Visualization of Caspase Activation in Human Monocyte-Derived Macrophages
Abstract
Source: Bolívar, B. E., et. al., Visualization of Inflammatory Caspases Induced Proximity in Human Monocyte-Derived Macrophages. J. Vis. Exp. (2022).
This video demonstrates the activation of caspase proteins in human macrophages using bimolecular fluorescence complementation reporter proteins. The pro-domains of caspase-1 are fused with non-fluorescent fragments of Venus proteins, which are recruited to inflammasome complexes. The proximity of inflammasomes induces the refolding and fluorescence of Venus fragments, confirming caspase activation in the macrophages.
Protocol
All procedures involving human participants have been performed in compliance with the institutional, national, and international guidelines for human welfare and have been reviewed by the local institutional review board.
1. Isolation of human monocytes and differentiation into macrophages
- Obtain anticoagulated blood from de-identified healthy individuals at a regional blood bank and isolate peripheral blood mononuclear cells (PBMCs) as indicated below.
NOTE: Perform all steps in a tissue culture laminar flow hood. Use sterile tubes only and wear gloves. Add 10% bleach to all blood-related products when disposing. Sterile phosphate-buffered saline (PBS) (1x) or Dulbecco's phosphate-buffered saline (DPBS) (without Ca2+ and Mg2+) can be used interchangeably.- Prepare the dilution buffer: Supplement 1x sterile PBS with 2% fetal bovine serum (FBS) and 0.5 mM ethylenediaminetetraacetic acid (EDTA).
- Prepare the culture medium: Supplement RPMI-1640 medium with FBS (10% (v/v)), glutamax (2 mM), and Penicillin/ Streptomycin (50 I.U./50 µg/mL)
- Precool the running buffer (Table of Materials) according to the manufacturer's protocol.
- Dilute whole blood with two volumes of dilution buffer. Using a serological pipet, transfer 15 mL of the anticoagulated blood to a 50 mL tube containing 30 mL of the dilution buffer. Mix gently by inversion.
- For each 10 mL of whole blood or 30 mL of diluted blood, add 15 mL of the density gradient medium to a 50 mL empty tube.
- Layer the density gradient medium from step 1.1.5 with 30 mL of diluted blood slowly and steadily using a 25 mL serological pipet. Keep the tip of the pipet against the wall of the tube and the tube at a tilted angle.
- Carefully transfer the tubes to a swinging bucket centrifuge. Avoid disturbing the two phases. Centrifuge the tubes at 400 x g at room temperature (RT) for 25 min with acceleration and deceleration set to the minimum value.
- Carefully remove the top (clear) plasma layer using a 10 mL pipet and dispose of it in a container with bleach (10%).
- Collect the interphase (white) layer of peripheral blood mononuclear cells (PBMCs, Figure 1) with a 10 mL pipet and transfer to a fresh 50 mL tube. Combine the white layer from different tubes of the same donor in a 50 mL tube up to 30 mL.
- Bring each tube to a total volume of 50 mL with the dilution buffer from step 1.1.1 and centrifuge at 300 x g and 4 °C for 10 min. Remove the supernatant with a 10 mL pipet and dispose of it in a container with bleach (10%).
- Resuspend each cell pellet in 1 mL of the pre-cooled running buffer from step 1.1.3 using a p1000 micropipette. Combine cell suspensions from the same donor in a new 15 mL tube. Bring the volume of each tube to 15 mL with pre-cooled running buffer and mix well by inversion.
- Take a 20 µL aliquot of the cell suspension from step 1.1.11 and prepare a 1:100 dilution using 1x sterile PBS. Determine the cell number using a hemocytometer.
- Centrifuge the cell suspension from step 1.1.11 at 300 x g and 4 °C for 10 min and remove the supernatant with a 10 mL pipet. If necessary, use a p200 micropipette to remove the supernatant completely.
- Resuspend the isolated PBMCs in 80 µL of pre-cooled MACS running buffer for each 1 x 107 cell, adding up to a maximum of 800 µL of the buffer.
- Add 20 µL of anti-human CD14 MicroBeads per each 1 x 107 cells or up to 100 µL per blood sample (~100 mL of undiluted blood). Mix well by inversion and place on a tube rotator for 20 min with continuous mixing at 4 °C.
- Remove the samples from the tube rotator, add 10 mL of pre-cooled running buffer to each tube, and centrifuge at 300 x g (acceleration = 5, deceleration = 5) and 4 °C for 10 min.
- Remove the supernatant with a 10 mL pipet and resuspend up to 1 x 108 cells in 500 µL of pre-cooled running buffer (2 x 108/mL).
- Perform the isolation of CD14-positive cells by magnetic cell sorting using a manual or automated system (Table of Materials) per the manufacturer's instructions.
- Take a 20 µL aliquot of the cell suspension from step 1.1.18 after CD14-positive selection and prepare a 1:100 dilution using 1x sterile PBS. Determine the cell number by counting the cells on a hemocytometer.
- Centrifuge the CD14-positive cells at 300 x g and RT for 10 min. Remove the supernatant using a 10 mL pipet or a vacuum system.
- Resuspend the cell pellet from step 1.1.20 in a pre-warmed culture medium from step 1.1.2 to a final cell density of 1 x 107 cells/mL.
- Seed the isolated CD14-positive monocytes at a cell density of 5 x 106 cells.
- On a 10 cm tissue culture dish, add 10 mL of culture medium from step 1.1.2 supplemented with 50 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF).
- Add 0.5 mL of the cell suspension from step 1.1.21 to the culture medium dropwise and gently swirl the plate. Incubate cells in a humidified tissue culture incubator (37 °C, 5% CO2) overnight.
- The next day, aspirate the medium using a vacuum system to remove cells that did not attach overnight. Add 10 mL of fresh culture medium supplemented GM-CSF (50 ng/mL) and incubate cells in a humidified tissue culture incubator (37 °C, 5% CO2) for 7 days to allow complete differentiation (see Figure 2A for the appearance of CD14+ monocytes at various stages of differentiation in GM-CSF ). Exchange the culture medium every 2-3 days and supplement with fresh GM-CSF (50 ng/mL) each time.
2. Preparation of electroporation components
NOTE: This protocol is designed for a 10 µL-Neon tip (Table of Materials). For each transfection, use 1-2 x 105 cells. It is recommended to seed transfected cells on a 48-well plate or 8-well chambered dish (10 µL-transfected cells per well). 1x sterile DPBS (without Ca2+ and Mg2+) can be used in place of PBS.
- On day 7, prepare an antibiotic-free medium by supplementing the RPMI-1640 medium with FBS (10 % (v/v)) and glutamax (2 mM).
- Place serum-free RPMI-1640 medium, trypsin-EDTA (0.25%) solution, 1x sterile PBS (without Ca2+ and Mg2+), and complete culture medium from step 1.1.2 in a 37 °C water bath.
- If using glass-bottomed dishes (for confocal microscopy) coat the dishes with poly-D-lysine hydrobromide.
- Coat an 8-well-chambered dish with 200 µL of poly-D-lysine hydrobromide (0.1 mg/mL in 1x sterile PBS) and incubate for 5 min at RT.
- Aspirate the poly-D-lysine solution and wash the glass once with 1x sterile PBS. Aspirate the PBS and proceed with step 2.4.
- Add 200 µL of antibiotic-free medium per well of the 48-well plate or 8 well-chambered dish and pre-incubate in a humidified tissue culture incubator (37 °C, 5% CO2) until ready to plate the transfected cells.
3. Preparation of cells for electroporation
NOTE: The yield of MDM from a 10 cm dish at the end of the 7-day differentiation period is approximately 1.5 x 106 cells. 1x sterile DPBS (without Ca2+ and Mg2+) can be used in place of PBS. This protocol was optimized so that most macrophages are detached from the plate with the maintenance of cell viability and integrity. MDM is difficult to detach from cell culture plates. Therefore, it may be necessary to perform steps 3.2 and 3.3 twice to dissociate the cells. Ensure that each incubation time with trypsin-EDTA (0.25%) does not exceed 5 min.
- Aspirate the media from fully differentiated macrophages on 10 cm dishes and wash the cell monolayer with warm serum-free RPMI-1640 medium. Make sure to completely remove the medium.
- Harvest the cells by adding 2 mL of warm trypsin-EDTA (0.25%) solution per 10 cm dish and incubate in a humidified tissue culture incubator (37 °C, 5% CO2) for 5 min.
- Complete the cell detachment by gently pipetting the trypsin-EDTA (0.25%) solution up and down over the entire dish area using a p1000 micropipette. Transfer the cell suspension to a 15 mL conical tube containing 5 mL of warm complete culture medium from step 1.1.2.
- Take the dish to a bright field microscope and check for cell detachment in various fields of view. If there is a considerable amount of cells still attached, repeat steps 3.2-3.3.
- Centrifuge the cell suspension at 250 x g for 5 min at RT.
- Aspirate the medium and resuspend the cells in 10 mL of 1x sterile PBS pre-warmed to 37 °C. Take a 20 µL aliquot to determine the cell number using a hemocytometer.
- Take 1-2 x 105 cells per intended transfection and place in a 15 mL tube. Bring to a final volume of 15 mL with pre-warmed 1x sterile PBS. Centrifuge at 250 x g for 5 min at RT.
- Aspirate the PBS and centrifuge one more time for 1 min at 250 x g. Remove any residual PBS from the cell pellet using a p200 micropipette.
4. Nucleofection of caspase BiFC components into human monocyte-derived macrophages
NOTE: This section of the protocol is performed using the Neon Transfection System (Table of Materials). This protocol outlines the steps to transfect 1-2 x 105 cells using a 10 µL Neon tip (Table of Materials). If using a 100 µL Neon tip, scale up accordingly. Avoid exposing cells to resuspension buffer R for more than 15 min, as this may decrease cell viability and transfection efficiency.
- Dilute the reporter plasmid (i.e., mCherry or dsRedmito at 100 ng/µL) in nuclease-free water or 0.5x TE buffer to visualize the transfected cells.
- Dilute the caspase BiFC plasmids to an appropriate concentration in nuclease-free water or 0.5x TE buffer, so that the total volume of plasmids does not exceed 30% of the total transfection volume of 10 µL (i.e., 300 ng/µL of C1 Pro-VC and 300 ng/µL of C1 Pro-VN).
- Prepare a 1.5 mL sterile microtube per intended transfection and add the appropriate amount of reporter plasmid (i.e., 50 ng or 0.5 µL) and caspase BiFC plasmids (i.e., 300 ng or 1 µL of C1 Pro-VC and 300 ng or 1 µL of C1 Pro-VN fragment). Keep the microtubes in the hood at all times.
- Place the pipette station, device, tips, electroporation tubes, and pipette in a sterile laminar flow hood.
NOTE: The pipette station, device, tips, electroporation tubes, and pipette are included in the Neon Transfection System. - Connect the high voltage and sensor connector on the pipette station to the rear ports on the device as per the manufacturer's instructions. Keep the pipette station close to the device.
- Connect the power cord to the rear AC inlet and proceed to connect the device to the electrical outlet. Press the power switch to turn on the device.
- Enter the transfection parameters in the startup screen displayed when the device is switched on and appropriately connected. Press on Voltage, enter 1000, and press on Done to set the voltage to 1000 V. Press on Width, enter 40, and press on Done to set the pulse duration to 40 ms. Lastly, press on # Pulses, enter 2, and press on Done to set the number of electrical pulses to 2.
- Take one of the electroporation tubes (provided in the kit) and fill it with 3 mL of electrolytic buffer E (for 10 µL tips and provided in the kit) at RT. Insert the electroporation tube into the pipette holder on the pipette station. Ensure that the electrode on the side of the tube is facing inwards and that a click sound is heard when the tube is inserted.
- Take the cell pellet from step 3.8 and add 10 µL of pre-warmed resuspension R buffer (provided in the kit) for each 1-2 x 105 cells. Mix gently with a p20 micropipette. Add 10 µL of the cell suspension to each tube set up in step 4.3 and mix gently with a p20 micropipette.
- Take the pipette and insert a tip by pressing the Push button on the second stop. Make sure the clamp completely picks up the mounting stem of the piston in the tip and that no gap is observed in the top head of the pipette.
- To aspirate the sample, press the Push button on the pipette to the first stop and dip it into the first tube containing the cell/plasmid DNA mixture. Slowly aspirate the mixture into the pipette tip.
NOTE: Avoid air bubbles as they can cause arcing during electroporation, and if detected by the device, can prevent delivery of the electric pulse. If air bubbles are observed, release the contents into the tube and try aspirating again. - Insert the pipette with the sample very carefully into the pipette holder. Make sure the pipette clicks and that it is properly placed.
- Press Start on the touchscreen and wait until the electric pulses are delivered. A message on the screen will indicate completion.
- Slowly remove the pipette from the station and immediately add the transfected cell suspension into the corresponding well with pre-warmed antibiotic-free medium from step 2.4 by slowly pressing the push-button to the first stop.
NOTE: This tip can be reused up to three times for the same plasmid; otherwise, discard it into a biohazard waste container by pressing the Push button to the second stop. - Repeat steps 4.10-4.14 for each tube containing a cell/plasmid DNA mixture.
- Gently rock the plate with transfected cells and incubate for 1-3 h in a humidified tissue culture incubator (37 °C, 5% CO2).
- Add to each well 200 µL of pre-warmed culture medium (complete medium) from step 1.1.2. Place the dish in the humidified tissue culture incubator (37 °C, 5% CO2) again. Allow at least 24 h for gene expression.
- The next day, inspect the cell viability and transfection efficiency using an epifluorescence microscope.
- Turn on the epifluorescence microscope and the fluorescent light source box per the manufacturer's instructions and place the culture dish on the microscope stage.
- Select the 10x or 20x objective and the 568 nm (RFP) filter.
- To estimate cell viability, press the Transmitted Light LED (TL) button to visualize all cells in the selected field. While looking into the microscope eyepiece, turn the focus knob until cells are observed and check for cell attachment in the selected field.
NOTE: Fully attached cells represent the viable cells while floating cells represent non-viable cells. If the confluency of the well is high, the presence of non-attached cells could be the result of an overestimation of cell number and not the result of low viability. However, low confluency accompanied by a high content of floating cells signifies low viability that may result from arcing during electroporation, plasmid toxicity, or overexposure to resuspension R buffer. Do not use wells that display the latter behavior. - To estimate transfection efficiency, focus on cells in the selected field under transmitted light as described above. Count the total number of cells in the selected field. With the Transmitted Light LED (TL) switched off, press the Reflected Light LED button (RL) to switch on.
- Fine focus on the reporter gene fluorescence (red cells) and count the total number of red fluorescent cells. Repeat these steps (4.18.4-4.18.5) for at least two more fields per well.
5. Treatment of transfected MDM and caspase BiFC data acquisition
NOTE: If planning to image the cells using an epifluorescence or confocal microscope, treatment with qVD-OPh (20 µM) for 1 h prior to treatment with the chosen stimulus is advised to prevent caspase-dependent cell death (predominantly apoptosis). This is used in imaging to prevent cells from lifting off due to apoptosis, making them very difficult to image as they move out of the focal plane. Note that caspase recruitment to the activation platform and the associated caspase BiFC is not dependent on the catalytic activity of the caspase, and consequently, caspase inhibition will not affect this step.
- Treat with chosen stimulus approximately 24 h after transfection and incubate for as long as necessary for each drug.
- Prepare the imaging medium by supplementing the culture medium from step 1.1.2 with Hepes (20 mM, pH 7.2-7.5) and 2-mercaptoethanol (55 µM).
- Add the desired concentration of stimulus to the pre-warmed imaging medium and mix gently.
- Remove the media from the cells carefully with a p1000 micropipette and add 500 µL of the stimulus solution from step 5.1.2 down the side of the well.
- To run untreated control wells, add an imaging medium without the stimulus.
- Incubate the cells in a humidified tissue culture incubator (37 °C, 5% CO2) for as long as indicated for each treatment.
- Visualize the cells using an epifluorescence or confocal microscope.
- Turn on the microscope and the fluorescent light source, following the manufacturer's instructions.
- Select the 10x or 20x objective and place the culture dish on the microscope stage.
- Using the microscope eyepiece, find cells under the 568 nm filter and focus on the cells expressing the dsRedmito/mCherry reporter (red cells).
- Count all the red cells in the visual field and record the number.
- While in the same visual field, change to the 488 or 512 filters (GFP or YFP), proceed to count the number of red cells that are also green (Venus-positive or BiFC-positive), and record the number.
- Count at least 100 dsRedmito/mCherry -positive cells from a minimum of three individual visual fields.
Representative Results
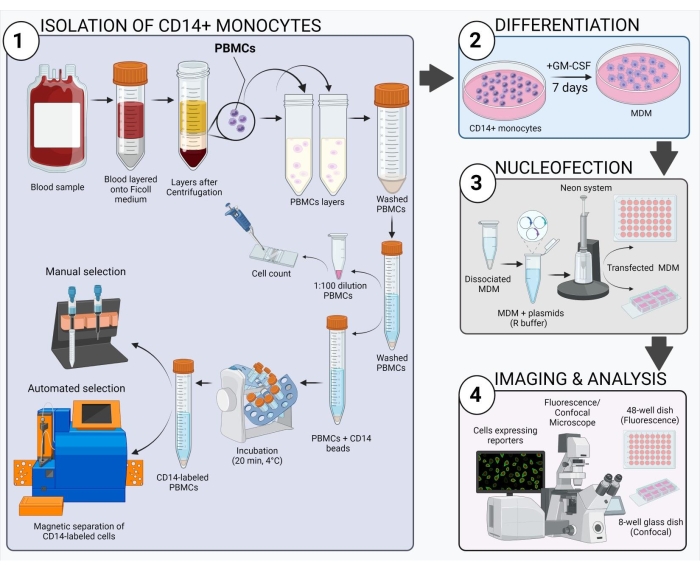
Figure 1: Schematic overview of the experimental workflow.
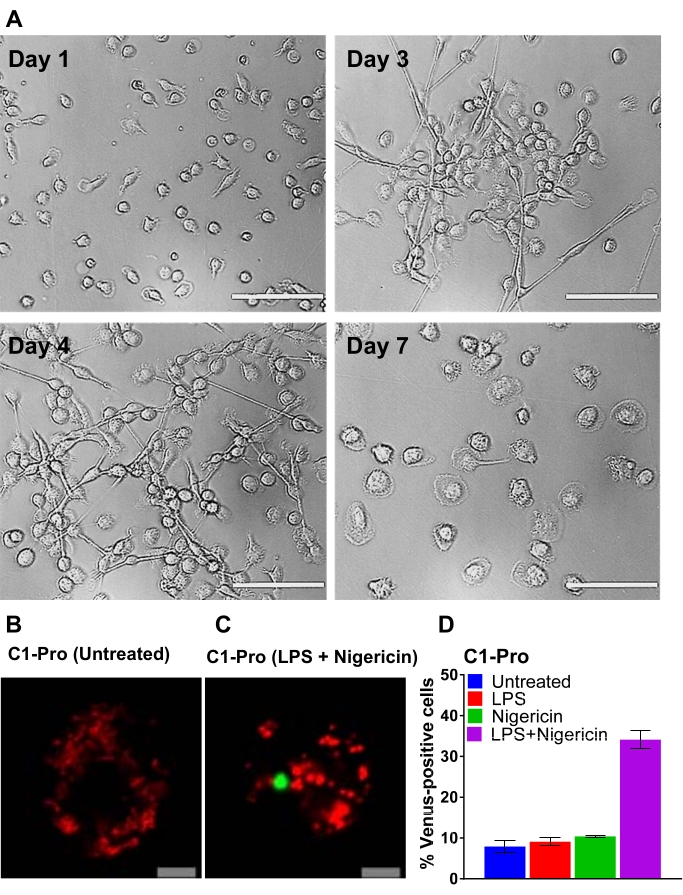
Figure 2: Differentiation of CD14-monocytes and transfection of human MDM. (A) Representative bright-field images of CD14+ monocytes from peripheral blood exposed to GM-CSF at days 1, 3, 4, and 7 of differentiation (Scale bar, 100 µm). (B-C) Human MDM was transfected with caspase-1 pro-BiFC pairs and the transfection reporter dsRedmito (50 ng, red). 24 h later, cells were primed with and without LPS (100 ng/mL) for 3 h and treated with nigericin (5 µM) in an imaging medium for 20 h. Representative confocal images of untreated and nigericin-treated cells are shown (Scale bar, 10 µm). The BiFC is shown in green. (D) Cells from (C) were assessed for the percentage of dsRedmito-positive cells (red, transfected cells) that were Venus-positive (green, caspase-1 BiFC complex) at 20 h. Error bars represent the SD of at least two independent counts per well.
Materials
| 48-well tissue culture2:34 plates | Genesee Scientific | 25-108 | |
| 10 cm Tissue Culture Dishes | VWR | 25382-166 | |
| 2 Mercaptoethanol 1000x | Thermo Fisher Scientific | 21985023 | |
| 8-well chambered coverglass with 1.5 HP coverglass | Cellvis | c8-1.5H-N | |
| AutoMACS columns | Miltenyi (Biotec) | 130-021-101 | For automated separation using AutoMACS Pro Separator only |
| AutoMACS Pro Separator | Miltenyi (Biotec) | 130-092-545 | For automated separation using AutoMACS Pro Separator only |
| AutoMACS Pro Washing Solution | Miltenyi (Biotec) | 130-092-987 | For automated separation using AutoMACS Pro Separator only |
| AutoMACS Rinsing Solution | Miltenyi (Biotec) | 130-091-222 | For automated separation using AutoMACS Pro Separator only |
| AutoMacs running buffer | Miltenyi (Biotec) | 130-091-221 | For manual or automated separation using QuadroMACS or AutoMACS pro Separator |
| Axio Observer Z1 motorized inverted microscope equipped with a CSU-X1A 5000 spinning disk unit | Zeiss | Any confocal microscope equipped with a laser module fitted with laser lines of 568 nm (RFP) and 488 or 512 nm (GFP or YFP) wavelengths can be used | |
| AxioObserver A1, Research-Grade Inverted Microscope | Zeiss | Any epifluorescence microscope with fluorescence filters capable of exciting 568 nm (RFP) and 488 or 512 nm (GFP or YFP) wavelengths can be used | |
| CD14+ MICROBEADS | Miltenyi (Biotec) | 130-050-201 | For manual or automated separation using QuadroMACS or AutoMACS pro Separator |
| DPBS without calcium chloride and magnesium chloride | Sigma | D8537-6x500ML | |
| DsRed mito plasmid | Clontech | 632421 | Similar plasmids that can be used as fluorescent reporters can be found on Addgene |
| Fetal Bovine Serum | Thermo Fisher Scientific | 10437028 | |
| Ficoll-Paque PLUS 6 x 100 mL | Sigma | GE17-1440-02 | |
| GlutaMAX Supplement (100x) | Thermo Fisher Scientific | 35050079 | |
| GM-CSF | Thermo Fisher Scientific | PHC2011 | |
| Hemin BioXtra, from Porcine, INSERT CHARACTR96.0% (HPLC) | Sigma | 51280-1G | |
| HEPES | Thermo Fisher Scientific | 15630106 | |
| Inflammatory caspase BiFC plasmids | Available by request from LBH lab | ||
| LPS-EB Ultrapure | Invivogen | TLRL-3PELPS | |
| LS Columns | Miltenyi (Biotec) | 130-042-401 | For manual separation using QuadroMACS Separator only |
| MACS 15 mL Tube Rack | Miltenyi (Biotec) | 130-091-052 | For manual separation using QuadroMACS Separator only |
| MACS MultiStand | Miltenyi (Biotec) | 130-042-303 | For manual separation using QuadroMACS Separator only |
| mCherry plasmid | Yungpeng Wang Lab | Similar plasmids that can be used as fluorescent reporters can be found on Addgene | |
| Neon Transfection System | Thermo Fisher Scientific | MPK5000 | Includes Neon electroporation device, pipette and pipette station |
| Neon Transfection System 10 µL Kit | Thermo Fisher Scientific | MPK1096 | Includes resuspension buffer R, resuspension buffer T, electrolytic buffer E, 96 x 10 µL Neon tips and Neon electroporation tubes |
| Nigericin sodium salt, ready made solution | Sigma | SML1779-1ML | |
| Penicillin-Streptomycin (10,000 U/mL) | Thermo Fisher Scientific | 15140122 | |
| Poly-D-Lysine Hydrobromide | Sigma | P7280-5mg | |
| QuadroMACS Separator | Miltenyi (Biotec) | 130-090-976 | For manual separation using QuadroMACS Separator only |
| qVD-OPh | Fisher (ApexBio) | 50-101-3172 | |
| RPMI 1640 Medium | Thermo Fisher Scientific | 11875119 | |
| Trypsin-EDTA (0.25%), phenol red | Thermo Fisher Scientific | 25200072 | |
| UltraPure 0.5 M EDTA, pH 8.0 | Thermo Fisher Scientific | 15575020 |

