Antigen Specific In Vivo Killing Assay using CFSE Labeled Target Cells
Summary
Many infections elicit a strong CTL response, but occasionally, the quantity of responding cells does not correlate to control of the pathogen1. One measure of CTL quality is their ability to kill specifically2. CFSE labeling of target cells can be used to investigate this CTL response quality in vivo3,4.
Abstract
Carboxyfluorescein diacetate succinimidyl ester (CFSE) can be used to easily and quickly label a cell population of interest for in vivo investigation. This labeling has classically been used to study proliferation and migration. In the method presented here, we have shortened the timeline after adoptive transfer to look at survival and killing of epitope specific CFSE labeled target cells4-6. The level of specific killing of a CD8 + T cell clone can indicate the quality of the response, as their quantity may be misleading. Specific CD8+ T cells can become functionally exhausted over time with a decline in cytokine production and killing7,8. Also, certain CD8 + T cell clones may not kill as well as others with differing TCR specificities9. For effective Cell Mediated Immunity (CMI), antigens must be identified that produce not only adequate numbers of responding T cells, but also functionally robust responding T cells. Here we assess the percent cell specific killing of two peptide specific T cell clones in BALB/c mice.
Protocol
1. Eliciting Target-Specific Effector CTLs by Immunization (Day 0)
- Before beginning, decide upon a specific effector:target system. In this example of a classical in vivo CTL assay, we will use a purified peptide immunization to elicit specific CD8 + T cells. The same peptide will be used to pulse the syngeneic target cells, creating a specific effector:target system. Alternatively, you may choose to use whole pathogen or protein to obtain your specific effectors and targets.
- Decontaminate the work surface inside of a biosafety hood to ensure sterility of the injections. Prepare the peptide immunization by first placing 50 μg of purified peptide into 100 μL 10% DMSO in PBS/mouse contained in a 2 mL tube. Next, add an equal amount of very cold Incomplete Freund’s Adjuvant and up to the .2 mL mark with 1 mm beads for mixing. From this point, keep immunogen on ice until injection. Secure lid with Parafilm.
- Use a bead beater set to maximum RPMs to mix the emulsion for 3 minutes, promptly placing the emulsion back on the ice after mixing. The resulting emulsion should be similar in consistency to mayonnaise. Allow emulsion to sit at 4° C overnight if time permits (up to a week), mixing further if any separation is noted.
- Use a 1 mL syringe and 20 or 20.5 gauge needle to pull up emulsion being careful to not pull up any beads (beads should be larger than gauge of needle, but still be aware).
- Secure mouse for subcutaneous injection at the base of the tail. We use a modified plastic 50 mL conical tube to safely and gently immobilize the mouse for injection. Inject the mouse with .2 mL of emulsion.
2. Harvesting Target Cells (Day 8)
- You will need naíve syngeneic mice for your target cells. Euthanize naíve mice according to AAALAC standards.
- Disinfect mouse by spraying 70% Ethanol to minimize the chance of contamination from mouse hair or other external contaminants.
- Using dissection scissors make small snip through the skin on the left side of the mouse. Pull skin flap over to uncover peritoneal sac and internal organs.
- Snip through the peritoneum, being careful not to compromise any of the underlying organs. The spleen is close to the surface and is similar in color to the liver (dark red) but smaller and more kidney bean in shape. Infected mice generally show notably larger spleens than uninfected controls.
- Carefully pull out the spleen, taking care to be gentle so as not to break the spleen into multiple pieces in order to recover maximum cell numbers. Immediately place the spleen into a 15 mL conical containing 10 mLs of prepared media on ice. Continue with remaining mice.
- Pour contents of the 15 mL conical tube (media and spleen) into a glass tissue grinder. Grind the spleen using plenty of upper arm strength. When all tissue has lost its color you may assume that you have recovered all cells out of the connective tissue.
- Pour contents of the grinder through a 70 μm cell filter into a 50 mL conical tube. Rinse grinder with fresh media and pour through a cell strainer.
- Centrifuge at 4° C, 1300 RPM for 7 minutes. Pour off supernatant.
- Resuspend in 10 mLs of prepared lysing reagent and incubate at 37° for 7 minutes.
- Add up to 50 mLs with fresh media and centrifuge, same as above.
- Resuspend in 50 mLs fresh media and centrifuge, same as above (wash step). *Note: At this time you can perform a magnetic cell separation for a specific cell type (i.e. macrophages) if a pure target cell population is desired for transfer into recipient mice. Also, at this time you could introduce an experimental treatment into the culture in order to study the affects of that treatment on cell specific killing.
- Count cells and plate 1 x 107 per well in 100μL in a 96 well round bottom tissue culture treated plate. *This is a high concentration of cells/well, but because the assay is a quick one, we have seen no loss in viability of the cells at this concentration.
3. Treatment and Labeling of Target Cells
- Pulse appropriate cells and replicates with pathogen specific peptide or irrelevant peptide control: Add 1 μg/mL peptide. Incubate at 4° C for 1.5 hours. Move to 37° C incubator for last 30 minutes. During this time prepare CFSE reagent.
- Prepare CFSE according to manufacturer specifications and dilute to a working concentration between 0.5 and 25 μM. Make “low” concentration 10x lower than “high” concentration CFSE. For example, for [low] add 0.5 μL CFSE to 10 mLs PBS and for [high], add 5 μL CFSE to 10 mLs PBS. Warm prepared CFSE to 37° C before labeling.
- Remove plate of cells from incubator and pellet in a high speed centrifuge at room temperature, 1300 RPM for 7 minutes. Flick to remove supernatant.
- Resuspend cells in either [low] or [high] CFSE by adding 200 μL of appropriate reagent to the corresponding wells and incubate for 15 minutes at 37° C.
- Remove plate of cells from incubator and again pellet at room temperature, 1300 RPM for 7 minutes. Flick to remove supernatant.
- Resuspend cells in 200 μL/well of warmed fresh media and incubate for 30 minutes at 37°C.
- Remove plate of cells from incubator and pellet at room temperature, 1300 RPM for 7 minutes. Flick to remove supernatant.
- Resuspend cells in 100 μL/well PBS.
- For adoptive transfer, add one well [low] labeled cells to one well [high] labeled cells per mouse. Resulting injection should be 50% specific targets and 50% irrelevant cells in .2 mLs.
- Don’t forget to set aside some labeled cells to act as your non-transferred controls during data analysis and for running on the flow cytometer (See Figure 1).
4. Adoptive transfer
- Anesthetize previously peptide immunized mice (From part #1 of this protocol) according to AAALAC standards.
- Inject .2 mLs of labeled target cells via retro-orbital route into recipient mice.
- Wait 6 hours or other predetermined optimal time before euthanizing the recipient mice.
5. Harvesting Labeled Cells
*Note: Steps 5.1 through 5.11 are the same as steps 2.1 through 2.11
- Count cells and plate out at 1 x 106 per well in 100 μL into 96 well round bottom tissue culture treated plate.
- Fix recovered and non-transferred cells by adding 100 μL 2% paraformaldehyde to a final concentration of 1% paraformaldehyde to all wells.
6. Flow Cytometry and Data Analysis
- Run samples on a flow cytometer and analyze the data.
7. Representative Results
A diminishing epitope specific peak on the resulting histogram is an indicator of cell specific killing (Figure 1). Use the following equations (applied in Table 1) to determine the actual value of cell specific lysis:
Ratio = Irrelevant Percentage : Epitope Specific Percentage ( [low] peak:[high] peak),
Percent Specific Lysis = [1-(Non-transferred control ratio/Experimental ratio)] x 100
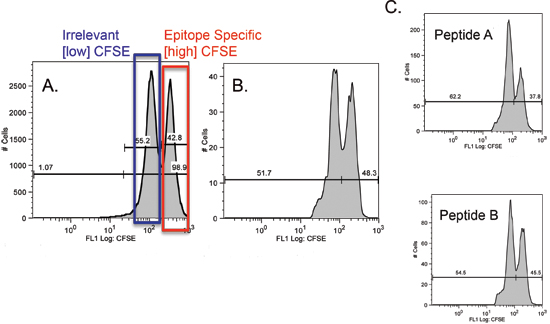
Figure 1. CFSE profile of recovered BALB/c mouse splenocytes. Numbers indicate the percentage of cells of the high or low CFSE phenotype. A) Non-transferred control. All [high] CFSE peaks represent epitope specific target cells, while [low] CFSE peaks represent targets pulsed with irrelevant peptide. B) CFSE labeled cells recovered from PBS immunized recipient mouse (Control). C) CFSE labeled cells recovered from two different peptide specific recipient mice, note the diminished epitope specific peaks.
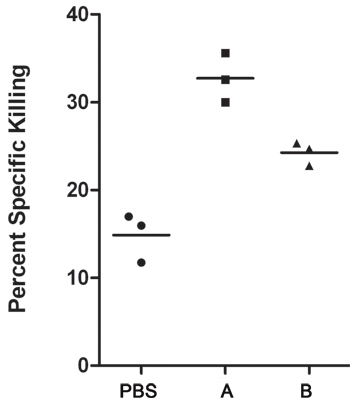
Figure 2. Graphical representation of analyzed data. Three data points shown for each group of mice, PBS, Peptide A, and Peptide B.
| Sample | CFSE HIGH % Epitope Pulsed | CFSE LOW % Irrelevant | Ratio Low:High | Percent Epitope Specific Killing |
| PBS | 42.8 | 57.2 | 1.34 | 16.97 |
| PBS | 43.1 | 56.9 | 1.32 | 15.94 |
| PBS | 44.3 | 55.7 | 1.26 | 11.74 |
| Peptide A | 39.7 | 60.3 | 1.52 | 32.56 |
| Peptide A | 38.6 | 61.4 | 1.59 | 35.61 |
| Peptide A | 40.6 | 59.4 | 1.46 | 29.99 |
| Peptide B | 38.7 | 61.3 | 1.58 | 24.68 |
| Peptide B | 38.5 | 61.5 | 1.60 | 25.32 |
| Peptide B | 39.3 | 60.7 | 1.54 | 22.76 |
| PBS NT Control | 47.4 | 52.6 | 1.11 | |
| Pep A NT Control | 49.4 | 50.6 | 1.02 | |
| Pep B NT Control | 45.6 | 54.4 | 1.19 |
Table 1. Analysis of representative data. “NT” is non-transferred.
Discussion
This assay can be modified to investigate the proliferative capacity of cells, including clonal expansion, because CFSE divides relatively equally among progeny10,11. It is also possible to use CFSE labeling to investigate cell migration6,12, though there are other cell tracing methods that may be more appropriate depending on your hypothesis, for example tetramers and bioluminescence13, which also may be combined with CFSE labeling.
It is important to note that CFSE bleeds over into multiple channels on the flow cytometer, so if you want to co-stain for a surface marker, PeCy7 would be a good choice of fluorophore, while PE might not work as well.
Disclosures
The authors have nothing to disclose.
Acknowledgements
I would like to thank Bianca Mothé, Carla Oseroff, and Marie-France DelGuercio for introducing me to immunological assays and mouse handling. Work in the lab of G. A. Splitter is funded by NIH grant 1-RO1-AI-073558, GLRCE grant 1-U54-AI-057153, and BARD US-3829-06.
Materials
| Material Name | Type | Company | Catalogue Number | Comment |
|---|---|---|---|---|
| 1 mm glass beads | Biospec Products | 11079110 | ||
| 70 μm cell strainer | BD Falcon | 352350 | ||
| 96-well plate | Costar | 3799 | ||
| Adjulite Incomplete Freund’s Adjuvant | Pacific Immunology | n/a | ||
| DMSO | Sigma | D-5879 | ||
| FBS | GIBCO | 16000-077 | ||
| FC 500 Flow Cytometer | Beckman Coulter | n/a | ||
| Kontes glass tissue grinder | Kontes | 885300-0015 | ||
| Mini Beadbeater | Biospec Products | n/a | ||
| Needle | B-D | 305175 | ||
| Parafilm “M” | Pechiney Plastic Packaging | PM992 | ||
| Paraformaldehyde | Electron Microscopy Sciences | 157-8 | ||
| PBS | GIBCO | 10010-023 | ||
| PharmLyse lysing reagent | BD Biosciences | 555899 | ||
| RPMI 1640 | GIBCO | 11875-093 | ||
| Syringe | B-D | 309602 | ||
| Vybrant CFSE Kit | Invitrogen (Molecular Probes) | V12883 |
References
- Blattman, J. N., Wherry, E. J., Ha, S. J., van der Most, R. G., Ahmed, R. Impact of epitope escape on PD-1 expression and CD8 T-cell exhaustion during chronic infection. J Virol. 83 (9), 4386-4394 (2009).
- Jenkins, M. R., Tsun, A., Stinchcombe, J. C., Griffiths, G. M. The strength of T cell receptor signal controls the polarization of cytotoxic machinery to the immunological synapse. Immunity. 31 (4), 621-631 (2009).
- Ingulli, E. Tracing tolerance and immunity in vivo by CFSE-labeling of administered cells. Methods Mol Biol. 380, 365-376 (2007).
- Durward, M. A., Harms, J., Magnani, D. M., Eskra, L., Splitter, G. A. Discordant Brucella melitensis antigens yield cognate CD8+ T cells in vivo. Infect Immun. 78 (1), 168-176 (2010).
- Lyons, A. B., Parish, C. R. Determination of lymphocyte division by flow cytometry. J Immunol Methods. 171 (1), 131-137 (1994).
- Weston, S. A., Parish, C. R. New fluorescent dyes for lymphocyte migration studies. Analysis by flow cytometry and fluorescence microscopy. J Immunol Methods. 133 (1), 87-97 (1990).
- Akondy, R. S. The yellow fever virus vaccine induces a broad and polyfunctional human memory CD8+ T cell response. J Immunol. 183 (12), 7919-7930 (2009).
- Wallace, P. K. Tracking antigen-driven responses by flow cytometry: monitoring proliferation by dye dilution. Cytometry A. 73 (11), 1019-1034 (2008).
- Labadi, A., Balogh, P. Differential preferences in serosal homing and distribution of peritoneal B-cell subsets revealed by in situ CFSE labeling. Int Immunol. 21 (9), 1047-1056 (2009).
- Suffner, J. Dendritic cells support homeostatic expansion of Foxp3+ regulatory T cells in Foxp3.LuciDTR mice. J Immunol. 184 (4), 1810-1820 .

