unknown or poorly described.
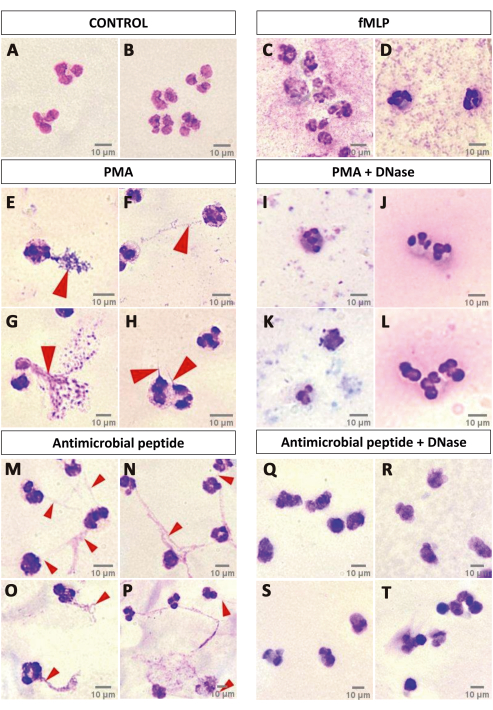
Figure 6: NET formation suggestive assay. A qualitative analysis of panoptic-stained neutrophils after 1 h of activation over a glass slide may indicate NET release. CTRL (A,B) and fMLP (C,D) groups did not show any indication of NET release, while activation with PMA (E–H) induced the formation of NET-like structures that were degraded by DNase I treatment (I–L). Investigation of the effects of an antimicrobial peptide on the neutrophils showed that such a stimulus might be able to elicit NET release, since the slides presented the NET-like structures (M–P) that were degraded by DNase I (Q–T). Red arrows point to NET-like structures. Scale bars: 10 µm. Please click here to view a larger version of this figure.
Discussion
Neutrophils are highly dynamic and responsive cells that are short-lived and cannot yet be cryopreserved19, making investigations into their biology challenging. Therefore, it is essential to follow careful steps to obtain viable, enriched, and resting neutrophils11,20. This study employed a density-based isolation technique that emphasizes gentle and minimal manipulation, as well as the use of low temperatures until the activation step. Additionally, blood processing must occur within 30 min after venipuncture and be stored at room temperature. The present work presents an option to diminish manipulation stress and experiment time. To further simplify the process, we introduced a new step (as an option) in the protocol, which involves removing RBCs by gentle resuspension and aspiration of the RBC layer after a single hemolysis procedure. This step reduces the manipulation stress and experiment time, as it removes the majority of RBCs with only one hypotonic hemolysis step. However, it should be noted that operator skills greatly affect the effectiveness of this step, and it may not always entirely remove RBCs. Furthermore, RBC aspiration can lead to a loss of neutrophils, but this does not affect the yield, as more neutrophils were initially recovered from the gradient when collecting closer to the RBC layer. Therefore, RBC aspiration is recommended if the assays do not require the highest possible purity, or if the operator has had consistently good results. For the screening tests presented here, a small amount of contaminant RBCs (<3%) would not interfere with the results.
To ensure that all the procedures are performed in due time, including reagent preparation, a workflow is presented (Figure 2) that details the timeline and division of tasks between two researchers. After all functional assays are finished, the remaining cells can be prepared for further molecular investigations, like omics studies by cell lysis with proper lysis buffer and storage.
ROS production
Respiratory burst is a hallmark of neutrophil priming and activation18,21, and evaluating ROS production is a common method used to estimate the activation status of neutrophils. This study evaluated ROS production using two approaches: optical microscopy and spectrophotometry.
The NBT test is a well-known approach to assess the respiratory burst in neutrophils22,23. Two commonly used methods are the optical microscopy-based (or slide test) and spectrophotometry-based assays24,25,26. Although optical microscopy allows for the visualization of details at the cell level, it is prone to user bias. Therefore, the NBT slide test serves as a qualitative complement to the spectrophotometric assay regarding phenotypic features, such as formazan formation patterns, intensity, and cell aggregation.
The critical steps for both the NBT slide and spectrophotometric tests are the NBT and formazan complete solubilization. Contrary to the manufacturer's recommendations, NBT does not dissolve well in water. However, homogenizing NBT in DMSO for at least 15 min and then adding water/HBSS and vortexing for 2 min provides complete solubilization. In addition, to analyze the absorbance of formazan, two critical steps must be achieved: (1) release of formazan crystals from cells by PMN lysis and (2) complete solubilization of formazan crystals. Both steps were achieved by treating the cell pellet with 10% SDS, as recently suggested27, followed by sonication and absorbance analysis of the supernatant at 570 nm.
Furthermore, the NBT results of the negative and positive control groups are the first step of the screening to be assessed. This step must show a remarkable difference, since ROS production evaluation through formazan indicates whether the isolation process might have been responsible for unwanted changes in the resting status of the cells either by stimulation or inhibition.
Other methods, such as cytochrome c reduction28 or flow cytometry29 analysis, are often used for ROS detection; however considering a screening approach, their application requires a longer experiment time, the use of specific markers, and expensive instruments. Chemiluminescence of luminol/isoluminol is a fast and cost-effective method for detecting ROS while also allowing the differentiation of intra- and extracellular ROS. However, a luminometer is required for this method30,31. Although instrument cost could be circumvented by the use of a facility, it would still be unfit for a screening analysis performed simultaneously with other assays.
Phagocytosis
The PMN/yeast incubation provides an overview of neutrophil phagocytosis capacity and efficacy by counting the number of neutrophils performing phagocytosis and the number of yeasts engulfed per neutrophil. However, as the yeast particle itself is an activation signal, comparing the control group with pre-treated PMN constitutes a dual stimulation system, which may not display significant changes, as shown in the CTRL versus fMLP-treated PMN (Figure 4). Nevertheless, this assay is useful in indicating whether a potential modulator would generate any impact on phagocytosis. A novel antimicrobial peptide was tested using this assay, and the results indicate an interesting potential response to this combination of stimuli, which has recently been discussed as needed for efficient activation32.
The critical step for this assay is the staining, as the analysis becomes unreliable if the slide is kept on panoptic No. 3 (hematoxylin) for ≥3 s. This is because the yeast cells and the neutrophil nuclear lobes cannot be distinguished from each other. Moreover, this approach allows the morphology analysis of neutrophils after exposure to modulators. Flow cytometry is another efficient technique to analyze phagocytosis33, although it has limitations through the difficulty in discriminating between membrane-bound and engulfed particles and other factors related to the proposed screening set, as discussed in the previous topic. The same limitation is observed in the method described herein, although this is to be considered as an initial screening to guide follow-up studies.
Real-time migration
The real-time screening protocol was optimized to evaluate the migration capacity of PMNs. Although a previous study suggested that coating the underside of the RTCA plate was needed for the migration assay to show a difference between the negative control and IL-8 treatment15, this study showed high confidence interval (CI) values for fMLP-treated cells without the addition of coating agents. The results presented high reproducibility with significant migration from 12 min onward. Furthermore, curve fitting analysis of the obtained migration data can reveal parameters, such as cell index maximum as well as increase and decrease slope, that are useful for understanding the dynamics of migration, and may depend on concentration or differ between conditions. The best fitting for neutrophil migration curves (Figure 5) was to the adjusted Gompertz function34, showing that fMLP increases the maximum cell index and the increased slope.
Special attention is required to avoid bubbles in the RTCA system as they can block the cells passing through the electrodes, compromising the experiment. A limiting factor is the high cost of the plates. Although cheaper techniques analyze cell migration, they lack good reproducibility or are difficult and time-consuming, such as the Boyden chamber35. Therefore, RTCA is a reliable migration analysis compatible with the other screening tests presented here.
NETs
Lastly, a promising, easy, and low-cost assay for a preliminary evaluation of NET formation was developed using optical microscopy. It was derived from the observation of phagocytosis slides, in which PMA, a specific NET-inducing stimulus tested for its effect on phagocytosis capacity, also induced a web-like structure formation compared to the CTRL and fMLP groups. This work hypothesized that those structures could indicate NET release. Such a hypothesis was tested by treating the samples with DNase I after activation, where no filamentous structures were found in PMA-activated neutrophils. The assumption that such structures are likely to be NETs is based on the fact that PMA is cited as a well-known inducer of NET formation36,37, the finding of similar structures by fluorescence microscopy using specific markers for NETs38,39 and the degradation of NETs catalyzed by DNase I40,41. It is important to emphasize that this is not a method for the confirmation of NET formation, but just a preliminary screening that suggests the presence of NETs. Although this test has limitations, as NETs can be confused with staining artifacts, it serves a relevant purpose in suggesting NET formation in a fast and low-cost screening that can be performed together with other functional tests. Once detected as possibly relevant to the study being screened, NETs can be further evaluated by confocal or electron microscopy42, as discussed later.
In our lab, novel antimicrobial peptides, also likely to have immunomodulatory activities, are frequently evaluated by this set of screening assays to better guide further analysis of the effects of some peptides on human neutrophils, as some show interesting ROS43, phagocytosis, migration and/or NET modulating properties.
In conclusion, the NeutroFun Screen offers a valuable tool for identifying potential modulators of the normal density neutrophil activity from a large number of untested compounds. However, it is important to note that this method only serves as a preliminary screening. After this initial screening, more expensive, advanced, and time-consuming methodologies can be directed to specific functions or pathways suggested by these first results for data validation. For ROS production, phagocytosis, and NET formation, there are alternative methods for data validation and to obtain further quantitative results. NET visualization and quantification can be performed through immunofluorescence microscopy analysis, which determines the presence and overlap of extracellular DNA and granule proteins, such as myeloperoxidase and neutrophil elastase, as described in detail recently42. ROS play different crucial roles in many biological processes, and therefore their study is highly widespread and diverse. Among the most established methodologies are those that make use of antibodies, such as the enzyme-linked immunosorbent assay (ELISA) and immunoblotting of luminescence, or fluorescence detection-such as flow cytometry of cells under different labeling-through DCFDA, EPR spectroscopy, and enzymatic activity measurements44. Similarly, robust phagocytosis evaluation is also done in several ways; the alternative methods include flow cytometry alone45,46 or combined with fluorescence microscopy47. The real-time cell migration described is a robust and sufficient assay that does not require further validation and presents parameters that other methodologies cannot contemplate. Other combinations of such methods might better suit specific scenarios, but in summary, this represents a fast and affordable combination that encompasses many neutrophil activities.
In summary, this study aims to provide a set of assays consisting of simple, fast, and low-cost methodologies to evaluate multiple neutrophil responses to novel molecules and conditions as a way to better direct efforts toward advanced methodologies. As major limitations, the results from ROS, NET, and phagocytosis assays should be considered preliminary and need further validation by more specific and elaborate assays, and those that rely on non-automated microscopy cell count are prone to unconscious bias.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors acknowledge the following funding agencies: FAPDF, CNPq, CAPES, UnB, FINEP, and FINATEC.
Materials
| CIM-Plate 16 | Agilent | 5665825001 | |
| CLARIOstar Plate Reader | BMG LABTECH | US Patent Number 9,733,124 Product details: MARS Data Analysis Software |
|
| Dimethyl sulfoxide | Dinâmica | 1582 | |
| DNAse I | Sigma – Aldrich | DN 25 | |
| Ethylenediaminetetraacetic acid disodium salt dihydrate | Sigma – Aldrich | E5134 | |
| Fast panoptic stain | Laborclin | 620529 | |
| Glass slide | Exacta | 7102 | |
| Hank’s Balanced Salt Solution with calcium, with magnesium, without phenol red. | Sigma – Aldrich | 55037C | |
| Hank’s Balanced Salt Solution without calcium chloride, magnesium sulfate and sodium bicarbonate. | Sigma – Aldrich | H4641 | |
| Heparin | Blau | 7896014655229 | |
| Laminar flow cabinet | Veco | VLFS-12 | |
| Microscope | Zeiss | 415501-0101-002 | Product details: Primostar 1 |
| Mixing Block | BIOER | MB-102 | |
| Neubauer improved bright-lined | New Optik | 1110000 | |
| N-formyl-methionyl-leucyl-phenylalanine | Sigma – Aldrich | F3506 | |
| Nitroblue tetrazolium | Neon | CAS 298-83-9 | |
| Percoll | Cytiva | 17089101 | separation media |
| Phorbol 12-myristate 13-acetate | Sigma – Aldrich | P8139 | |
| Phosphate buffered saline tablet | Sigma – Aldrich | P4417 | |
| ROTOFIX 32 A | Hettich | 1206 | |
| Saccharomyces cerevisiae | Fleischmann | ||
| Safranin | Sigma – Aldrich | 50240 | |
| Sodium dodecyl sulfate | Cytiva | 17-1313-01 | |
| Sonicator | Qsonica | Q125 | |
| Trypan blue solution | Vetec | C.I. 23850 | |
| Vortex Genie 2 | Scientific Industries, Inc. | 0K-0500-902 | |
| xCELLigence Real-Time Cell Analysis (RTCA) DP (dual purpose) | Agilent | 380601050 | Product details: RTCA system composed of detection hardware, cell plates and software |
References
- Nauseef, W. M., Borregaard, N. Neutrophils at work. Nature Immunology. 15 (7), 602-611 (2014).
- Groeneweg, L., Hidalgo, A. Emerging roles of infiltrating granulocytes and monocytes in homeostasis. Cellular and Molecular Life Sciences. 77 (19), 3823-3830 (2020).
- Rosales, C., Lowell, C. A., Schnoor, M., Uribe-Querol, E. Neutrophils: their role in innate and adaptive immunity 2017. Journal of Immunology Research. 2017, 9748345 (2017).
- Castro, M., et al. Proteome analysis of resting human neutrophils. Protein & Peptide Letters. 13 (5), 481-487 (2006).
- Li, Y., et al. The regulatory roles of neutrophils in adaptive immunity. Cell Communication and Signaling. 17, 147 (2019).
- de Oliveira, S., Rosowski, E. E., Huttenlocher, A. Neutrophil migration in infection and wound repair: going forward in reverse. Nature Reviews Immunology. 16 (6), 378-391 (2016).
- Burn, G. L., Foti, A., Marsman, G., Patel, D. F., Zychlinsky, A. The neutrophil. Immunity. 54 (7), 1377-1391 (2021).
- El-Benna, J., et al. Priming of the neutrophil respiratory burst: role in host defense and inflammation. Immunological Reviews. 273 (1), 180-193 (2016).
- Castro, M. S., Cilli, E. M., Fontes, W. Combinatorial synthesis and directed evolution applied to the production of alpha-helix forming antimicrobial peptides analogues. Current Protein & Peptide Science. 7 (6), 473-478 (2006).
- Mihaila, A. C., et al. Transcriptional profiling and functional analysis of N1/N2 neutrophils reveal an immunomodulatory effect of S100A9-blockade on the pro-inflammatory N1 subpopulation. Frontiers in Immunology. 12, 708770 (2021).
- Kuhns, D. B., Priel, D. A. L., Chu, J., Zarember, K. A. Isolation and functional analysis of human neutrophils. Current Protocols in Immunology. 111 (1), 7-23 (2015).
- Paulíková, E., Kociková, A., Sabol, M. Modification of a panoptic method of staining isolated cells. Bratislavske Lekarske Listy. 94 (12), 638-640 (1993).
- Strober, W. Trypan blue exclusion test of cell viability. Current Protocols in Immunology. 111 (1), 1-3 (2015).
- Libério, M. S., et al. Anti-proliferative and cytotoxic activity of pentadactylin isolated from Leptodactylus labyrinthicus on melanoma cells. Amino Acids. 40 (1), 51-59 (2011).
- Cano, P. M., Vargas, A., Lavoie, J. P. A real-time assay for neutrophil chemotaxis. BioTechniques. 60 (5), 245-251 (2016).
- Stefanowicz-Hajduk, J., Adamska, A., Bartoszewski, R., Ochocka, J. R. Reuse of E-plate cell sensor arrays in the xCELLigence Real-Time Cell Analyzer. BioTechniques. 61 (3), 117-122 (2016).
- Björkstén, B., Nyström, K., Lindqvist, B. The nitroblue tetrazolium (NBT) test in endemic benign (epidemic) nephropathy. Acta Medica Scandinavica. 199 (1-6), 147-150 (1976).
- Aquino, E., et al. Proteomic analysis of neutrophil priming by PAF. Protein & Peptide Letters. 23 (2), 142-151 (2016).
- Blanter, M., Gouwy, M., Struyf, S. Studying neutrophil function in vitro: cell models and environmental factors. Journal of Inflammation Research. 14, 141-162 (2021).
- Hsu, A. Y., Peng, Z., Luo, H., Loison, F. Isolation of human neutrophils from whole blood and buffy coats. Journal of Visualized Experiments. (175), e62837 (2021).
- Moghadam, Z. M., Henneke, P., Kolter, J. From flies to men: ROS and the NADPH oxidase in phagocytes. Frontiers in Cell and Developmental Biology. 9, 628991 (2021).
- Pattan, S. S., Bhat, K. G., Pattar, G. D., Kuntagi, M. Comparison of three different techniques for isolation of neutrophils from blood and their utility in performing nitroblue tetrazolium test. International Journal of Basic and Applied Physiology. 8 (1), 41 (2019).
- Gooty, J. R., Shashirekha, A., Guntakala, V. R., Palaparthi, R. Estimation of phagocytic activity of polymorphonuclear leukocytes in chronic and aggressive periodontitis patients with nitroblue tetrazolium test. Journal of Indian Society of Periodontology. 23 (4), 316 (2019).
- Langer, S., et al. Clinical and laboratory profiles of 17 cases of chronic granulomatous disease in north India. Indian Journal of Hematology and Blood Transfusion. 37 (1), 45-51 (2021).
- Oualha, R., et al. Infection of human neutrophils with Leishmania infantum or Leishmania major strains triggers activation and differential cytokines release. Frontiers in Cellular and Infection Microbiology. 9, 153 (2019).
- Zilinskas, J., Zekonis, J., Zekonis, G., Valantiejiene, A., Periokaite, R. The reduction of nitroblue tetrazolium by total blood in periodontitis patients and the aged. Stomatologijal. 9 (4), 105-108 (2007).
- Benov, L. Improved formazan dissolution for bacterial MTT assay. Microbiology Spectrum. 9 (3), e01637 (2021).
- Chen, Y., Junger, W. G. Measurement of oxidative burst in neutrophils. Methods in Molecular Biology. 844, 115-124 (2012).
- Richardson, M. P., Ayliffe, M. J., Helbert, M., Davies, E. G. A simple flow cytometry assay using dihydrorhodamine for the measurement of the neutrophil respiratory burst in whole blood: comparison with the quantitative nitrobluetetrazolium test. Journal of Immunological Methods. 219 (1-2), 187-193 (1998).
- Jancinová, V., et al. The combined luminol/isoluminol chemiluminescence method for differentiating between extracellular and intracellular oxidant production by neutrophils. Redox Report. 11 (3), 110-116 (2006).
- Nosál, R., et al. Pharmacological intervention with oxidative burst in human neutrophils. Interdisciplinary Toxicology. 10 (2), 56-60 (2017).
- Mol, S., et al. Efficient neutrophil activation requires two simultaneous activating stimuli. International Journal of Molecular Sciences. 22 (18), 10106 (2021).
- Schneider, L., et al. Flow cytometry evaluation of CD14/CD16 monocyte subpopulations in systemic sclerosis patients: a cross sectional controlled study. Advances in Rheumatology. 61 (1), 27 (2021).
- Akin, E., Pelen, N. N., Tiryaki, I. U., Yalcin, F. Parameter identification for gompertz and logistic dynamic equations. PLoS One. 15 (4), e0230582 (2020).
- Guy, J. B., et al. Evaluation of the cell invasion and migration process: A comparison of the video microscope-based scratch wound assay and the boyden chamber assay. Journal of Visualized Experiments. (129), e56337 (2017).
- Brinkmann, V., et al. Neutrophil extracellular traps kill bacteria. Science. 303 (5663), 1532-1535 (2004).
- de Bont, C. M., Koopman, W. J. H., Boelens, W. C., Pruijn, G. J. M. Stimulus-dependent chromatin dynamics, citrullination, calcium signalling and ROS production during NET formation. Biochimica et Biophysica Acta. Molecular Cell Research. 1865, 1621-1629 (2018).
- Masuda, S., et al. Measurement of NET formation in vitro and in vivo by flow cytometry. Cytometry Part A. 91 (8), 822-829 (2017).
- Zharkova, O., et al. A flow cytometry-based assay for high-throughput detection and quantification of neutrophil extracellular traps in mixed cell populations. Cytometry Part A. 95 (3), 268-278 (2019).
- Hosseinnejad, A., et al. DNase I functional microgels for neutrophil extracellular trap disruption. Biomaterials Science. 10 (1), 85-99 (2022).
- Chrysanthopoulou, A., et al. Neutrophil extracellular traps promote differentiation and function of fibroblasts. The Journal of Pathology. 233 (3), 294-307 (2014).
- Tong, M., Abrahams, V. M. Visualization and quantification of neutrophil extracellular traps. Methods in Molecular Biology. 2255, 87-95 (2021).
- Santana, C. J. C., et al. Biological properties of a novel multifunctional host defense peptide from the skin secretion of the chaco tree frog, boana raniceps. Biomolecules. 10 (5), 790 (2020).
- Murphy, M. P., et al. Guidelines for measuring reactive oxygen species and oxidative damage in cells and in vivo. Nature Metabolism. 4 (6), 651-662 (2022).
- Boero, E., et al. Use of flow cytometry to evaluate phagocytosis of staphylococcus aureus by human neutrophils. Frontiers in Immunology. 12, 635825 (2021).
- Karsten, C. B., et al. A versatile high-throughput assay to characterize antibody-mediated neutrophil phagocytosis. Journal of Immunological Methods. 471, 46-56 (2019).
- Smirnov, A., Solga, M. D., Lannigan, J., Criss, A. K. Using imaging flow cytometry to quantify neutrophil phagocytosis. Methods in Molecular Biology. 2087, 127-140 (2020).

