An In Vitro Technique to Study T Cell Interaction with Supported Planar Lipid Bilayers
Abstract
Source: Steblyanko, M., et al. Assessment of the Synaptic Interface of Primary Human T Cells from Peripheral Blood and Lymphoid Tissue. J. Vis. Exp. (2018).
This video demonstrates an in vitro technique to study immune synapse formation between primary human T cells and supported lipid bilayers (SLB). T cell surface proteins interact with respective ligands immobilized on SLB to form an immune synapse, followed by the movement of lytic granules towards the synapse, which is visualized using fluorescence microscopy.
Protocol
All procedures involving sample collection have been performed in accordance with the institute's IRB guidelines.
1. Isolation of CD4+ T Cells for an Image Analysis
- Thaw a 1 mL aliquot containing 107 frozen peripheral blood mononuclear cells (PBMCs) or lymph node mononuclear cells (LNMCs) from collected samples. In a sterile hood, add the thawed cells to 9 mL of RPMI supplemented with penicillin/streptomycin and glutamine.
- Centrifugate the cells for 10 min at 300 x g at 4 °C, aspirate the supernatant and resuspend the cells in 5 mL of supplemented RPMI containing 10% FBS (complete medium). Incubate the cells overnight in a CO2 incubator at 37 °C.
- The next day, purify CD4+ T cells by negative immunomagnetic sorting using a commercially available kit according to the manufacturer's instructions.
- To measure the number of freshly purified CD4+ T cells, mix 5 µL of the cell suspension with an equal volume of a trypan blue solution. Load a hemocytometer with a cell-trypan blue mixture and count the live cells inside the 5 sections of the hemocytometer.
- Take the average of the cell number and determine the number of cells in the original cell suspension: the number of cells/1 mL = the average count x 2 x 104. If the total number of isolated cells is too small, use the cells as is without counting.
- Centrifuge the cells at 300 x g for 10 min and resuspend them in an assay buffer (20 mM HEPES, pH 7.4, 137 mM NaCl, 2 mM Na2HPO4, 5 mM D-glucose, 5 mM KCl, 1 mM MgCl2, 2 mM CaCl2, and 1% human serum albumin) at 105 cells/50 µL or less and keep the cells at 4 °C (for 1–2 h) until ready to use in the experiments.
- Dispose of all biological waste according to the relevant institutional guidelines.
- If desired as a control cell population, prepare activated CD8+ T cells from cord blood PBMC, place 107 cells in 5 mL of complete medium in a T25 culture flask covered with a mixture of anti-CD3 and anti-CD28 antibodies at 10 µg/mL and 1 µg/mL, respectively.
- The next day, remove the activated cord blood cells from the flask, wash them 1x with fresh complete medium, and expand the cells in the presence of recombinant IL-2 (100 U/mL) for 2 weeks.
- Purify the cord-blood CD8+ T cells by negative immunomagnetic sorting using the commercially available kit according to the manufacturer's instructions. Count the cells and exchange the media to the assay buffer as described in steps 1.3–1.6 for LN and PB CD8+ T cells.
2. Components for the Preparation of the Planar Lipid Bilayers
- Prepare 3 kinds of liposomes as described elsewhere: (a) 0.4 mM DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine) liposomes, (b) 0.4 mM DOPC liposomes containing 33 mol% DOGS-NTA (1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (ammonium salt)) lipids, (c) 0.4 mM DOPC liposomes containing 4 mol% Biotinyl-Cap-PE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(cap biotin) (sodium salt)).
- Prepare 5% casein solution as previously described.
- Dissolve 5 g of casein powder in 100 mL of ultrapure water and add 350 µL of 10 M sodium hydroxide. Stir everything on a regular magnetic stirrer at a slow speed according to the available scale, at room temperature for 2 h, and then overnight at 4 °C. Adjust the pH to 7.3 and ultracentrifuge the solution for 2 h at 100,000 x g at 4 °C. Filter the supernatant with a 0.22 µm sterile filter and store the solution in aliquots at -80 °C.
CAUTION: Sodium hydroxide solution can cause chemical burns and may induce permanent blindness upon contact with eyes. Use rubber gloves, safety clothing, and eye protection when handling this chemical or its solutions.
- Dissolve 5 g of casein powder in 100 mL of ultrapure water and add 350 µL of 10 M sodium hydroxide. Stir everything on a regular magnetic stirrer at a slow speed according to the available scale, at room temperature for 2 h, and then overnight at 4 °C. Adjust the pH to 7.3 and ultracentrifuge the solution for 2 h at 100,000 x g at 4 °C. Filter the supernatant with a 0.22 µm sterile filter and store the solution in aliquots at -80 °C.
- Label anti-CD3 antibodies with biotin to produce mono-biotinylated antibody molecules with a previously described approach.
- Prepare a solution of Biotin-PEO4-NHS in dimethyl sulfoxide (DMSO) at 0.1 mg/mL. Add 3.7 µL of the Biotin-PEO4-NHS solution to 1 mg of antibody in 0.5 mL of phosphate-buffered saline (PBS) containing 100 mM sodium bicarbonate.
- Incubate the mixture for 2 h at room temperature. Prepare a solution of Alexa Fluor 488 NHS ester at 10 mg/mL in DMSO. Add the Alexa Fluor 488 NHS ester solution to the antibody labeled by Biotin-PEO4-NHS at a 10-fold molar excess.
- Incubate the mixture for 1 h at room temperature with slow stirring on a regular magnetic stirrer. Separate the unbound dye using size-exclusion chromatography.
- Determine the antibody concentration by measuring the optical density of the antibody solution at 280 nm (A280). Measure the optical density of the labeled antibodies at 577 nm (A577).
- Determine the dye-to-antibody ratio using the following equation:
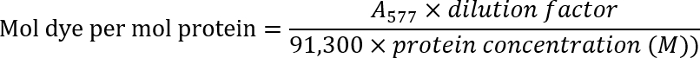
NOTE: Find additional details in the manufacturer's protocol.
- Express a recombinant soluble Intercellular Adhesion Molecule 1 (ICAM-1) protein in a Drosophila expression system as described previously.
- Clone, with cDNA encoding, the ectodomain of ICAM-1 into Drosophila expression vector pMT/V5-His with an inducible metallothionein promoter to produce a recombinant protein appended with a His6 tag on the C-terminal end.
- Co-transfect S2 cells with the resulting ICAM-1 containing plasmid and a geneticin (G418) expression vector. Select stable transfectants using Schneider's Drosophila Media supplemented with 10% fetal calf serum (FCS) and 0.5 mg/mL of G418 for 3 weeks. Expand the cells in serum-free insect medium and induce a protein expression with 0.5 mM CuSO4 for 3 d.
- Concentrate the culture supernatant 10x and dialyze against PBS via a tangential flow concentrator as previously described.
- Apply the concentrated culture supernatant to a column containing Sepharose with a covalently immobilized anti-ICAM-1 monoclonal antibody and elute the bound ICAM-1 with a 50 mM glycine buffer, pH 3.0. Immediately neutralize the eluted ICAM-1 protein with a 2 M Tris buffer, pH 8.0.
- Dialyze the eluted material against PBS, pH 8.0, and add the dialyzed material to a column containing Ni-NTA (Nickel-Nitriloacetic acid) agarose. Elute soluble ICAM-1 with 200 mM imidazole, pH 8.0. Dialyze the eluted material against a PBS buffer, pH 8.0.
- Label the purified ICAM-1 with Cyanine5 N-Hydroxysuccinimide (Cy5 NHS) ester according to the manufacturer's instructions.
NOTE: The best final dye-to-protein ratio is 1:1.
- Produce Fab fragments from an anti-CD107a antibody by papain digestion and purify the Fab fragments by ion exchange chromatography as described previously. Label the Fab fragments with Alexa Fluor 568 NHS ester according to the manufacturer's instructions.
3. Formation of Glass-supported Planar Lipid Bilayers
- Prepare a fresh acidic piranha solution by mixing 140 mL of concentrated sulfuric acid and 60 mL of 30% hydrogen peroxide. Wash the glass coverslips for the flow slides by soaking them in the acid piranha solution for 30 min. Hold the glass coverslip with polypropylene scissor-type forceps.
CAUTION: Piranha solution is an extremely strong oxidizer. Remember to wear safety glasses or goggles or a full-face shield together with thick rubber gloves at all times while handling the solution. Only work with piranha solution under a fume hood. Avoid heating, transporting, or shaking it at any time during use, as it may explode. Collect the piranha waste into a glass bottle with a lead-containing hole. Contact the Institutional Safety Committee about proper waste utilization. - Rinse the washed coverslips 7x with ultrapure water by transferring them sequentially into beakers containing fresh water. Set the wet coverslips aside to let the remaining water roll off the clean glass, leaving the dry glass behind.
- Alternatively, use a pipette tip attached to a vacuum pump to carefully remove the remaining water droplets from the coverslips.
- In the sterile hood, perform dilutions of various lipids to produce the liposome mix for making bilayers. First, combine 37 µL of DOPC liposomes and 3 µL of Biotinyl-Cap-PE liposomes. Second, mix 14 µL of DOPC liposomes and 15 µL of DOGS-NTA liposomes. Third, add 1 µL of the first mix to 29 µL of the second mix to fabricate the final liposome mixture.
- In the sterile hood, set up a workspace with dry coverslips close by. Aliquot 2 µL of the final liposome mixture (see step 3.3) precisely in the center of a self-adhesive slide channel. Immediately and very precisely align a clean and dry coverslip with the slide and gently lower the coverslip on the sticky side of the slide.
- If preparing more than one slide, work on one slide at a time since the liposome mixture evaporates rapidly. Turn the slide over and use the outer ring of polypropylene scissor-type forceps to apply gentle pressure to the peripheral contact of the coverslip with the slide, making sure that the slip is tightly attached to the slide to preclude leakage.
NOTE: Do not press against the channels of the slide to avoid breaking or cracking the coverslip. - Turn the slide over again and mark the position of the formed bilayer, which looks like a drop between the coverslip and the channel slide, by drawing 4 dots with a permanent marker around the bilayer on the external slide side of the assembly.
- If preparing more than one slide, work on one slide at a time since the liposome mixture evaporates rapidly. Turn the slide over and use the outer ring of polypropylene scissor-type forceps to apply gentle pressure to the peripheral contact of the coverslip with the slide, making sure that the slip is tightly attached to the slide to preclude leakage.
- Before the first injection of a liquid into the channel, designate one port of the channel as the entry port and the other as the exit port and maintain this designation throughout the experiment.
- To avoid forming bubbles, insert the end of the pipet tip directly into the entry port of the channel. Slowly fill the channels of the slide with 50 µL of warm (at least room temperature) assay buffer (see step 1.5 for the buffer composition).
- Prepare a 0.5 M nickel(II) chloride solution. Thaw a 2 mL aliquot of casein solution in a water bath at 37 °C for 30 min and supplement it with a nickel chloride solution at a final concentration of 200 µM.
- Wash the bilayers by first injecting 100 µL of the casein solution in the entry port of the channel and then immediately removing 100 µL out of the exit port on the slide by pipetting. Block the bilayers with the same solution by injecting 100 µL of the casein solution into the entry port of each channel and incubating the slide for 45 min at room temperature.
- Thaw aliquots of Cy5-ICAM-1-His6 and streptavidin proteins. Combine the proteins in the assay buffer at the final concentration of 2 µg/mL each. Centrifuge the solution for 30 min at 20,000 x g and 4 °C to remove any aggregates.
- Remove the rest of the blocking solution from the exit port of the slide channel by pipetting. Inject 100 µL of the solution containing ICAM-1 and streptavidin into the entry port.
- Incubate the slide for 45 min at room temperature. Remove any excess of the protein solution from the exit port. Wash the bilayer 2x by first injecting 100 µL of the assay buffer in the entry port of the channel and then immediately removing 100 µL out of the exit port.
- Dilute an Alexa-Fluor-488-labelled anti-CD3 antibody with the assay buffer to a final concentration of 2 µg/mL. Inject 100 µL of the antibody solution into the entry port of the slide and incubate it for 45 min at room temperature. Remove any excess of the protein solution from the exit port. Wash the bilayer 2x with 100 µL of the assay buffer as in step 3.11.
4. Imaging of the T Cells Interaction with the Planar Bilayer
- Preheat the stage and the objective of a confocal or total internal reflection fluorescence (TIRF) microscope until the temperature is equilibrated at 37 °C. Set up the slide with the bilayer(s) on the heated stage. Move the stage to an appropriate position according to the ink marks and focus on the bilayer employing fluorescence of Cy-labelled ICAM-1 molecules.
- Use a 61X objective for the confocal microscope, or a 100X objective for the TIRF microscope, with appropriate filter settings.
- For granule release imaging by TIRF microscopy, add Alexa-Fluor-568-labeled anti-CD107a antibody Fab fragments to the cell suspension at a final concentration of 4 µg/mL before injecting the cells into the entry ports.
- Resuspend the prepared CD4+ T cells isolated from LN PB or cord blood and inject 50 µL of the cell suspension into the entry port of the slide channel containing the bilayer.
- Choose the desired number of fields and record images of each field 1x every 2 minutes for 30 min after the injection.
- Exploit bright-field, reflected light, and fluorescent channels (Alexa 488 and Cy5) of the confocal microscope to acquire the images. Use TIRF mode for Alexa-Fluor-488 and Alexa-Fluor-568 fluorescence and widefield for Cy5 fluorescence, as well as bright-field imaging, on the TIRF microscope.
Disclosures
The authors have nothing to disclose.
Materials
| CD4 T cell isolation kit, human | Miltenyl Biotec | 130-096-533 | |
| CD8 T cells Isolation Kit, human | Miltenyl Biotec | 130-096-495 | |
| DOPC | Avanti Polar Lipids | 850375C | |
| DOGS NTA | Avanti Polar Lipids | 790528C | |
| Biotinyl Cap PE | Avanti Polar Lipids | 870273C | |
| Human Serum Albumin | Octapharma USA | NDC 68982-643-01 | |
| sticky-Slide VI 0.4 | ibidi | 80608 | |
| Coverslips for sticky-Slides | ibidi | 10812 | |
| Bioptech FCS2 Chamber | Bioptech | 060319-2-03 | |
| anti-CD3 antibody | Thermo Fisher Scientific | 16-0037-81 | OKT3 clone, hybridoma cells are available from ATCC |
| anti- CD28 antibody | Genetex | GTX14664 | 9.3 clone |
| Casein | Sigma | C5890 | |
| Biotin-PEO4-NHS | Thermo Fisher Scientific | 21329 | |
| DMSO | Sigma | D2650-5 | |
| Alexa Fluor 488 protein labeling kit with column for labeled protein purification | Thermo Fisher Scientific | A10235 | |
| Alexa Fluor 568 protein labeling kit with column for labeled protein purification | Thermo Fisher Scientific | A10238 | |
| Amersham Cy5 NHS Ester | GE Life Science | PA15101 | |
| pMT/V5-His A, B, C Drosophila Expression Vectors | Thermo Fisher Scientific | V412020 | |
| pcopneo, G418 Drosophila expression vector for positive selection | ATCC | 37409 | |
| Serum free Drosophial media Insect-XPRESS | Lonza | 12-730Q | |
| Hybridoma YN1/1.7.4 | ATCC | CRL1878 | The hybridoma secrets antibody against ICAM-1. |
| Cyanogen bromide-activated-Sepharose 4B | Sigma-Aldrich | C9142 | Utilized for preparation of Sepharose with covelently bound anti-ICAM antibody. |
| MasterFlex tangential flow concentrator | Cole-Parmer | 77601-60 7592-40 | Used for ICAM-1 containing supernatant concentration and dialysis of ICAM-1 containing supernant |
| Centramate Lab Tangential Flow Systems | Pall Laboratory | FS002K10 OS010T12 FS005K10 | Used for ICAM-1 containing supernatant concentration and dialysis of ICAM-1 containing supernant |
| Ni-NTA Agarose | QIAGEN | 30210 | |
| Dialysis tubing | Spectra/Por | 131384 | |
| Papain from papaya latex | Sigma | P3125 | |
| mouse anti-human antibody against CD107a | BD Bioscences | 555798 | Clone H4A3 |
| Ansell Natural Blue Gloves | Fisher Scientific | 19-014-539 | |
| Nalgene Polypropylene Scissor-Type Forceps | Thermo Fisher Scientific | 6320-0010 | |
| Streptavidin | ProZyme | SA10 | |
| Confocal microscope | Nikon | Nikon TiE inverted microscope equipped with PFS for long-term image stability control, 60x oil objectives, 4 lasers with excitation lines at 405, 458, 488, 514, 561, and 640 nm, 2 GaAsP detectors and 2 high sensitivity PMTs, DIC transmitted light, Programmable X,Y,Z stage for multiple positions and stitching of large areas, time lapse functions, Tokai-Hit temperature and CO2-controlled chamber for live imaging, and anti-vibration isolation table | |
| TIRF microscope | Andor | Andor Revolution XD system equipped with Nikon TIRF-E illuminator, Lasers with 405,488,561 and 640 lines, DIC transmitted light, Yokogawa CSU-X1 spinning disk head for confocal imaging, 100/1.49 NA objective, Andor iXon X3 EM-CCD camera, objective heater, and a piezoelectric motorized stage with Perfect Focus System (PFS) |

