A Snap Chip Technology for a Cross-Reactivity-Free Multiplexed Sandwich Immunoassay
Abstract
Source: Li, H. et al., Snap Chip for Cross-reactivity-free and Spotter-free Multiplexed Sandwich Immunoassays. J. Vis. Exp. (2017)
The video showcases the implementation of snap chip technology for cross-reactivity-free multiplexed sandwich immunoassays. The simple action of snapping two microarray slides, each containing different reagents, ensures reagent transfer without cross-contamination and helps to detect multiple proteins using sandwich immunoassay.
Protocol
All procedures involving sample collection have been performed in accordance with the institute's IRB guidelines.
1. Fabrication and storage of snap chips
- Single transfer method (Figure 1a)
- Spot cAb solutions containing 400 µg/mL antibodies and 20% glycerol in phosphate-buffered saline (PBS) onto a nitrocellulose (or a functionalized glass) assay slide with an inkjet microarray spotter at a relative humidity of 60% (1.2 nL for each spot) with 800 µm center-to-center spacing. Make sure the slide is fixed on the spotter deck according to one corner (here, the bottom left corner was used).
- Incubate the spotted assay slide at room temperature for 1 h with a relative humidity of 60%.
- Clamp a slide module gasket with 16 compartments on the assay slide to divide it into 16 wells. Rinse the slide three times by adding 80 µL of PBS containing 0.1% Tween-20 (PBST) into each well, and shaking at 450 rpm on a shaker, 5 min each time.
- Add 80 µL of blocking solutions to each well and shake for 1 h at 450 rpm.
- Remove the gasket and dry the assay slide with a stream of nitrogen.
- Fix a transfer slide on the spotter deck and push the bottom left corner of the slide against the bottom left corner of the spotter deck. Inkjet spots an array of alignment marks (a solution of polystyrene micro-beads) with the same layout as that for the capture antibodies (cAbs).
- Let the alignment marks dry. Flip the transfer slide and put it back onto the spotter deck, fixing it against the bottom left corner.
- Prepare the detection antibody (dAb) spotting solutions containing 20 µg/mL antibodies, 20% glycerol, and 1% bovine serum albumin (BSA).
- Using the spotter's camera, take a picture of an alignment mark. Implement this picture into the spotting program as a fiducial and get the image recognition system of the inkjet spotter to identify the top left mark. Use its coordinate as the first spot of the dAb array. Spot 8 nL per droplet with a center-to-center spacing of 800 µm.
- Double transfer method (Figure 1b)
- Place a transfer slide 1 on the inkjet spotter deck against the bottom left corner. Spot cAbs solutions containing 400 µg/mL antibodies, 20% glycerol, and 1% BSA in PBS onto the slide with an inkjet microarray spotter at a relative humidity of 60% (0.4 nL for each spot and ~ 200 µm in diameter). The center-to-center spacing is 400 µm.
- Snap the transfer slide with a nitrocellulose coated (or a functionalized glass) assay slide using a snap apparatus (Figure 2) to transfer the cAb droplets onto the assay slide.
- Turn all six buttons on the side of the snap apparatus by 90 degrees to pull and lock all the plungers in the open position. Insert the transfer slide into the slide holder in its receptacle with the clipped corner facing the fixed pogo pin. Turn the first set of three buttons to release the plungers with the golden pogo pin and make sure the slides are pushed against the alignment pins.
- Insert a nitrocellulose-coated slide in the snap apparatus upside-down sitting on the 4 pogo pins. Turn the second set of three buttons to release plungers with the silver pin and make sure the slides are pushed against the alignment pins.
- Close the snap apparatus by placing the top shell on the slide holder using the alignment pillars and holes for a precise positioning.
- Insert the closed snap apparatus into its cage and push the closure tab completely down to bring the microarrays face-to-face while applying the appropriate pressure. Keep closed for 1 min.
- Separate the slides. Incubate the assay slide at room temperature for 1 h with a relative humidity of 60%. Wash, block, and dry the assay slide as described in steps 1.1.3 – 1.1.5.
- Place another transfer slide on the inkjet spotter deck and push against the bottom left corner. Spot dAb solutions containing 50 or 100 µg/mL of antibodies (see Table 1), 20% glycerol, and 1% BSA in PBS. Ensure that each spot is 0.8 nL and the center-to-center spacing between spots is 400 µm.
- Store the assay and the transfer slide. Seal both assay and transfer slides in an airtight bag containing desiccant and put in a -20 °C freezer.
2. Multiplexed immunoassays with snap chips
- Retrieve the assay slide from the freezer. Leave the bag sealed for 30 min until the slide comes to room temperature.
- Prepare 7-point serial diluted sample solutions by spiking proteins in PBS containing 0.05% Tween-20.
NOTE: Here, a 5-fold dilution factor is used. The starting concentrations for each protein are listed in Table 1. - Prepare sample solutions as appropriate. For example, dilute human serum 4 times in PBST buffer.
- Clamp a 16-compartment gasket on the assay slide.
- Fill one column of 8 wells with the 7 protein dilution solutions and a protein-free PBST buffer solution by pipetting. Pipette the sample solutions in the other 8 wells on the same slide.
NOTE: 80 µL solutions can be fit in each well. More slides can be used to measure additional samples when necessary. - Incubate the samples on a shaker at 450 rpm either for 1 h at room temperature or for overnight at 4 °C. Wash the slide three times with PBST on the shaker at 450 rpm, 5 min each time. Remove the gasket and dry the slide under a stream of nitrogen gas.
- Retrieve the transfer slide with dAbs from the freezer. Keep the bag sealed for 30 min at room temperature. Then, incubate the slide in a closed chamber (e.g. empty tips box) containing 60% humidity stabilization beads for 20 min for rehydration.
- Snap the transfer slide with the assay slide using a snap apparatus (Figure 2) to transfer the dAb droplets. See section 1.2.2 for the operation of the snap apparatus.
- Separate the slides. Incubate the assay slide in a closed chamber containing 60% humidity stabilization beads for 1 h. Clamp the assay slide with a 16-compartment gasket and rinse the slide 4 times using PBST on a shaker at 450 rpm, 5 min each time.
- Pipette 80 µL of solutions containing 2.5 µg/mL streptavidin fluorophore in PBS into each well. Incubate for 20 min on a shaker at 450 rpm.
- Rinse the slide 3 times with PBST and once with distilled water on the shaker at 450 rpm. Remove the gasket and dry the slide using nitrogen gas.
3. Slide scanning and data analysis
- Scan the assay slide with a fluorescence microarray scanner using the 635-nm-laser.
- Extract the net intensity of each spot using an analysis software (e.g. array-pro analyzer).
- Calculate the limit of detection (LOD) of each protein using a statistics software and determine the protein concentrations in the samples.
NOTE: The LOD is defined as the Y-intercept of the standard curve incremented by three times the standard deviation of three independent assays.
Table 1. Protein concentrations and LODs in buffer from the 50-plex assay. The LOD for CA 15-3 is in U/ml (*).
| Protein name | Starting concentration (ng/ml) | LOD (pg/ml) | Protein name | Starting concentration (ng/ml) | LOD (pg/ml) |
| ANG2 | 500 | 1.3 × 104 | IL-6b | 1 | 2.1 × 102 |
| BDNF | 500 | 1.0 × 102 | IL-5 | 50 | 77 |
| CA 15-3* | 1 | 4.9 × 103 | IL-4 | 1000 | 1.5 × 104 |
| CEA | 1000 | 5.4 × 103 | IL-2 | 50 | 76 |
| CXCL10/IP-10 | 50 | 1.7 × 102 | LEP | 200 | 4.0 × 102 |
| CRP | 200 | 44 | MIG | 500 | 11 × 102 |
| ENG | 1000 | 6.2 × 102 | CCL3/MIP-1α | 50 | 3.3 |
| EGF | 50 | 1.8 × 102 | CCL4/MIP-1β | 50 | 12 |
| EGFR | 200 | 1.9 × 102 | MMP-3 | 500 | 1.0 × 102 |
| FAS-L | 500 | 4.5 × 102 | M-CSF | 500 | 8.2 × 103 |
| FGF | 500 | 1.0 × 103 | MMP-9 | 200 | 3.1 × 102 |
| G-CSF | 500 | 39 | CCL2/MCP-1 | 50 | 55 |
| GM-CSF | 50 | 3.8 | NCAM-1 | 500 | 1.7 × 103 |
| GRO-α | 50 | 3.0 × 102 | β-NGF | 200 | 1.4 × 103 |
| HER2 | 500 | 3.5 × 103 | NT-3 | 200 | 5.8 × 102 |
| PDGF-BB | 200 | 73 | OPN | 500 | 1.9 × 103 |
| IL-1β | 500 | 7.9 × 102 | RBP4 | 200 | 1.2 ×102 |
| IL-1ra | 200 | 1.1 × 103 | SPARC | 1000 | 4.6 ×104 |
| IL-15 | 50 | 8.2 × 102 | TNF-α | 50 | 4.4 |
| IL-12 | 1 | 30 | TNF-RI | 50 | 1.3 × 102 |
| IL-11 | 500 | 1.2 × 103 | TNF-RII | 50 | 12 |
| IL-10 | 500 | 6.1 ×102 | FAS/TNFRSF6 | 200 | 2.8 ×102 |
| IL-8 | 50 | 6.6 | uPA | 200 | 24 |
| IL-7 | 2.5 | 26 | uPAR(CD87) | 200 | 76 |
| IL-6a | 1000 | 84 | VEGF | 200 | 6.7 × 102 |
Representative Results

Figure 1. Schematic of assay procedure comparing (a) single and (b) double transfer methods.
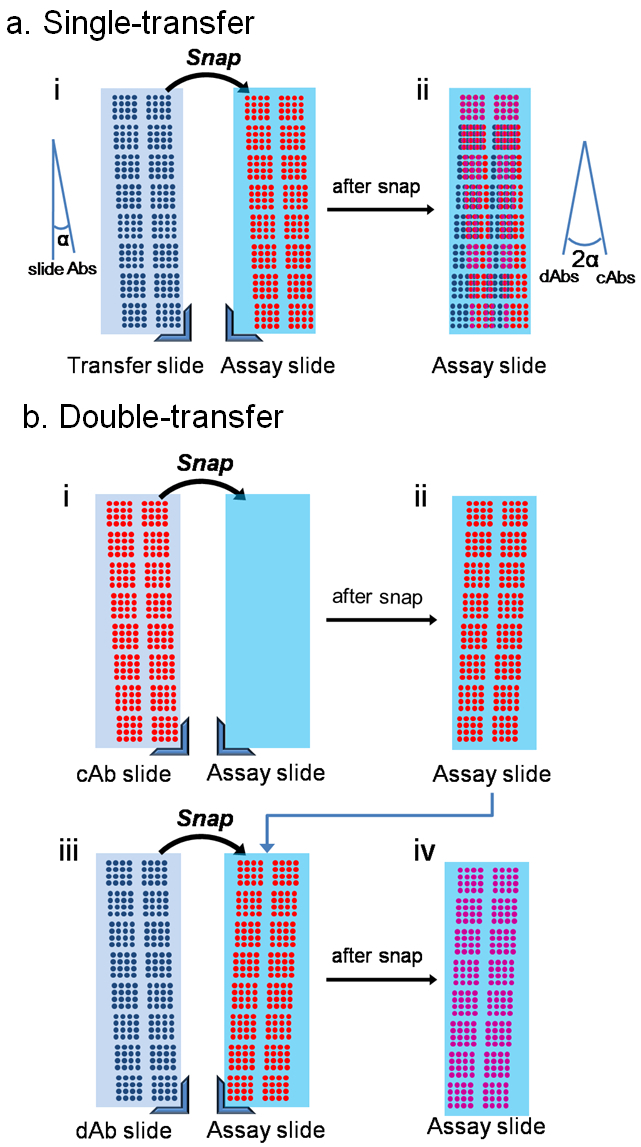
Figure 2. Schematic showing differences between single- and double-transfer. (a) mirroring of the transfer reagents amplifies the angular misalignment between the slide and the inkjet XY stage. (b) double-transfer method overcomes the angular misalignment.
Disclosures
The authors have nothing to disclose.
Materials
| Phosphate buffered saline tablet | Fisher Scientific | 5246501EA | |
| Streptavidin-conjugated Cy5 | Rockland | s000-06 | |
| Tween-20 | Sigma-Aldrich | p1379 | |
| Bovine serum albumin | Jackson ImmunoResearch Laboratories, Inc | 001-000-162 | |
| Glycerol | Sigma-Aldrich | G5516 | |
| Blocking solution: BSA-free StabilGuard Choice Microarray Stabilizer | SurModics, Inc | SG02 | |
| Nitrocellulose coated slides | Grace Bio-Laboratories, Inc | 305116 | |
| Aminosilane coated slides | Schott North America | 1064875 | |
| Snap Device | Parallex BioAssays Inc. | PBA-SD01 | |
| Inkjet microarray spotter | GeSiM | Nanoplotter 2.0 | |
| Slide module gasket | Grace Bio-Laboratories, Inc | 204862 | |
| Humidity Stabilization Beads | Parallex BioAssays Inc. | PBA-HU60 | |
| Array-Pro Analyzer software | Media Cybernetics | Version 4.5 | |
| Fluorescence microarray scanner | Agilent | SureScan Microarray Scanner | |
| Biostatistics software | GraphPad Software | GraphPad Prism 6 | |
| Endoglin capture antibody | R&D Systems | MAB10972 | |
| Endoglin protein | R&D Systems | 1097-EN | |
| Endoglin detection antibody | R&D Systems | BAF1097 | |
| IL-6a (see Table 1) | R&D Systems | ||
| IL-6b (see Table 1) | Invitrogen |

