Tracking miRNA Release into Extracellular Vesicles using Flow Cytometry
Summary
Here, we describe a straightforward protocol that enables in vitro assessment of the abundance of fluorescently labeled microRNAs to study the dynamics of microRNA packaging and export into extracellular vesicles (EVs).
Abstract
Extracellular vesicles (EVs) are important mediators of cellular communication that are secreted by a variety of different cells. These EVs shuttle bioactive molecules, including proteins, lipids, and nucleic acids (DNA, mRNAs, microRNAs, and other noncoding RNAs), from one cell to another, leading to phenotypic consequences in the recipient cells. Of all the various EV cargo, microRNAs (miRNAs) have garnered a great deal of attention for their role in shaping the microenvironment and in educating recipient cells because of their clear dysregulation and abundance in EVs. Additional data indicates that many miRNAs are actively loaded into EVs. Despite this clear evidence, research on the dynamics of export and mechanisms of miRNA sorting is limited. Here, we provide a protocol using flow cytometry analysis of EV-miRNA that can be used to understand the dynamics of EV-miRNA loading and identify the machinery involved in miRNA export. In this protocol, miRNAs predetermined to be enriched in EVs and depleted from donor cells are conjugated to a fluorophore and transfected into the donor cells. The fluorescently tagged miRNAs are then verified for loading into EVs and depletion from cells using qRT-PCR. As both a transfection control and a tool for gating the transfected population of cells, a fluorescently labeled cellular RNA (cell-retained and EV-depleted) is included. Cells transfected with both the EV-miRNA and cell-retained-miRNA are evaluated for fluorescent signals over the course of 72 h. The fluorescence signal intensity specific for the EV-miRNAs diminishes rapidly compared to the cell-retained miRNA. Using this straightforward protocol, one could now assess the dynamics of miRNA loading and identify various factors responsible for loading miRNAs into EVs.
Introduction
MicroRNAs (miRNAs) are one of the best-characterized subsets of small noncoding RNAs, which are known for their critical role in post-transcriptional gene regulation. The expression and biogenesis of most miRNAs follow a coordinated series of events that begins with transcription of the primary miRNAs (pri-miRNAs) in the nucleus. Following nuclear processing by the microprocessor complex into precursor-miRNAs (pre-miRNAs), the pre-miRNAs are exported to the cytoplasm, where they undergo further processing by the RNase III endonuclease, dicer into 21-23 nucleotide mature miRNA duplexes1. One of the strands of the processed mature miRNA binds to target messenger RNAs (mRNAs), leading to degradation or translational repression of the targets2. Based on the pleiotropic role of miRNAs in simultaneously regulating multiple diverse target mRNAs, it is not surprising that miRNA expression is tightly regulated3. Indeed, inappropriate expression contributes to various disease states, especially in cancer. The aberrant expression of miRNAs not only represents a disease-specific signature but has also emerged as a target for prognostic and therapeutic potential4,5,6. In addition to their intracellular roles, miRNAs also have non-autonomous roles. For example, miRNAs can be selectively packaged into EVs by donor cells and exported to recipient sites where they elicit diverse phenotypic responses in normal and disease physiology7,8,9.
Despite clear evidence that EV-associated miRNAs are functional biomolecules, it is not completely understood how specific subsets of miRNAs are dysregulated in disease states such as cancer and how cellular machinery selects and sorts miRNAs into Evs10,11. Given the potential role of EV-miRNAs in modulating the microenvironment, it is critical to identify the mechanisms involved in the export of select miRNAs to fully elucidate the role of EVs in intercellular communication and disease pathogenesis. Understanding the process of miRNA release into EVs will not only highlight important mediators of miRNA export but can also provide insights into potential therapeutics. To achieve this level of knowledge, tools need to be adapted to faithfully address these new experimental questions. Indeed, studies that evaluate EVs are growing exponentially due to the introduction of new techniques and controls12,13. In relation to evaluating the abundance of exported miRNAs into EVs, next-generation sequencing and quantitative reverse transcription polymerase chain reaction (qRT-PCR) have been the current standard tools. While these tools are useful for evaluating the abundance of miRNAs in EVs, there are limitations to their sensitivity and specificity, and using these static approaches for evaluating dynamics is insufficient. Delineating the dynamics of miRNA export into EVs requires a combination of specialized tools. Here, we present a comprehensive protocol using fluorescently conjugated miRNAs, to analyze the dynamics of miRNA release from donor cells and incorporation into EVs. We also discuss the advantages and limitations of the method and provide recommendations for optimal use. The versatile method presented in this paper will be useful to researchers interested in studying miRNA release into EVs and their potential roles in cellular communication and disease.
Protocol
NOTE: As a prerequisite to using this technique, identification and validation of selectively exported miRNAs is required. Since different cell lines vary based on the miRNA cargo sorted into their EVs, it is recommended that the cell line of interest and associated EVs be evaluated for miRNAs prior to use. Additionally, lipofectamine-based transfection is one of the critical steps in the protocol; predetermining transfection efficiency prior to setting up the experiment is recommended.
1. Growing cells in culture and transfecting cells with fluorophore-conjugated EV-miRNA
- Plate 200,000 to 400,000 cells in triplicate wells in a 6-well plate in an appropriate culture medium. Gently swirl the plate to ensure uniform cell distribution. Incubate the cells until they reach 70%-80% confluency, approximately 24-48 h.
- For each well to be transfected, dilute the transfection reagent in 100µL of serum-free medium in a microcentrifuge tube according to the manufacturer's instructions.
NOTE: The optimal concentration of lipid should be determined for the cell line of interest prior to this setup. - For each well to be transfected, dilute the fluorophore-conjugated EV-miRNA (2nM) and cell-miRNA (2nM) separately in a microcentrifuge tube in 100 µL of serum-free media.
NOTE: The total volume should be calculated for the total number of wells transfected. In this protocol, the fluorescence was evaluated at 7 time points; therefore, the total volume was 700 µL for step 1.2 and 700 µL for step 1.3. Additionally, to confidently capture the positive fluorescent population of cells, the user should transfect cells with a non-fluorescent scrambled miRNA and set up a gate excluding these cells. - Add the diluted RNA mix (from step 1.3) to the diluted transfection reagent (from step 1.2) and mix gently by inverting the tube 3-4 times. Let the combined lipid-RNA mix incubate for 30 min at room temperature (RT).
- Remove the regular culture media from the cells and add 800 µL of serum-free media to each well. Gently add 200 µL of the lipid-RNA mix (from step 1.4) dropwise to the cells.
NOTE: Opti-MEM media was used for transfecting the cell line of interest (Calu6 cells). Try to add the lipid-RNA mix directly to the culture media, avoiding adding the mixture to the sides of the plate.
2. Harvesting transfected cells at various time points for analysis of fluorescence
- Incubate the transfected cells at 37 °C for 7 h.
NOTE: To validate consistent transfection of both miRNAs (EV-miRNA and cell-miRNA), cells are collected from one well 3 h post-transfection to analyze the fluorescence using the flow cytometer. - After the 7 h incubation period, remove the transfection complex from the wells and replace it with complete media.
- To evaluate the 7-h time-point, harvest the cells and wash the cells three times with 1x PBS.
- If working with adherent cells, add 200 µL of trypsin to one well and wait for the cells to detach from the plate. Transfer the detached cells to a microcentrifuge tube and pellet by centrifugation at 200xg for 5 min.
- Discard the supernatant carefully to avoid disrupting the cell pellet. For the first PBS wash, resuspend the pellet in ~500 µL of 1x PBS and repeat the pelleting using the centrifugation steps above. Repeat the PBS wash an additional two times.
- Resuspend the cell pellet in 200 µL of 2% paraformaldehyde (PFA) solution and evaluate fluorescence.
NOTE: After adding 2% PFA, the cell pellet can be stored at 4 °C for 3-4 days.
- Harvest the remaining time points following the steps in section 2.3. Collect all of the stored cell pellets and proceed with the steps in section 3.
3. Flow cytometry analysis of cell-miRNA and EV-miRNA signal in cells
- Prepare cells for flow cytometry analysis. Filter the cell suspension from section 2 through 35 µm strainer caps that are part of the round bottom polystyrene FACS tubes to enable the removal of debris and cell aggregates that might interfere with fluorophore analysis.
- Set up the instrument following calibration instructions. Turn on the flow cytometer and allow it to warm up for at least 30 min before use. Prime the fluidics system with sheath fluid, and check and adjust the laser alignment.
- Run quality control to confirm instrument fluidics. Load ultrapure filtered water into the flow cytometer and run at an appropriate flow rate for adequate acquisition time.
NOTE: The appropriate flow rate and acquisition time will depend on the cell type of interest, sample complexity, and desired data quality. - Open the flow cytometry data analysis software and confirm the performance of the detector and lasers. Set up threshold and voltage parameters for detection based on the cell line of interest and fluorophores of choice.
NOTE: For all downstream experiments, Calu6 cells were used.- Set the threshold for capturing the signal for Calu6 cells at FSC 5000. Perform the analysis for 1 x 104 cells for each experimental repeat.
- To account for the spectral overlap between different fluorophores, use un-transfected cells and cells independently transfected with only one of the fluorophores to set up compensation controls.
- For detection of miRNA export, use candidate EV-miRNAs (miR-451a and miR-150) conjugated to Alexa-fluor 488, and a control cell-miRNA (cel-miR-67) conjugated to Cy3.
- For each EV-miRNA, conjugate the fluorophore to the 5′ end of the 5p strand. Before transfection, anneal the fluorophore-conjugated strand to its 100% complementary 3p strand.
- For the expression analysis in transfected cells, analyze the samples using the FSC, SSC, FITC, and PE channels set to 258, 192, 269, and 423 voltage units, respectively (Figure 1A, B).
- Introduce the negative control sample into the flow cytometer. Capture cell signals after defining forward scatter (FSC) and side scatter (SSC) intensities in a custom worksheet.
- In the plot, choose FSC for the X-axis, SSC for the Y-axis, and gate for the population of interest by drawing a polygon around it (see Figure 1B, bottom panel).
- Create a new plot using the gated population.
- Set the X-axis to fluorescence signal 1 (FS1) for cell-miRNA detection and the Y-axis to fluorescence signal 2 (FS2) for EV-miRNA detection. Set up compensation parameters for the individual fluorophores using cells independently transfected with each of the fluorescently tagged miRNAs. In the new plot, gate on the population of interest.
- For detection of EV-miRNA, use the FITC channel. For the evaluation of cell-miRNA signal, use the PE channel. In the software, create a gate around the cells that were positive for both FITC and PE by drawing a polygon around the double fluorescence population when FITC was selected for the X-axis and PE was selected for the Y-axis.
- Record the statistics for the gated population.
- Record the percentage of cells in the gated population that are positive for both FITC and PE, as well as for those that are positive for either FITC or PE.
- Repeat the above steps for any additional samples or controls in the experiment.
4. Isolating EVs from transfected cells
- For cells cultured in serum-containing media, deplete EVs from the serum to prevent cross-contamination.
- To generate EV-depleted serum, dilute the serum to 50% in cell culture media to reduce viscosity and centrifuge the resulting 50% serum at 110,000 x g for 18 h.
- Collect the supernatant (EV-free serum) carefully and filter it using a 0.2 µm filter assembly. Use the resulting 50% EV-free serum to generate complete media, in this case, RPMI plus 10% serum.
- To collect EVs, add the EV-depleted complete media to the cells and collect the EVs 48 h after incubation.
NOTE: The protocol outlined below has been modified from Kowal et al.14.
- Grow cells in 15 cm tissue culture dishes to 80% confluency. Transfect the cells separately with cell-miRNA and EV-miRNA in 10 mL of Opti-MEM following the transfection protocol in section 1 of the protocol.
NOTE: Not all transfection protocols scaleup linearly. It may be essential to first adapt and evaluate the transfection conditions in 15 cm plates. - After 7 h, aspirate the transfection media and replace it with the EV-depleted media (15 mL). Grow cells in the EV-depleted media for 48 h.
- Remove the conditioned media from the cells and dispense it into a 50 mL centrifuge tube inside the biosafety cabinet. Centrifuge the conditioned media at 2000 x g for 10 min at 4 °C to remove dead cells and cell debris. Transfer the resulting supernatant to a fresh tube.
- Centrifuge the supernatant at 10,000 x g for 40 min at 4 °C. Remove the supernatant and filter it using a 0.2 µm filter assembly.
- Transfer the filtered cell culture media to a sterile ultracentrifuge tube inside the biosafety cabinet. Weigh and balance the tubes against each other. Spin the samples at 110,000 x g for 90 min at 4 °C. Discard the supernatant.
- Resuspend the pellet in an appropriate volume of 1x PBS to wash and centrifuge at 110,000 x g for 90 min 4 °C.
NOTE: The EV pellet was resuspended in 60 mL of 1x PBS based on the maximum volume allowed for the fixed-angle rotor tubes used in this study. - Aspirate PBS and suspend the EV pellet in an appropriate volume (50-100 µL) of fresh 1x PBS and vortex. Store EVs at -80 °C for later use.
5. Flow cytometry analysis of cell-miRNA and EV-miRNA signal in EVs
- To prepare EV suspension for flow cytometry analysis, dilute EVs in an appropriate volume of ultrafiltered 1x PBS.
NOTE: The loading volume for the flow cytometry analysis of EVs should be determined based on the number of EVs in samples. Roughly, if there are ~30,000 particles in the EV suspension, the final volume should be ~500-600 µL in 1x PBS. A clean sample has around 35-50 particles/µL. - Use the nano-flow cytometer or an alternative equipment powerful enough to analyze nano-sized particles.
NOTE: Here, Attune NxT nano-flow cytometer was used. - Open the software and set the instrument up following the recommended calibration instructions. Perform the Performance Test using performance nano-beads. If the nano-beads are detected in the standard size range, the performance is optimal. Determine the appropriate flow rate for the detection of EV particles.
- Run quality control using ultrapure filtered water to confirm instrument fluidics and performance of detector and lasers.
- Using calibration beads suitable for the experiment, set the flow cytometer to accurately detect different-sized beads and measure fluorescence signals.
NOTE: For the setup of EV analysis by flow cytometry, ApogeeMix beads were used. These beads contain a mixture of non-fluorescent silica beads and fluorescent polystyrene beads ranging in sizes from 80 nm to 1300 nm.- Set up the parameters to assess the sensitivity and resolution of different populations in the bead mix according to the manufacturer's protocol. Use the appropriate gating to resolve the bead population from the instrument noise.
- Vortex the samples thoroughly to evenly distribute EVs before loading for analysis.
- Open the flow cytometry data analysis software and introduce ultra-filtered PBS. Set the gating up for EV particles using ultra-filtered PBS while ensuring that the gating corresponds to the appropriate size of particles based on the beads in step 5.5.
- Load the negative control sample to analyze EVs as they pass through the flow cytometer. Capture EV signals while plotting FSC on the X-axis and SSC on the Y-axis.
NOTE: The SSC threshold for capturing EVs was set at 300. The acquisition volume was set at 95 µL from a total of 145 µL drawn volume. The flow rate was set at 12.5 µL/min. The analysis was performed with 1 x 106 EV particles. The EV population is heterogeneous and will result in some debris isolated along with EVs. Overall particle detection in the FSC vs. SSC plot highlights particles in a diverse range, which is expected when using crude EV preparations. - Create a new plot using the gated population.
- This time, choose fluorescence signal 1 (FS1) for cell-miRNA detection on the X-axis and fluorescence signal 2 (FS2) for EV-miRNA detection on the Y-axis.
- Set up compensation parameters for the fluorophores using the EV samples isolated from cells transfected with a single miRNA. In the new plot, gate on the EV population of interest.
- For the expression analysis of fluorophore-conjugated miRNAs in EVs isolated from the transfected cells, analyze the EV samples using the FSC, SSC, BL1, and YL1 channels set to 370, 370, 400, and 400 voltage units, respectively (Figure 2A, B).
- Record the statistics for the gated population and visualize them using histograms and dot plots.
- Repeat the above steps for any additional samples or controls in the experiment.
6. Validating the release of miRNAs in EVs through qRT-PCR
- Extract RNA from EVs and parent cells using miRVana RNA isolation kit following the manufacturer's instructions.
NOTE: Quantification of RNA isolated from EVs is not possible because the total amount of RNA isolated from EVs does not typically cross the minimal quantity threshold of a standard Nano-drop. This is due to the depletion of ribosomal RNA from EV samples, which can be validated using the Agilent Bioanalyzer. Thus, the integrity score for EV-RNA quantification is usually not reliable and is not the true representation of EV-RNA quantity. Therefore, for qRT-PCR quantification of the EV-RNA and cell-RNA, equal volumes of RNA isolates following isolation from an equal number of cells were used. For normalization of qRT-PCR, miRNAs ubiquitously present across multiple samples as determined from previously acquired RNA sequencing data were used. - Convert the extracted RNA to cDNA using a reverse transcriptase kit specific for small RNAs.
- Set up the qRT-PCR reaction using a cDNA template and miRNA-specific primers following the manufacturer's instructions.
NOTE: For validation experiments, miRCURY LNA qRT PCR kit and corresponding primers from the manufacturer were used to evaluate the expression of cell-miRNA and EV-miRNA.
Representative Results
Here, we utilize flow cytometry as a powerful tool to investigate the release of miRNA from the cells into EVs. Using this protocol, flow cytometry analysis of cells transfected with cell-miRNA and EV-miRNA revealed a sequential decrease of fluorescence signal corresponding to EV-miRNA, while the signal corresponding to the cell-miRNA was retained in the cells. To ensure that fluorophore conjugation does not interfere with miRNA release into EVs, miR-451a was conjugated to two separate fluorophores (i.e., Alexa fluor 488 and Alexa fluor 750), and cell retention was evaluated. The results indicate minimal variability between the detection of the separate fluorophore signals post-transfection, validating the reliability of this approach (Figure 3A,B).
Because various reasons could explain the loss of fluorescently tagged miR-451a from the cells, including RNA degradation, it was critical to verify that cellular loss coincided with EV abundance. Using flow cytometry, accumulation of miR-451a conjugated to Alexa fluor-488 in EVs was observed in a time-dependent manner, providing evidence of efficient packaging and eventual release of the miRNA into EVs. In contrast, the fluorescence signal corresponding to the cell retained miRNA, cel-miR-67 was not detected in the EVs (Figure 4). These findings using this new approach were validated using qRT-PCR, which revealed a significantly higher EV/cell expression ratio for miR-150, an additional EV-miRNA, when compared to the cell-miRNA, cel-miR-67 (Figure 5).
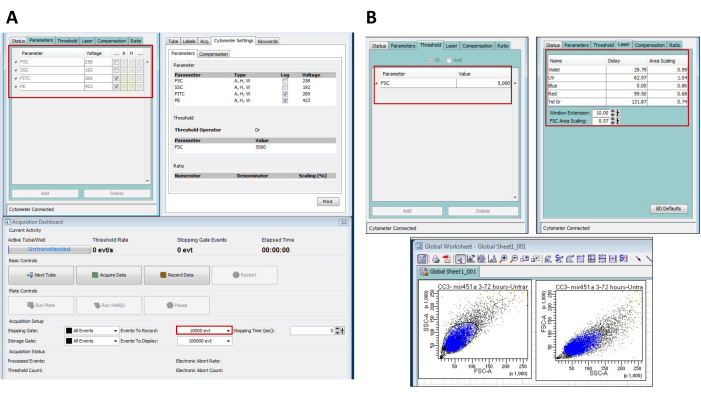
Figure 1: Set up of flow cytometer parameters for detection of signals in cells. (A) For the analysis of cell fluorescence, using the software, the highlighted voltages were set up for the parameters indicated. The instrument was set up to record 10,000 cells per sample. The Inspector View confirms the parameters set up for detection and analysis. (B) For the analysis of cell fluorescence, the Threshold for the FSC parameter was set at 5,000 under the cytometer settings tab of the software. The top right panel shows the laser settings for the data capture. The bottom panels show an active global worksheet during the progress of the detection of cells. The samples were run at a medium flow and captured while establishing FSC-A and SSC-A as x-axis and y-axis parameters, respectively. The gating was set using a drawing polygon tool around the population of interest. The gating might be different for another cell line of interest depending upon the diversity of the cell population. Please click here to view a larger version of this figure.
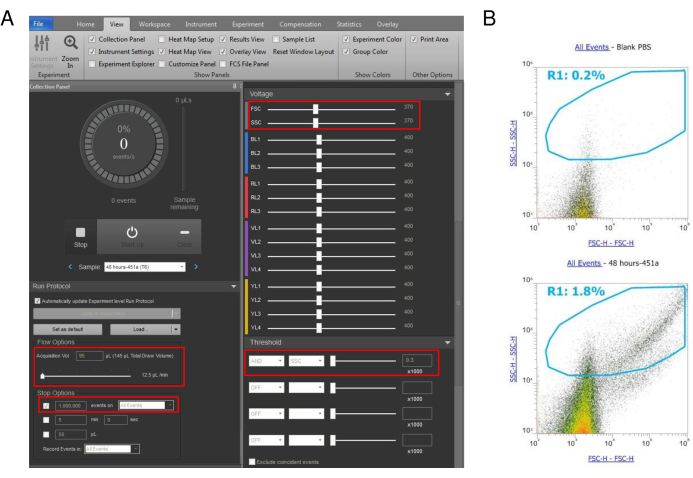
Figure 2: Set up of flow cytometry parameters for signal detection in EVs. (A) For the detection and downstream analysis of EVs, the acquisition volume was set at 95 µL with the flow speed set at 12.5 µL/min. The chosen threshold for EV detection was 0.3 x1000 under the instrument settings tab of the software based upon the calibration beads used. Voltages for FSC, SSC, BL1, and YL1 were 370, 370, 400 and 400, respectively. (B) Setup of gating for capturing heterogenous EV population and distinguishing EVs from the background noise of the instrument. A blank PBS sample (top) was used as a negative control to resolve background noise from the EV population. EV sample (bottom) shows that the population of interest is only 1.8% of the total population because the sample is heterogeneous with overlapping particles in the size range. However, stringent gating was chosen to ensure the detection of the EV population of interest. Please click here to view a larger version of this figure.
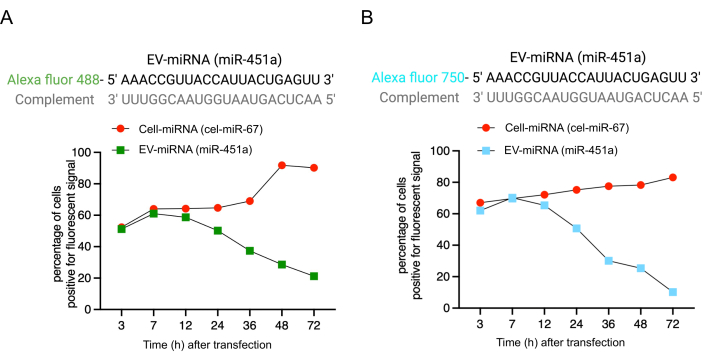
Figure 3: Signal detection in cells through flow cytometry. (A) Fluorescence corresponding to EV-miRNA and cell-miRNA was detected in cells at times indicated between 7 h and 72 h following transfection. The signal corresponding to EV-miR-451a (Alexa fluor-488) diminished with time in comparison to the signal from the cell-retained-miRNA, cel-miR-67. (B) Fluorescence corresponding to EV-miR-451a labeled with Alexa fluor-750 showed the same trend as the Alexa Fluro-488 labeled miR-451a, indicating that the fluorophore does not impact the retention of miRNAs by the cells. Overall, the representative results indicate that the transfected miRNA behaves similarly to the endogenous miRNA based on the export of the miRNA into EVs. Please click here to view a larger version of this figure.
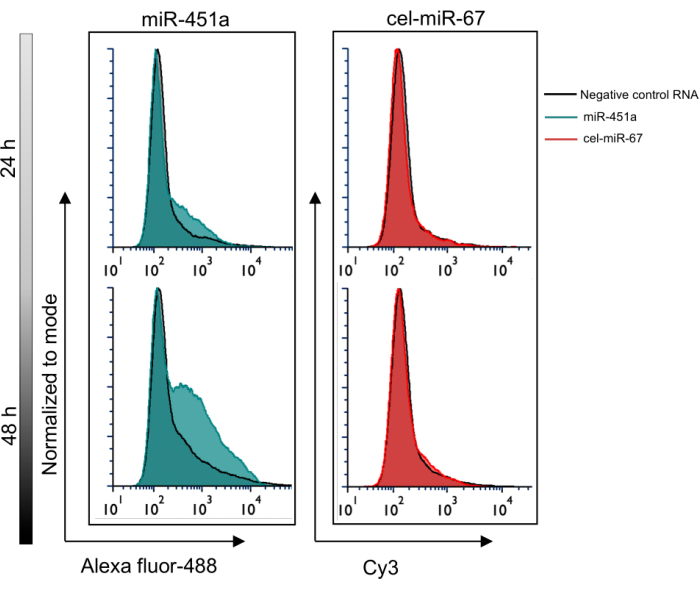
Figure 4: Signal detection in EVs through flow cytometry. Fluorescence corresponding to EV-miR-451a and cell-cel-miR-67 was detected in EVs 24 h and 48 h post-transfection. The signal corresponding to EV-miR-451a (Alexa fluor-488) was captured in EVs 48 h following transfection. In contrast, the signal for cel-miR-67 was not detected in EVs. A blank PBS sample was used to set up the gating for this experiment, as shown in Figure 2B. EV samples isolated from cells transfected with non-fluorescent scrambled RNA were used as a negative control. Please click here to view a larger version of this figure.
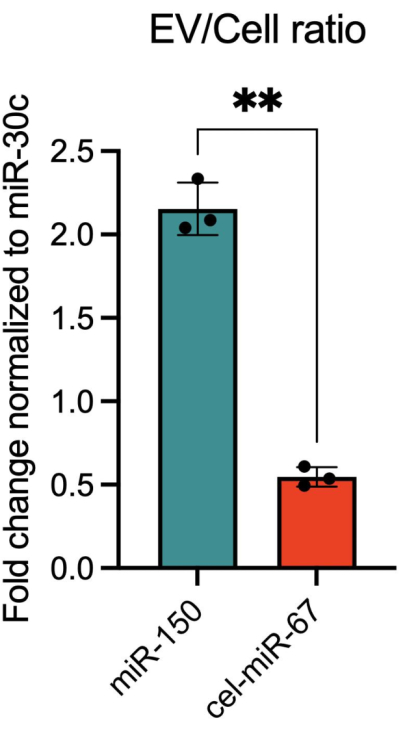
Figure 5: Quantification of EV-miRNA and cell-miRNA in cells and EVs using qRT-PCR. Calu6 cells were transfected with 2 nM of miR-150 and cel-miR-67 each. EVs were collected 48 h after transfection. EV enrichment was calculated as the ratio of EV expression divided by cellular expression in transfected Calu6 cells for miR-150 and cel-miR-67. Based upon RNA sequencing results comparing the expression of miRNAs in Calu6 cells and their corresponding EVs, miR-30c was used as an endogenous control and a normalizer for fold change computation. The data presented are from one biological replicate, with each data point presenting triplicate qRT-PCR reactions. Data are expressed as mean ± SD, and p-values were computed using an unpaired t-test with Welch's correction (**p < 0.01). Please click here to view a larger version of this figure.
| Strengths | Weaknesses |
| Allows for simultaneous assessment of selective export of multiple miRNAs in cells and respective EVs | Require additional procedures to differentiate between active and passive export phenomenon |
| Assessment of crude heterogenous population is possible and does not require prior separation of EV populations. Can be used to analyze different types of EVs NOTE: Threshold for detection of different types of particles will vary |
Low sensitivity does not allow for detection of limited concentrations of miRNAs in cells or EVs. This can be overcome by incorporating downstream qRT-PCR analysis following RNA isolation from EVs |
| Requires a short amount of time for set up and analysis in comparison to standard methods used for miRNA quantification | Lack of standardization protocols for different types of equipment, cell-lines, and various fluorophores. |
Table 1: The major strengths and weaknesses of the protocol.
Discussion
The newly established protocol enables capturing kinetics of miRNA release into EVs post-transfection of EV-miRNAs. The approach allows simultaneous analysis of multiple EV-miRNAs and cell-miRNAs, subject to the capabilities of the cytometer. Moreover, while flow cytometry analysis can provide valuable insights into EV miRNA biology, it is not without limitations. Albeit some of the limitations can be overcome when used in conjunction with other techniques for a more comprehensive understanding.
An emerging area of interest in EV biology is comprehending the selective export of miRNAs into EVs15,16. While this protocol offers a powerful tool to allow studying selective export of miRNA, one of the critical challenges is distinguishing between active and passive export. Caution must be exercised since overexpressing a miRNA at a high enough concentration could alter the export of miRNA, potentially resulting in inappropriate passive loading. Therefore, experimentation to determine the optimal transfection concentrations of miRNAs is crucial while studying selective export.
Moreover, the protocol, when combined with complementary techniques such as nanoparticle tracking analysis (NTA), enables the researcher to assess the EV particle count while also evaluating the miRNA release and has the potential to enable distinguishing between alterations in EV biogenesis pathways and miRNA export mechanisms. This is especially important due to the overlapping mediators of these processes17. Considering that miRNAs can impact various phenotypes, including some involved in EV biogenesis, it would be prudent to evaluate the particle count following transfection of miRNAs18. To overcome this hurdle, another option would be to incorporate an EV-miRNA that remains unaltered under the experimental conditions, thereby ruling out factors involved in EV biogenesis while highlighting the pathways involved in EV-miRNA export.
Several studies have quantified miRNAs using qRT-PCR, a standard method for endogenous miRNA quantification10,11,16. However, it lacks the capability to dynamically capture the release of miRNAs into EVs. When comparing flow cytometry to qRT-PCR, they differ greatly based on the assay sensitivity and design. While qRT-PCR is a highly sensitive technique and theoretically capable of quantifying minimal miRNA copy counts, distinguishing two samples requires at least a 2-fold (100%) difference in the copy number of miRNAs of interest. Flow cytometry, on the other hand, captures signal-based differences based on the abundance of fluorescently labeled miRNA and can differentiate small percentage changes in signals. Nonetheless, its low sensitivity restricts the detection of miRNAs with limited copy numbers. The two techniques synergistically complement each other when used in conjunction, as indicated in the protocol.
In summary, this protocol offers a valuable tool to investigate selective miRNA export dynamics. Despite the aforementioned challenges, the strength of this protocol lies in its ability to analyze multiple miRNAs simultaneously, contributing to a better understanding of miRNA biology, their export, and their role in EV biology. The major strengths and weaknesses of the protocol that need to be considered are listed in Table 1.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We acknowledge support and advice from Dr. Jill Hutchcroft, Director of the Flow Cytometry Core Facility at Purdue University. This work was supported by R01CA226259 and R01CA205420 to A.L.K., an American Lung Association Innovation Award (ANALA2023) IA-1059916 to A.L.K., a Purdue Shared resource facility grant P30CA023168, and a flagship Fulbright Doctoral Scholarship awarded by the Department of the State, USA to H.H.
Materials
| 1.5 mL microcentrifuge tubes | Fisher | 05-408-129 | |
| 15 mL Falcon tubes | Corning | 352097 | |
| 6-well clear flat bottom surface treated tissue culture plates | Fisher Scientific | FB012927 | |
| ApogeeMix 25 mL, (PS80/110/500 & Si180/240/300/590/880/1300 nm) | Apogee Flow Systems | 1527 | To set up gating and parameters for particle detection |
| Attune Nxt Flow Cytometer | ThermoFisher Scientific | N/A | For fluorescence analysis in EVs |
| BD Fortessa Cell Analyzer | BD Biosciences | N/A | For fluorescence analysis in cells |
| Cell culture incubator | N/A | N/A | Maintaining temperature of 37 °C and 5% CO2 |
| Cell Culture medium | N/A | N/A | specific for the cell-line of interest |
| Cell line of interest | N/A | N/A | Any cell line tested and evaluated for EV and cellular abundance of miRNAs on interest. |
| Cell-miRNA (miRIDIAN microRNA Mimic Red Transfection Control) | Horizon discovery | CP-004500-01-05 | miRNAs predetermined to be retained by the cell-line of interest and not selectively exported out into EVs |
| EV-miRNA (Fluorophore conjugated miRNAs) | Integrated DNA Technologies (IDT) | miRNAs predetermined to be selectively sorted into EVs | |
| Fluorescence microscope | N/A | N/A | |
| Gibco Opti-MEM Reduced Serum Medium | Fisher Scientific | 31-985-070 | |
| Hausser Scientific Hemocytometer | Fisher | 02-671-54 | |
| Hyclone 1x PBS (for cell culture) | Fisher | SH30256FS | |
| Lipofectamine RNAimax | Fisher Scientific | 13-778-150 | |
| miRCURY LNA Reverse Transcriptase | Qiagen | 339340 | |
| miRCURY LNA SYBR Green PCR | Qiagen | 339347 | |
| mirVana RNA Isolation | Qiagen | AM1561 | |
| Nuclease free water | Fisher Scientific | 4387936 | |
| Paraformaldehyde, 4% in PBS | Fisher | AAJ61899AK | |
| Reagent reservoir nonsterile | VWR | 89094-684 | |
| Thermo ABI QuantiStudio DX Real-Time PCR | ThermoFisher Scientific | N/A | |
| Trypsin 0.25% | Fisher | SH3004201 | |
| Ultracentrifuge | N/A | N/A |
References
- Kasinski, A. L., Slack, F. J. MicroRNAs en route to the clinic: Progress in validating and targeting microRNAs for cancer therapy. Nature Reviews Cancer. 11 (12), 849-864 (2011).
- Treiber, T., Treiber, N., Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nature Reviews Molecular Cell Biology. 20 (1), 5-20 (2019).
- Lu, J., et al. MicroRNA expression profiles classify human cancers. Nature. 435 (7043), 834-838 (2005).
- Orellana, E., Tenneti, S., Rangasamy, L., Lyle, L., Low, P., Kasinski, A. FolamiRs: Ligand-targeted, vehicle-free delivery of microRNAs for the treatment of cancer. Science Translational Medicine. 9 (401), eaam9327 (2017).
- Orellana, E., et al. Enhancing microRNA activity through increased endosomal release mediated by nigericin. Molecular Therapy- Nucleic Acids. 16, 505-518 (2019).
- Abdelaal, A. M., et al. A first-in-class fully modified version of miR-34a with outstanding stability, activity, and anti-tumor efficacy. Oncogene. , (2023).
- Crescitelli, R., et al. Distinct RNA profiles in subpopulations of extracellular vesicles: Apoptotic bodies, microvesicles and exosomes. Journal of Extracellular Vesicles. 2, (2013).
- Hasan, H., et al. Extracellular vesicles released by non-small cell lung cancer cells drive invasion and permeability in non-tumorigenic lung epithelial cells. Scientific Reports. 12 (1), 972 (2022).
- Costa-Silva, B., et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nature Cell Biology. 17 (6), 816-826 (2015).
- Villarroya-Beltri, C., et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nature Communications. 4, 2980 (2013).
- Cha, D., et al. KRAS-dependent sorting of miRNA to exosomes. elife. 4, e07197 (2015).
- Gandham, S., et al. Technologies and standardization in research on extracellular vesicles. Trends in Biotechnology. 38 (10), 1066-1098 (2020).
- Théry, C., et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles. 7 (1), 1535750 (2018).
- Kowal, J., et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proceedings of the National Academy of Sciences of the United States of America. 113 (8), E968-E977 (2016).
- Sork, H., et al. Heterogeneity and interplay of the extracellular vesicle small RNA transcriptome and proteome. Scientific Reports. 8 (1), 10813 (2018).
- Mittelbrunn, M., et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nature Communications. 2, 282 (2011).
- Chiou, N. T., Kageyama, R., Ansel, K. M. Selective export into extracellular vesicles and function of tRNA fragments during T cell activation. Cell Reports. 25 (12), 3356-3370 (2018).
- Guzewska, M. M., Witek, K. J., Karnas, E., Rawski, M., Zuba-Surma, E., Kaczmarek, M. M. miR-125b-5p impacts extracellular vesicle biogenesis, trafficking, and EV subpopulation release in the porcine trophoblast by regulating ESCRT-dependent pathway. The FASEB Journal. 37 (8), e23054 (2023).

