An In Vitro Technique to Study Antimicrobial Treatments on Bacterial Aggregates
Abstract
Source: Gannon, A. D., et al. Tools for the Real-Time Assessment of a Pseudomonas aeruginosa Infection Model. J. Vis. Exp. (2021).
This video demonstrates an in vitro technique to study the impact of antimicrobial treatment on Pseudomonas aeruginosa (Pa) aggregates. Fluorescently-labeled Pa cells are grown as aggregates, mimicking the growth pattern during infection. Upon challenging the aggregates with a sublethal concentration of the antibiotic colistin, the real-time assessment of the bacteria-antibiotic interaction is performed via fluorescence microscopy.
Protocol
1. Prepare synthetic cystic fibrosis medium (SCFM2)
NOTE: Preparation of SCFM2 comprises three main stages outlined below (Figure 1).
- Sterilization of porcine mucin
- Prepare sterile mucin at a final concentration of 5 mg/mL in SCFM2. For example, for a 5 mL volume of SCFM2, weigh 25 mg of Type II mucin in a sterile Petri dish, and place it into an ultraviolet (UV) sterilizer for 4 h, gently agitating every hour.
- After 4 h, transfer the UV-treated mucin into autoclaved 1.7 mL tubes under sterile conditions, and store at -20 °C.
- To confirm complete sterilization, dissolve a sample of mucin in a sterile liquid, such as water or Luria-Bertani (LB) broth, and observe under a microscope.
NOTE: Sterilized mucin can be stored at -20 °C for up to 6 months.
- Preparation of buffered base
- Prepare salt and amino acid stock solutions by adding the appropriate amounts by weight to deionized water as listed in Table 1. Filter-sterilize all stock solutions using a 0.22 μm filter, wrap in foil to protect from light degradation, and store at 4 °C for up to one month.
- Prepare the buffered base by combining 190 mL of deionized water with amino acid and salt stock solutions by volumes listed in Table 1. Adjust the solution to pH 6.8, and increase to a final volume of 250 mL. Filter-sterilize using a 0.22 μm filter, and store at 4 °C for up to 30 days.
- On the evening before the experiment, aliquot the desired amount of buffered base into a glass culture flask, and add mucin (5 mg/mL as described in step 1.1.1) and purified salmon sperm DNA (0.6 mg/mL). Agitate gently, wrap in foil and leave at 4 °C overnight to allow mucin and DNA to dissolve into solution.
NOTE: Salmon sperm DNA aliquots should be thawed on ice, vortexed, and added to buffered base and mucin. Tryptophan, asparagine, and tyrosine stock solutions must be prepared in solutions of NaOH (see Table 1 for concentrations) instead of deionized water. Keep buffered base and all stock solutions wrapped in foil to protect from light exposure. Most stocks will be stable for up to a month. Stocks that become discolored should not be used and should be replaced before use.
- Addition of supplemental stocks
- On the day of the experiment, add the stocks listed in Table 2 to the buffered base containing mucin and DNA.
NOTE: Prepare a fresh FeSO4 solution on the day of the experiment, but all other stocks can be made ahead of time and stored for 30 days at 4 °C. 1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC) contains chloroform. Handle with caution, and do not use near open flames. After the addition of DOPC, incubate SCFM2 at 37 °C with shaking (250 rpm) for at least 20 min (for 5 mL culture). This incubation period allows the chloroform in the DOPC to evaporate. The flask should not be airtight; instead, cover the flask opening loosely with foil.
- On the day of the experiment, add the stocks listed in Table 2 to the buffered base containing mucin and DNA.
2. Real-time assessment of antimicrobial tolerance in bacterial aggregates
- Prepare overnight cultures
- On the evening before the experiment, inoculate 5 mL of LB broth with several colonies of Pa PAO1-pMRP9-113 from an LB agar plate containing antibiotic (carbenicillin 300 μg/mL). Grow overnight at 37 °C with agitation at 250 rpm.
NOTE: Grow overnight cultures with the addition of antibiotics required for the selection of required plasmids (here, the green fluorescent protein (GFP) expression plasmid, pMRP9-1). Note that Pa cells are to be washed before SCFM2 is inoculated. LB may be substituted for other rich laboratory media for overnight cultures. All bacterial isolates should be handled using appropriate BSL-2 guidelines throughout this protocol.
- On the evening before the experiment, inoculate 5 mL of LB broth with several colonies of Pa PAO1-pMRP9-113 from an LB agar plate containing antibiotic (carbenicillin 300 μg/mL). Grow overnight at 37 °C with agitation at 250 rpm.
- Inoculate SCFM2
- On the day of the experiment, back-dilute overnight cultures of Pa 1:10 (culture: liquid media) by inoculating 500 μL into 5 mL of fresh LB broth. Grow cells until log phase (60-90 min) at 37 °C with agitation at 250 rpm.
- Centrifuge log phase cultures at 10,000 × g for 5 min. Wash the cells by removing the supernatant and resuspending them in 3 mL of filter-sterilized phosphate-buffered saline (PBS, pH 7.0). Repeat twice, and resuspend the pellet in a final volume of 1 mL of PBS.
- Measure the absorbance of the washed cells using a spectrophotometer at 600 nm (OD600), and calculate the volume of culture required for a starting OD600 of 0.05 in 5 mL of SCFM2. Inoculate Pa into the SCFM2 and vortex gently to distribute cells throughout. Pipette 1 mL of the inoculated SCFM2 into each chamber of a 4-well, glass bottom, optical dish, and incubate for 4 h statically at 37 °C.
NOTE: The doubling time of Pa cultures is dependent on the strain and the availability of oxygen. In SCFM2, under the conditions described here, the doubling time of Pa is ~1.4 h.
3. Visualizing aggregates during antibiotic treatment with confocal laser scanning microscopy (CLSM)
NOTE: This section describes the use of confocal laser scanning microscope and image capture software for the imaging of Pa aggregates in SCFM2. The goal is to observe and characterize the remaining (tolerant) bacterial biomass after treatment with antibiotics. The steps outlined can be performed with success on other confocal microscopes, although the instrument operating manual should be referenced for specific guidance.
- Image Pa cultures using either a heated chamber or a heated microplate fitted on the microscope stage to maintain an ambient temperature of 37 °C. Start the Incubation module at least 2 h prior to the beginning of the experiment to allow all apparatus to reach the desired temperature, and reduce further expansion and movement during data collection.
- After 4 h, transfer SCFM2 cultures containing Pa cells to the heated microscope stage. Designate 3 out of 4 wells as technical replicates for antibiotic treatment, and consider the 4th well as a no-treatment control, containing only Pa cells in SCFM2, without antibiotics. Identify aggregates using brightfield microscopy within the Locate tab prior to any excitation of fluorescent reporters. Define an area for imaging within each well, and store its position (x-y-z coordinates) using the Positions module in the imaging software.
- Use a 63x oil-immersion objective to visualize Pa cultures containing the GFP expression plasmid pMRP9-1 in SCFM2 with an excitation wavelength of 488 nm and emission wavelength of 509 nm. Take images using the z-stack option within the Acquisition module at 1 μm intervals (total of 60 slices). Use the Line-averaging module to reduce background fluorescence in the GFP channel within the total volume of the 60 μm z-stack images (1,093.5 mm3). Take control images of uninoculated SCFM2 by using identical settings to determine the background fluorescence for image analysis.
NOTE: Images are acquired by producing 512 pixels x 512 pixels (0.26 μm x 0.26 μm pixels) 8-bit z-stack images that are 60 μm from the base of the coverslip. - Use the time series option within the imaging software to capture 60 slices at each position (well) at 15-min intervals over a period of 18 h. Use the definite focus strategy within the software to store a focal plane for each position, which is returned to at each time point throughout the experiment.
- After a total 4.5 h of incubation, image each position using the above settings to determine the aggregate biomass within each of the four wells before the addition of antibiotic.
- After 6 h of total incubation, add antibiotic at 2x-below minimum inhibitory concentration (MIC) to each replicate. Pipette directly and gently into the middle of the well, just below the air-liquid interphase. Maintain all cultures in the heated confocal chamber.
NOTE: Here, colistin sulfate at a concentration of 140 μg/mL was used. - Begin imaging post-antibiotic treatment by clicking on the Start Experiment option within the imaging program.
NOTE: Antibiotic concentration used is dependent on the antibiotic, the Pa isolate, and whether the user would like to examine killing or tolerance effects. This experiment uses a single dose. Additional doses can be added without disrupting cultures if needed.
4. Propidium iodide staining of Pa aggregates
NOTE: Propidium iodide (PI) is commonly utilized as a staining reagent to identify non-viable (dead) bacterial cells in culture. Here, it is used to identify aggregates sensitive to antibiotic treatment applied in section 3. Throughout this protocol, the expression and detection of GFP in Pa cells is used as the main proxy for cell viability. This final step allows confocal imaging to be used once more to identify the spatial positioning of live/dead aggregates in relation to each other. Additionally, aggregates are identified as live/dead for further downstream cell sorting in section 3.
- After 18 h, add PI to each well of the four-chambered optical bottom dish containing SCFM2 cultures. Follow the manufacturer's instructions for the volume of PI and incubation time (e.g., ~2 μL per mL of culture, ~20-30 min).
5. Isolating live cells from aggregates using a Fluorescence-activated cell sorting (FACS) approach
NOTE: FACS presents a powerful platform to sort and isolate groups of cells according to a fluorescently tagged phenotype. Here, FACS is used to isolate live (antibiotic-tolerant) aggregates from non-viable aggregates.
- After staining with propidium iodide, remove the cultures from incubation, and transfer to a FACS instrument in an insulated container to maintain 37 °C.
- Run 1 mL aliquots of SCFM2 containing Pa aggregates at the lowest flow rate.
NOTE: Each aliquot will contain ~15,000 aggregates. - To detect GFP, illuminate the cells with a 488 nm laser, and record the fluorescent signal height at 530/30 nm. Visualize the PI staining by excitation with a 561 nm laser, and record the fluorescent signal height at 610/20 nm. Perform sorting using a 70-u nozzle.
NOTE: Sorted aggregates can be pooled in multiple ways depending on the user's application. In this case, FACS was used to sort viable Pa aggregates for downstream RNA sequencing.
Table 1: Preparation of salt, amino acid, DNA, and mucin stocks required for buffered base of synthetic cystic fibrosis sputum medium, SCFM2.
| Chemical | Stock Concentration (M) | Final concentration (mM) | Notes |
| NaH2PO4 | 0.2 | 1.3 | |
| Na2HPO4 | 0.2 | 1.25 | |
| KNO3 | 1 | 0.35 | |
| K2SO4 | 0.25 | 0.27 | |
| NH4Cl | 2.28 | Add solid directly to buffered base | |
| KCl | 14.94 | Add solid directly to buffered base | |
| NaCl | 51.85 | Add solid directly to buffered base | |
| MOPS | 10 | Add solid directly to buffered base | |
| Ser | 0.1 | 1.45 | |
| Glu HCl | 0.1 | 1.55 | |
| Pro | 0.1 | 1.66 | |
| Gly | 0.1 | 1.2 | |
| Ala | 0.1 | 1.78 | |
| Val | 0.1 | 1.12 | |
| Met | 0.1 | 0.63 | |
| Ile | 0.1 | 1.12 | |
| Leu | 0.1 | 1.61 | |
| Orn HCl | 0.1 | 0.68 | |
| Lys HCl | 0.1 | 2.13 | |
| Arg HCl | 0.1 | 0.31 | |
| Trp | 0.1 | 0.01 | Prepared in 0.2 M NaOH |
| Asp | 0.1 | 0.83 | Prepared in 0.5 M NaOH |
| Tyr | 0.1 | 0.8 | Prepared in 1.0 M NaOH |
| Thr | 0.1 | 1.07 | |
| Cys HCl | 0.1 | 0.16 | |
| Phe | 0.1 | 0.53 | |
| His HCl H2O | 0.1 | 0.52 | |
| Salmon Sperm DNA | 0.6 mg/mL | ||
| Porcine Mucin | 5 mg/nL |
Table 2: Preparation of additional stocks required for buffered base of synthetic cystic fibrosis sputum medium, SCFM2.
| Chemical | Stock concentration (M) | Final concentration (mM) | Notes |
| Dextrose (D-glu2cose) | 1 | 3 | |
| L-lactic acid | 1 | 9.3 | Adjust to pH 7.0 with NaOH |
| CaCl2*H2O | 1 | 1.75 | |
| MgCl2*6H2O | 1 | 0.61 | |
| FeSO4*7H2O | 1 mg/mL | 0.0036 | Make fresh daily |
| N-acetylglucosamine | 0.25 | 0.3 | |
| 1,2-dioleoyl-sn-glycero-3-phosphocoline (DOPC) | 25 mg/mL | 100 µg/mL | Incubate for at least 20 min at 37 °C after addition |
Representative Results
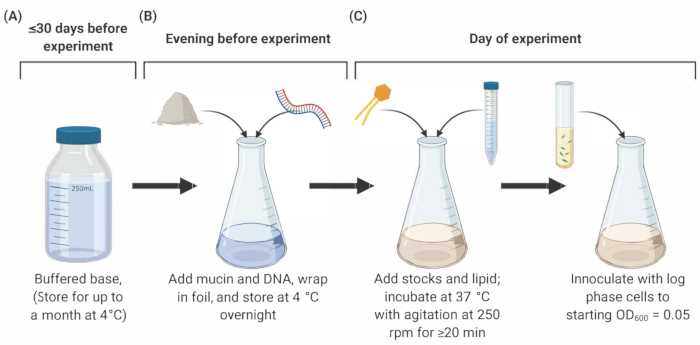
Figure 1: Preparation and inoculation of SCFM2 medium. (A) Buffered base is prepared using salts and amino acids listed in Table 1 and Table 2. Buffered base can be stored at 4 °C for up to 30 days, but must be protected from light exposure. (B) Mucin and DNA are added to an aliquot of buffered base and dissolved into solution overnight at 4 °C. (C) Lipid and additional stocks are added to the overnight solution and incubated at 37 °C with agitation at 250 rpm for 20 min. SFCM2 is then inoculated with washed, log phase cells at an OD600 = 0.05. Abbreviations: SCFM2 = synthetic cystic fibrosis sputum medium.
Disclosures
The authors have nothing to disclose.
Materials
| Amino acids | |||
| Alanine | Acrs Organics | 56-41-7 | |
| Arginine HCl | MP | 1119-34-2 | |
| Asparagine | Acrs Organics | 56-84-8 | Prepared in 0.5 M NaOH |
| Cystine HCl | Alfa Aesar | L06328 | |
| Glutamic acid HCl | Acrs Organics | 138-15-8 | |
| Glycine | Acrs Organics | 56-40-6 | |
| Histidine HCl H2O | Alfa Aesar | A17627 | |
| Isoleucine | Acrs Organics | 73-32-5 | |
| Leucine | Alfa Aesar | A12311 | |
| Lysine HCl | Alfa Aesar | J62099 | |
| Methionine | Acrs Organics | 63-68-3 | |
| Ornithine HCl | Alfa Aesar | A12111 | |
| Phenylalanine | Acrs Organics | 63-91-2 | |
| Proline | Alfa Aesar | A10199 | |
| Serine | Alfa Aesar | A11179 | |
| Threonine | Acrs Organics | 72-19-5 | |
| Tryptophan | Acrs Organics | 73-22-3 | Prepared in 0.2 M NaOH |
| Tyrosine | Alfa Aesar | A11141 | Prepared in 1.0 M NaOH |
| Valine | Acrs Organics | 72-18-4 | |
| Antibiotic | |||
| Carbenicillin | Alfa Aesar | J6194903 | |
| Day-of Stocks | |||
| CaCl2* 6H2O | Fisher Chemical | C79-500 | |
| Dextrose (D-glucose) | Fisher Chemical | 50-99-7 | |
| 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) | Fisher (Avanti Polar Lipids) | 4235-95-4 | Shake 15-20 min at 37 °C to evaporate chloroform |
| FeSO4 * 7H2O | Acrs Organics | 7782-63-0 | This stock equals 1 mg/mL, MUST make fresh |
| L-lactic acid | Alfa Aesar | L13242 | pH stock to 7 with NaOH |
| MgCl2 * 6H2O | Acrs Organics | 7791-18-6 | |
| N-acetylglucosamine | TCI | A0092 | |
| Prepared solids | |||
| Porcine mucin | Sigma | M1778-100G | UV-sterilize |
| Salmon sperm DNA | Invitrogen | 15632-011 | |
| Stain | |||
| Propidium iodide | Alfa Aesar | J66764MC | |
| Salts | |||
| K2SO4 | Alfa Aesar | A13975 | |
| KCl | Alfa Aesar | J64189 | Add solid directly to buffered base |
| KNO3 | Acrs Organics | 7757-79-1 | |
| MOPS | Alfa Aesar | A12914 | Add solid directly to buffered base |
| NaCl | Fisher Chemical | S271-500 | Add solid directly to buffered base |
| Na2HPO4 | RPI | S23100-500.0 | |
| NaH2PO4 | RPI | S23120-500.0 | |
| NH4Cl | Acrs Organics | 12125-02-9 | Add solid directly to buffered base |
| Consumables | |||
| Conical tubes (15 mL) | Olympus plastics | 28-101 | |
| Conical tubes (50 mL) | Olympus plastics | 28-106 | |
| Culture tubes w/air flow cap | Olympus plastics | 21-129 | |
| 35 mm four chamber glass-bottom dish | CellVis | NC0600518 | |
| Luria-Bertani (LB) broth | Genessee Scientific | 11-118 | |
| Phosphate-buffered saline (PBS) | Fisher Bioreagents | BP2944100 | |
| Pipet tips (p200) | Olympus plastics | 23-150RL | |
| Pipet tips (p1000) | Olympus plastics | 23-165RL | |
| Serological pipets (5 mL) | Olympus plastics | 12-102 | |
| Serological pipets (25 mL) | Olympus plastics | 12-106 | |
| Serological pipets (50 mL) | Olympus plastics | 12-107 | |
| Ultrapure water (RNAse/DNAse free); nanopure water | Genessee Scientific | 18-194 | Nanopure water used for preparation of solutions in Table 1 |
| Syringes (10 mL) | BD | 794412 | |
| Syringes (50 mL) | BD | 309653 | |
| 0.22 mm PES syringe filter | Olympus plastics | 25-244 | |
| PS cuvette semi-mico | Olympus plastics | 91-408 | |
| Software | |||
| Biorender | To prepare the figures | ||
| FacsDiva6.1.3 | Becton Dickinson, San Jose, CA | ||
| Imaris | Bitplane | version 9.6 | |
| Zen Black | |||
| Equipment | |||
| FacsAriallu | Becton Dickinson, San Jose, CA | ||
| LSM 880 confocal laser-scanning microscope | Zeiss |

