Natural Killer Cell-Mediated Cytotoxicity Assay Sample Preparation for Flow Cytometric Analysis
Abstract
Source: McBride, J. E., et al. Flow Cytometric Analysis of Natural Killer Cell Lytic Activity in Human Whole Blood. J. Vis. Exp. (2017).
This video demonstrates the lytic activity of natural killer cells against fluorescently-stained target cells. The method enables NK cell-induced cytotoxicity measurement by staining dead cells and using a flow cytometer to quantify live and dead target cells.
Protocol
1. Preparation of Controls
- Transfer the following into separate and appropriately labeled 1.5 ml tubes:
- Add 500 µL of fresh IMDM containing resuspended DiO-labeled K562 cells into the "Double Positive" labeled 1.5 mL tube.
- Add 500 µL of fresh IMDM containing resuspended DiO-labeled K562 cells into the "DiO only" labeled 1.5 mL tube.
- Add 500 µL of fresh IMDM containing resuspended unlabeled K562 cells into the "Propidium Iodide (PI) only" labeled 1.5 mL tube.
- Place the Double Positive and PI-only tubes in a 55 °C water bath for 5 min.
- After the 5 min has elapsed, remove the tubes and wipe them down with 70% ethanol.
- Add 10 L of PI to the Double Positive and PI only 1.5 mL tubes.
- Place all three K562 target cell controls in the incubator at 37 °C for 30 min.
- After the 30-minute incubation has elapsed, centrifuge all three K562 target cell controls for 2 min at 163 x g.
- Carefully remove the supernatant without disturbing the cell pellet.
- Resuspend each control with 20 µL of fresh IMDM cell culture media, and leave in the 37 °C incubator with 5% CO2 for at least 30 min for optimal DiO signal.
NOTE: Controls are now ready to be run through the imaging flow cytometer.
2. Cytotoxicity Assay Sample Preparation
- Prepare and label 1.5 mL tubes for each sample/participant accordingly.
- Pipette desired the ratio of NK cells and DiO-labeled K562 cells in each tube.
NOTE: For example, the desired ratio of K562 target cells and NK effector cells is 1:5. - Centrifuge for 5 min at 135 x g.
- Carefully remove the supernatant without disturbing the cell pellet.
- Resuspend NK-DiO-labeled K562 cell mixture in 500 µL of NK cell media without interleukin-2 (IL-2) and 2-mercaptoethanol (2-ME) (incomplete NK cell culture media).
NOTE: The incomplete NK cell culture media is Minimum Essential Medium Eagle with sodium bicarbonate, without L-glutamine, ribonucleosides, and deoxyribonucleosides. - Add 5 µL of PI to each tube.
- Centrifuge for 2 min at 163 x g.
- Incubate cells at 37 °C for 2 h.
- Following 2 h incubation, centrifuge for 2 min at 163 x g.
- Carefully remove the supernatant without disturbing the cell pellet.
- Resuspend cells in 25 µL incomplete NK cell culture media.
3. Preparation of Spontaneous ("S") Sample
- Pipette 500 µl of DiO-labeled K562 cells (concentration of 1 x 106 cells/mL) into a 1.5 mL tube.
- Add 10 µL of PI to each tube.
- Centrifuge tube for 2 min at 163 x g.
- Incubate cells at 37 °C for 2 h.
- Following 2 h incubation, centrifuge for 2 min at 163 x g.
- Carefully remove the supernatant without disturbing the cell pellet.
- Resuspend cells in 25 µl incomplete alpha-minimum Essential Medium (α-MEM) cell culture media.
4. Data Acquisition with Imaging Flow Cytometer
- Press the green button inside the front door of the imaging flow cytometer to turn on the instrument.
- Turn on all computers associated with the imaging flow cytometer.
- Launch the imaging flow cytometer software.
- Click the "Startup" button to flush the system and prepare the sample line.
- Once the "Startup" is complete, close out the "Calibrations" window.
- Assign channels: on the top left-hand side, click on each channel in order to assign them.
- On the right-hand side, click on the scatter plot button to create 4 scatterplots: Raw Max Pixel _MC_6 vs Area_M06, Raw Max Pixel _MC_2 vs Area_M02, Raw Max Pixel _MC_5 vs Area_M05 and FieldArea_M01 vs AspectRatio_M01.
- Begin analyzing samples by first using the "Double Positive" control.
- Click on "Load."
- Place the 1.5 ml tube with the "Double Positive" sample from Steps 1.4 to 1.8 into the sample loader.
- Select the 40X objective under the "Magnification" tab.
- Turn on the 405 mW and 642 mW lasers.
- Turn on the "Brightfield" channel.
- Click on "Select Intensity."
- Based on the "Double Positive" control sample, determine the desired intensity for the 405 mW laser so the detector is not overloaded.
Note: For example, the optimal intensity for this experiment was set at 11 mW.
- After the desired set-up is achieved, acquire data.
- Under the "File Acquisition" tab, enter a custom filename text. Select a folder for saving the data file(s).
- Enter the number of cells to acquire next to "Collect". Typically this number varies between 1,000 to 10,000.
- Click on "Acquire."
NOTE: Once the desired number of cells is acquired, the data file is automatically saved in the previously selected folder.
- After acquisition finishes, load the next control sample – DiO only control.
- Click on "Load."
- Place the 1.5 mL tube with the "DiO only" sample into the sample loader.
- Leave the 40X objective under the "Magnification" tab selected.
- Leave the 405 mW laser turned on.
- Turn OFF the 642 mW laser and the "Brightfield" channel.
NOTE: Now that the desired set-up has been achieved, data can be acquired. - Under the "File Acquisition" tab, enter a custom filename text and select a folder for saving the data file(s). Enter the number of cells to acquire next to "Collect.". Typically this number is 1,000.
- Click on "Acquire."
NOTE: Once the desired number of cells is acquired the data file is automatically saved in the previously selected folder.
- Repeat step 4.10 for the "PI only" control sample. The experimental samples are now ready to be collected.
- Handle the remaining experimental samples, including the "Spontaneous 'S' Samples" as follows:
- Leave the 40X objective under the "Magnification" tab selected.
- Turn ON the 405 mW and 642 mW lasers.
- Turn ON the "Brightfield" channel.
- Click on "Set Intensity."
- Under the "File Acquisition" tab, enter a custom filename text and select a folder for saving the data file(s). Enter a custom filename text.
- Enter the number of cells to acquire next to "Collect."
- Click on the "Acquire" button.
NOTE: Once the desired number of cells is acquired the data file is automatically saved in the previously selected folder.
- Repeat step 4.12 for all experimental samples.
- After all experimental data and files have been collected, click the "Shutdown" button to sterilize the system.
5. Imaging Flow Cytometer Sample Analysis
- Open the imaging flow cytometer analysis software application.
- Under "File", open an experimental.RIF file.
- Build a new matrix using a single color.RIF files (DiO-only control and PI-only control, created during steps 4.10 and4.11) by selecting "Create a New Matrix" under the Compensation tab in the imaging flow cytometer software.
NOTE: The software will prompt for the selection of the single color files and merge them to create a matrix file (.ctm file extension), that is to be selected to apply channel compensation. - Create dot plots by using the "building blocks" function of the software.
- Create a dot-plot (BrightFieldGradient_RMS vs frequency) to gate the focused cells. Call the gate "Focus" (Figure 1A).
- Using the "Focus" gate, create a dot plot of Bright Field Area vs Bright Field Aspect Ratio to gate the singlet cells. Call the gate "Single" (Figure 1B).
- Using the "Single" gate, create a dot plot of Intensity_MC_Ch02 vs Intensity_MC_Ch5. Use this plot to identify and gate DiO-positive only cells (targets, alive) and PI-DiO double-positive cells (targets, dead) (Figure 1C).
NOTE: All the plots described in steps 5.4.1, 5.4.2, and 5.4.3 can be created by using the "building blocks" function of the software.
- Click on the statistics function of the dot plot to access the cell numbers of each gate.
- Calculate the percentage of dead targets in the spontaneous sample and experimental samples using the following formula:
% dead targets in sample = (#dead targets x 100)/(#live targets + #dead targets) - Calculate cytotoxicity using the following formula:
% cytotoxicity = [(Experimental dead-Spontaneous dead)/(100-Spontaneous dead)]x100
Representative Results
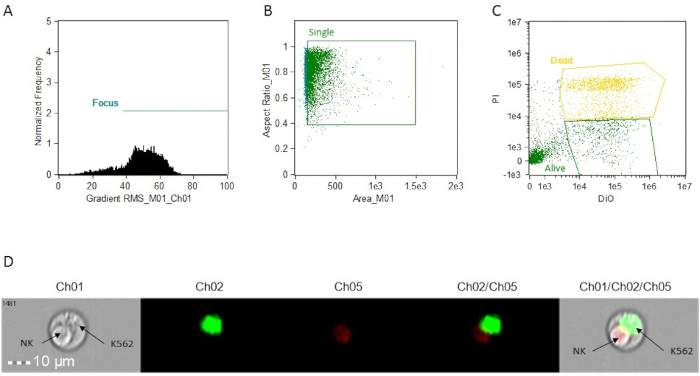
Figure 1: Representative histograms, scatter plots, and images for cytotoxic activity analysis. (A) focus cell analysis. (B) single cell analysis. (C) target cell staining analysis. All determinations are made using the image attached to each event. This can be accessed in analysis software by simply clicking on the event on the graphs. (D) representative image of a doublet event, showing an apoptotic NK cell and a live K562 target. Ch01, Brightfield. Ch02, DiO. Ch05, PI.
Disclosures
The authors have nothing to disclose.
Materials
| K-562 lymphoblasts | ATCC | CCL-243 | |
| Iscove's Modified Dulbecco's Media | ATCC | 30-2005 | High glucose, with L-Glutamine, with HEPES, Sterile-filtered |
| Alpha Minimum Essential medium | ATCC | CRL-2407 | Without ribonucleosides and deoxyribonucleosides but with 2 mM L-glutamine and 1.5 g/L sodium bicarbonate |
| Propridium Iodide Staining Solution | BD Pharmingen | 51-66211E | |
| Vybranto DiO cell-labeling solution | Vybranto | V-22886 | |
| ImageStream X Mark II Imaging Flow Cytometer | |||
| Whole Blood CD56 MicroBeads, human | Miltenyi Biotec | 130-090-875 | |
| ImageStream X Mark II Imaging Flow Cytometer | EMD Millipore | ||
| Speedbeads | Amnis Corporation | 400030 | |
| INSPIRE Software | EMD Millipore | Version Mark II, September 2013 | |
| Ideas Application Software | EMD Millipore | Version 6.1, July 2014 |

