An In Vitro Assay to Measure Antibody-Dependent Cellular Phagocytosis by Phagocytes
Abstract
Source: Powell, R. L., et al. Isolation of Leukocytes from Human Breast Milk for Use in an Antibody-dependent Cellular Phagocytosis Assay of HIV Targets. J. Vis. Exp. (2019).
This video demonstrates an assay to measure the antibody-dependent cellular phagocytosis (ADCP) by breast milk phagocytes. Fluorescently labeled microspheres conjugated to an antigen-antibody complex, are combined with a cell suspension isolated from breast milk to induce antibody-dependent cellular phagocytosis by the phagocytic leukocytes. Upon labeling the cells with antibodies against common leukocyte antigen (CD45), flow cytometry is used to measure the fluorescence intensity of microsphere-positive viable phagocytic leukocytes, indicating the level of ADCP activity.
Protocol
All procedures involving sample collection have been performed in accordance with the institute's IRB guidelines.
1. Target Microsphere preparation
- Select a relevant target antigen.
NOTE: In this example, the recombinant protein V1V2-2F5K was used, which was designed to mimic the trimeric apex of native HIV envelope. - Perform biotinylation using a commercial kit (Table of Materials) according to the manufacturer's protocol.
- Calculate mmol of the biotin reagent to add to the reaction for a 5-fold molar excess using the formula: mmol biotin = mL protein x (mg protein/mL protein) x (mmol biotin/mg protein) x (5 mmol biotin/mmol protein). Then calculate µL of the biotin reagent to add using the formula: µL biotin = mmol biotin x (1,000,000 µL/L) x (L/10 mmol).
- Equilibrate biotin to room temperature before opening. Dissolve protein in 0.5−2.0 mL of phosphate-buffered saline (PBS) according to the calculation made above.
- Prepare a 10 mM solution of biotin reagent in dimethylsulfoxide (DMSO) and add the appropriate volume of 10 mM biotin reagent to the protein solution, and incubate reaction on ice for 2 h or at room temperature for 30 min.
- Remove excess biotin using protein concentrators (polyethersulfone [PES] membranes, 3 kDa molecular weight cut-off [MWCO], 0.5 mL; Table of Materials) according to the manufacturer's instructions.
- Deposit sample into the upper chamber of spin column and add PBS up to 400 µL. Cap, then insert this sample chamber into a collection tube. Centrifuge at 12,000 x g at room temperature for 30 min.
- Discard flow-through and add PBS to 400 µL. Repeat centrifugation. Discard flow-through and add PBS to 100 µL. Measure protein concentration by a spectrophotometer.
NOTE: Protein can be aliquoted and frozen at -80 °C until used.
2. Antibody-Dependent Cellular Phagocytosis (ADCP) Assay Plate Preparation
- Conjugate biotinylated protein to 1 µm microspheres (‘beads'; Table of Materials) according to the manufacturer's instructions.
- Per plate of conjugated beads, incubate 6 µg of protein with 12 µL of stock beads in 200 µL of 0.1% bovine serum albumin (BSA)-PBS at room temperature for 1 h, vortexing gently every 20 min.
- Centrifuge at 13,000 x g for 5 min. Discard supernatant, vortex gently and resuspend in 1200 µL of 0.1% BSA-PBS. Repeat spin and wash step 2x. Resuspend in 1200 µL of 0.1% BSA-PBS.
- Aliquot 10 µL of bead solution per well in a round bottom 96-well plate. Prepare dilutions of antibody or immune sera of interest in 12 µL of 2% HSA HBSS, typically starting at 50 µg/mL of antibody or a 1/100 serum dilution.
NOTE: In the sample data, monoclonal antibody (mAb) 830A was used. - Add 10 µL of titrated antibody/sera to the bead plate and incubate for 2 h at 37 °C. Add 200 µL of 2% HSA HBSS to each well and centrifuge plate at 2,000 x g for 10 min.
- Carefully remove supernatant by a rapid overturning and decanting of liquid from the plate wells into a sink to avoid disturbing the invisible bead pellet.
3. Breast milk cell isolation
- Obtain human breast milk from healthy lactating women, expressed using double electronic or manual pumps. Isolate cells within ~4 h of expression, keeping milk at room temperature.
- Using 50 mL tubes, centrifuge 50 mL milk at 800 x g for 15 min. Carefully pour off the skim milk and fat while leaving the cell pellet undisturbed. Wipe the inside of the tube with a lint-free wipe to remove all fat from the tube wall.
- Add 10 mL of 2% human serum albumen in Hank's balanced salt solution (2% HSA HBSS [without Ca2+ or Mg+]). Resuspend the pellet by gentle pipetting to avoid cell activation and apoptosis. Transfer to a 15 mL tube and centrifuge at 450 x g for 10 min.
- Pour off supernatant and repeat step 1.3. Then, gently resuspend cells in 1−2 mL of 2% HSA HBSS depending on expected cell number, and count cells by a hemocytometer or an automated cell counter, noting also the cell viability.
4. ADCP assay
NOTE: Methods described here are adapted from Ackerman et al.
- Add 50,000 freshly isolated BM cells to each ADCP assay plate well in 200 µL of 2% HSA-HBSS. Incubate for 2 h at 37 °C.
- For control experiments, pre-incubate cells at 37 °C with 10 µg/mL actin inhibitor (cytochalasin-D), 50 µg/mL Fc receptor blocking agent (FcBlock), or a combination of both prior to their addition to the plates.
- Centrifuge plate at 930 x g for 10 min. Add 200 µL of 2% HSA HBSS and repeat centrifugation. Carefully remove supernatant as in step 2.7 and repeat wash.
- Carefully remove supernatant and stain cells for viability using 0.5 µg/mL (final concentration) fixable viability stain 450 per well in 50 µL of 2% HSA HBSS. Incubate 20 min at room temperature in the dark. Centrifuge plate at 930 x g for 10 min and remove supernatant as in step 2.7. Add 200 µL of 2% HSA HBSS and centrifuge plate again followed by removal of supernatant as in step 2.7.
- After viability staining, stain cells for leukocyte markers of interest, minimally including a CD45-specific stain such as PE-mouse anti-human CD45 (clone HI30) at an optimized concentration (1 µg/µL in 50 µL of 1% BSA HBSS in the example data).
NOTE: Any lineage-specific markers of interest can be included. - Incubate 20 min at room temperature in the dark. Centrifuge plate at 930 x g for 10 min and remove supernatant as in step 2.7. Add 200 µL of 1% BSA HBSS and repeat centrifugation. Remove supernatant. Fix cells in 200 µL of 0.5% formaldehyde in the dark at room temperature for 30 min or overnight at 4 °C. Refrigerate in the dark until analysis.
5. Analysis by flow cytometry
- Perform initial gating to eliminate doublets on a forward scatter (FSC) vs. side scatter (SSC) plot and debris (material smaller than FSC = 5000) (see Figure 1). Use an SSC vs. viability stain (V450 in this case) plot to eliminate dead cells (those that are positive for viability stain).
- Use an SSC vs. CD45 plot to differentiate the major leukocyte classes (granulocytes, monocytes, lymphocytes) as extensively described.
NOTE: This classification is only suggestive and lineage-specific markers are needed to confirm cell type. - For all CD45+ cells, or for each leukocyte subset of interest, measure ADCP activity by gating with a marker on the bead-positive cells in a histogram of the fluorescein isothiocyanate (FITC) channel, where the fluorescent beads are detected.
NOTE: The negative control wells in which beads were not added will indicate where the bead-positive cells are apparent in the histogram and therefore where to place the gating marker. - Calculate ADCP scores as (median fluorescence intensity [MFI] of bead-positive cells) x (% of total CD45 + cells in the positive population). Use graphics software to plot scores at each Ab/serum concentration and to perform an area-under-the-curve (AUC) analysis.
NOTE: ADCP is considered positive if the AUC is greater than 3x standard deviation of the ADCP score AUC of a non-specific negative control mAb (in this case, 3865).
Representative Results
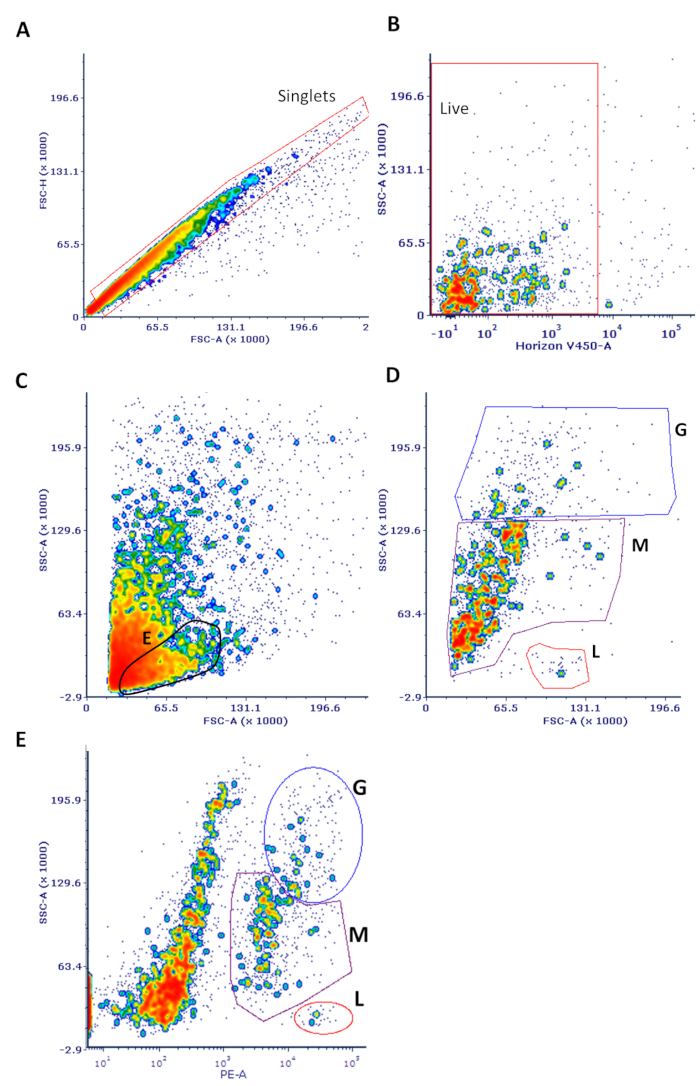
Figure 1: Sample flow cytometry data of cells isolated from breast milk. Cells were processed and stained as described in the protocol. (A) Single cells were gated on to eliminate doublets in an FSC-H vs. FSC-A plot as shown, also gating out the small debris <5,000 in FSC-A. (B) This population was used to gate on live cells (which do not stain with the viability dye) in an SSC vs. V450 (viability stain) plot. (C) These live cells were used in an FSC vs. SSC plot. The expected position of non-leukocytes, likely to be predominantly mammary epithelial cells, is highlighted ("E"). (D) The same FSC vs. SSC plot is shown only with CD45+ cells. The major leukocyte subsets noted are only purported identities based on well-established and expected SSC parameters (G = granulocytes; M = monocytes; L = lymphocytes). (E) Viable cells were used for an SSC vs. CD45 plot with the major leukocyte subsets noted. Back-gating from this plot yielded the data shown in panel D. Note that this classification is only suggestive and that lineage-specific markers are needed to confirm cell type.
Disclosures
The authors have nothing to disclose.
Materials
| 1 µm FluoSpheres NeutrAvidin-Labeled Microspheres | Thermo Fisher | F8776 | |
| BD Pharmingen PE Mouse Anti-Human CD45 | BD | 560975 | |
| Bovine serum Albumin | MP Biomedicals | 8810025 | |
| Corning V-bottom polystyrene 96-well plate | Corning | 3894 | |
| Cytochalasin D | Sigma | 22144-77-0 | |
| EZ-Link NHS-LC-LC-Biotin kit | Thermo Fisher | 21338 | |
| Falcon 15mL Conical Centrifuge Tubes | Corning | 352196 | |
| Falcon 50mL Conical Centrifuge Tubes | Corning | 352070 | |
| Fixable Viability Stain 450 | BD | 562247 | |
| Formaldehyde solution | Sigma | 252549 | |
| HBSS without Calcium, Magnesium or Phenol Red | Life Technologies | 14175-095 | |
| Human BD Fc Block | BD | 564219 | |
| Human Serum Albumin | MP Biomedicals | 2191349 | |
| Kimtech Science Kimwipes Delicate Task Wipers | Kimberly-Clark Professional | 34120 | |
| PBS 1x pH 7.4 | Thermo Fisher | 10010023 | |
| Polystyrene 10mL Serological Pipettes | Corning | 4488 | |
| Protein Concentrators PES, 3K MWCO, 0.5 mL | Pierce | 88512 |

