A Comparative Approach to Characterize the Landscape of Host-Pathogen Protein-Protein Interactions
Summary
This article focuses on the identification of high-confident interaction datasets between host and pathogen proteins using a combination of two orthogonal methods: yeast two-hybrid followed by a high-throughput interaction assay in mammalian cells called HT-GPCA.
Abstract
Significant efforts were gathered to generate large-scale comprehensive protein-protein interaction network maps. This is instrumental to understand the pathogen-host relationships and was essentially performed by genetic screenings in yeast two-hybrid systems. The recent improvement of protein-protein interaction detection by a Gaussia luciferase-based fragment complementation assay now offers the opportunity to develop integrative comparative interactomic approaches necessary to rigorously compare interaction profiles of proteins from different pathogen strain variants against a common set of cellular factors.
This paper specifically focuses on the utility of combining two orthogonal methods to generate protein-protein interaction datasets: yeast two-hybrid (Y2H) and a new assay, high-throughput Gaussia princeps protein complementation assay (HT-GPCA) performed in mammalian cells.
A large-scale identification of cellular partners of a pathogen protein is performed by mating-based yeast two-hybrid screenings of cDNA libraries using multiple pathogen strain variants. A subset of interacting partners selected on a high-confidence statistical scoring is further validated in mammalian cells for pair-wise interactions with the whole set of pathogen variants proteins using HT-GPCA. This combination of two complementary methods improves the robustness of the interaction dataset, and allows the performance of a stringent comparative interaction analysis. Such comparative interactomics constitute a reliable and powerful strategy to decipher any pathogen-host interplays.
Introduction
The increasing amount of data collected to generate protein-protein interaction maps opens perspectives to further understand pathogen infections. As global understanding of pathogen infections begins to emerge, it provides access to the range of perturbations induced by pathogens proteins when connecting the human proteome 1. It thereby offers a way to comprehend how pathogens manipulate the host cell machinery. In particular, the mapping of several viral-host interaction networks revealed that viral proteins preferentially target host proteins that are highly connected in the cellular network (hubs), or that are central to many paths in a network (bottlenecks proteins) 2-4. These interactions allow viruses to manipulate important cellular processes, which is instrumental to replicate and produce infectious progeny. Comparative interactions mapping were recently conducted on related viruses with the goal to extract information relative to pathogenesis 4. Furthermore, studies of the host-pathogens interactions landscape have been extended to many pathogens 5. The targeted cell factors identification provides insights into the strategies used by pathogens to infect cells and allows detection of potential pathogenic markers.
The yeast two-hybrid system is the most popular method to identify binary interactions since genetic screening is an efficient and sensitive tool for high-throughput mapping of protein-protein interactions. The approach proposed here further improves individual Y2H screenings by assessing a protein of multiple pathogens strain variants, thus providing access to a comparative overview of pathogen-host protein-protein interactions. Moreover, a major limitation of yeast two-hybrid screening lies in a high false-negative rate, since it recovers about 20% of total interactions 6. This implies that interactions detected with only a subset of pathogens variants might have escaped detection with the others. Therefore, individual partners emerging from all the two-hybrid screenings are further challenged for interaction with the complete range of strains studied, providing comparative interaction datasets. Since it was demonstrated that combining different methodologies strongly increases the robustness of protein-protein interaction datasets 7, this validation is performed in mammalian cells by a newly developed protein-fragment complementation assay called HT-GPCA 8. This cell-based system allows the detection of protein interactions by complementation of the Gaussia princeps split luciferase and has been designed to be compatible with a high-throughput format. Thanks to its luminescence-based technology and its low background noise compared with other fluorescence-based assay, HT-GPCA shows a strikingly high sensitivity.
Overall, this approach constitutes an efficient tool to generate a comprehensive mapping of host-pathogen interplays, which represents the first step toward a global understanding of the host cell hijacking.
Protocol
1. Yeast Two-hybrid Screenings
- Make the following required solutions
- Complete Synthetic Drop Out (SD): dissolve 26.7 g of minimal SD base with glucose in 1 L of water. Add 2 g of amino acid mixture prepared as follows: 2 g Adenine hemisulfate, 2 g Arginine HCl, 2 g Histidine HCl, 2 g Isoleucine, 4 g Leucine, 2 g Lysine HCl, 2 g Methionine, 3 g Phenylalanine, 2 g L-Serine, 2 g L-Threonine, 3 g Tryptophan, 2 g Tyrosine, 1.2 g Uracil, 9 g Valine.
- Selective Drop Out SD-L/-W/-H: SD containing every essential amino acids except those specified (-L: -Leucine ; -W: -Tryptophan; -H: -Histidine).
- YPGLU: For 1 L, weight 10 g of yeast extract, 10 g of Bacto Peptone and 20 g of glucose. Complete with 25 g Bacto agar for solid media.
- YCM: Same as YPGLU, but pH adjusted to 4.5.
- Mating and spreading
- Use frozen aliquots of pre-transformed Y187 yeast strain (mating type α) containing a cDNA library cloned in pACT2. Check cell viability by plating serial dilutions on SD-L plates.
- Clone the pathogen ORF into pGBKT7 vector, and transform the recombinant plasmid using a standard procedure into AH109 yeast strain (mating type a).
- Spread pGBKT7-transformed AH109 yeast on SD-W plates. Incubate at least two days 30 °C in a humidified incubator. Plates can be stored at 4 °C until the two-hybrid screening.
- Use 3-aminotriazole (3-AT), a competitive inhibitor of the HIS3 enzyme, to counteract the potential self-transactivation of the HIS3 reporter gene by the pathogens’ proteins. Perform an auto-activation test by determining the concentration of 3-AT, which prevents growth of pGBKT7-transformed yeast on SD-W/-H plates.
- Seed 30 ml of SD-W with few colonies of pGBKT7-transformed AH109. Incubate 30 hr at 30 °C with rotation.
- Seed 200 ml of SD-W with the 30 ml of the AH109 cultures. Incubate with rotation around 20 hr at 30 °C.
- Incubate thawed cDNA library-transformed Y187 in 20 ml YPGLU, incubate 10 min at 30 °C with rotation.
- Centrifuge a volume of pGBKT7-transformed AH109 culture corresponding to an equivalent amount of viable Y187 yeast cells for 5 min at 3,500 rpm at 20 °C. Carefully withdraw the supernatant and resuspend pellet in 10 ml YPGLU.
- Mix the resuspended AH109 yeast with Y187 containing library. Transfer in a 50 ml tube. Centrifuge for 5 min at 3,500 rpm at 20 °C, carefully withdraw supernatant (fragile pellet).
- Resuspend pellet in YPGLU to get a total of 1.5 ml.
- Spread yeast on to 3 YCM-Agar 150 mm plates, 0.5 ml/plate. Incubate 4.5 hr at 30 °C.
- Collect mated yeast from YCM plates by scraping with a rake glass in 10 ml of SD-L/-W/-H. Rinse the plates two times with 5 ml SD-L/-W/-H medium and pool.
- Centrifuge 5 min at 3,500 rpm, 20 °C. Discard supernatant and resuspend the yeast pellet in 5 ml SD-L/-W/-H medium.
- Spread mated yeast on to 10 plates (0.5 ml/plate) of SD-L/-W/-H Agar supplemented with the appropriate concentration of 3-aminotriazole (3-AT) determined in the auto-activation test. Incubate at 30 °C in a humidified atmosphere for 6-10 days.
- Determine the diploid (2n) rate in order to evaluate mating efficiency by spreading 250 μl of serial dilutions (10-4 to 10-6) of the mated yeast suspension on SD-L/-W plates. Incubate plates at 30 °C for two days. Count colonies grown on SD-L/-W, and deduce the total diploid number present in the initial volume of mated yeast. Report this diploid number to viable library-containing yeast number to calculate the diploid rate. It should range around 10-25%.
- 6-10 days after incubation at 30 °C, pick mated yeast colonies in fresh SD-L/-W/-H + 3-AT plates. [Tip: order the yeast in a scheme of 8 lanes of 12 wells reproducing a 96 well plate so that it will be easier afterward for sequencing]. Let yeast colonies grow for 4-5 days at 30 °C in a humidified atmosphere.
- Screening of his3 positive clones by PCR and sequencing
The goal is to PCR amplify the ORF contained in the pACT2 vectors of yeast colonies grown on selective SD-L/-H/-W plates.- Put a 96 well PCR plate on ice.
- To lyse yeast cells, add 50 μl per well of a Zymolase 20T solution (2.5 mg/ml in 10 mM HEPES Buffer pH 7.7).
- Gently resuspend one colony per well.
- Incubate 15 min at 37 °C and 15 min at 95 °C.
- Put the plate back on ice. Add 50 μl of distilled water per well. Mix well by pipetting up and down.
- Centrifuge for 5 min at 3,500 rpm and 4 °C.
- PCR amplify 10 μl to detect an insert using primers annealing either sides of the ORF derived from cDNA in pACT2 vectors. The protocol is given for amplification of 96 lysed yeast colonies with the ExTaq polymerase. Prepare the following mix: 525 μl 10X ExTaq buffer, 420 μl dNTPs mix (2.5 mM), 10.5 μl Forward Primer (1 μg/μl), 10.5 μl Reverse Primer (1 μg/μl), 31.5 μl ExTaq (5 U/μl) and 3202.5 μl H20. Distribute 40 μl of the mix per well in a 96 well PCR plate. Add 10 μl of lysed yeast per well. Perform amplification as follows: 94 °C for 3 min, then 35 cycles at 94 °C for 30 sec, 60 °C for 1 min, 72 °C for 4 min, and end with 72 °C for 7 min. Run 3 μl of the PCR products on a 1.2% agarose gel to detect PCR-positive clones. Note that using pre-cast 96 well agarose gels is recommended.
[Tip: plate can be stored at -20 °C for a few months and re-used for new PCR] - Sequence the PCR amplified fragments isolated from HIS3 positive clones. Perform a BLAST analysis to identify cellular ORF. Low-confidence alignments (E value >10-10, identity <80%, peptide length <20 amino acids) are discarded as well as frameshifted and premature STOP codon containing sequences.
2. Matrix building for HT-GPCA
HT-GPCA is a mammalian cell-based assay where both pathogen and host proteins are transiently expressed fused to complementary fragments of the split Gaussia princeps luciferase. Upon interaction of the partners, this enzyme is reconstituted allowing the measure of its enzymatic activity. The interaction intensity is thus deduced from a Normalized Luminescence Ratio (see below and Figure 1)
- Selection of host cell partners
- Select proteins that are identified more than three times in the Y2H screenings for further validation in mammalian cells to increase the dataset confidence 9.
- Add known interacting partners of the pathogen proteins as positive controls.
- Transfer into HT-GPCA compatible vectors
- Clone each pathogen protein ORF in a pSPICA-N2 vector to generate pathogen proteins with amino acids 110 to 185 of the humanized Gaussia princeps luciferase fused at their N-terminus (GL2-pathogen protein fusions).
- PCR amplify cellular protein ORF directly from the Y2H positive clones or from other ORF resources (Human ORFeome http://horfdb.dfci.harvard.edu/) and transfer them into pSPICA-N1 vectors to generate proteins fused at the N-terminus with the amino acids 18 to 109 of humanized Gaussia princeps luciferase (GL1-host protein fusions).
[Tip: For large-scale cloning, the Gateway cloning system is recommended. In that case, for PCR amplification, use primers designed to contain Gateway compatible sites]. - Sequence all expression vectors. Sequences of the pSPICA-N1 and pSPICA-N2 can be found in reference 8.
Note that plasmid constructs can be inversed, i.e. pathogens proteins in pSPICA-N1 and host protein into pSPICA-N2.
3. High-Throughput Gaussia princeps Luciferase-Based Protein Complementation Assay (HT-GPCA)
- Cell Transfection
- Seed 293T cells at 350,000 cells/ml of DMEM with 10% Fetal Bovine Serum (FBS) without antibiotic. Distribute 100 μl/well in sterile white culture plates. Do not exceed 10 plates in a single experiment. Let cells grow for 24 hr at 37 °C with 5% CO2.
[Tip: Since the pSPICA plasmids contain an SV40 origin, the use of 293T cells allows their replication in transfected cells and thus leads to overexpression of the two luciferase fragments.] - The following day, transfect cells using any suitable transfection protocol. Note that for certain transfection reagents, the number of cells seeded may have to be adapted. Use 100 ng per well of both the pathogen and host ORF plasmid constructs. Co-transfect 10 ng per well of a of CMV-Firefly luciferase plasmid to normalize for transfection efficiency. Each point is tested in triplicate.
[Tip: DNA mixes can be performed the day before transfection and stored at -20 °C with adhesive seals.] - Transfer transfection mix to the plates of cultured 293T cells. Be careful to gently deposit the DNA mix onto the cells, as 293T are not strongly adherent and easily detach from the bottom of the well. Incubate in a cell culture incubator for 24-30 hr at 37 °C with 5% CO2.
- Seed 293T cells at 350,000 cells/ml of DMEM with 10% Fetal Bovine Serum (FBS) without antibiotic. Distribute 100 μl/well in sterile white culture plates. Do not exceed 10 plates in a single experiment. Let cells grow for 24 hr at 37 °C with 5% CO2.
- Cell lysis and Gaussia luciferase dosage
- Wash cells with PBS (100 μl/well). Add 40 μl 1X Renilla lysis buffer. Incubate for 30 min at room temperature under agitation.
- Save 10 μl of cell lysates in another white plate for subsequent dosage of the Firefly luciferase activity. Store at 4 °C or alternatively at -20 °C with sealing.
- Measure the Gaussia activity on the remaining 30μl of cell lysates by adding 100 μl of Renilla luciferase assay reagent and counting luminescence for 10 sec with a luminometer. Dosage of a 96 well plate takes about half an hour. Perform measurement with fresh cell extracts since freezing or long-term storage may affect interaction-mediated Gaussia activity.
- Measurement of Firefly activity
- Measure the Firefly activity on the saved 10 μl of cell lysates by adding 50 μl of Firefly luciferase assay reagent and counting luminescence for 10 sec. Dosage of a 96 well plate takes about half an hour. [Tip: The dosage of Firefly luciferase can be performed the day after on frozen lysates.]
- NLR Calculations
- For each interaction, calculate the ratio between Gaussia luciferase activity and Firefly luciferase activity to obtain a normalized Gaussia luminescence value taking into account transfection efficiency and cell viability. Calculate the mean of the triplicates.
- To monitor the interaction, estimate a Normalized Luminescence Ratio (NLR) for each pathogen-host pair. To do so, divide the normalized Gaussia luminescence activity detected in presence of both pathogen and host proteins by the sum of the activities measured in control wells (Figure 1 adapted from reference 8). The NLR cut-off value for positive interactions has been estimated to be around 3.5 8.
- Data analysis
- Perform hierarchical clustering using the R statistic package. First, group the NLR data into categories, then calculate an interaction dendrogram using the complete linkage method. Visualize this dendrogram with JavaTreeView 10. Calculate the distance between interaction dendrogram and phylogenetic tree using a pearson correlation coefficient.
- To generate interaction networks, select the positive interactions from the HT-GPCA dataset and use the Cytoscape software 1. Extract the degrees of the host proteins and plot their distribution to access the network local dynamics. Host-pathogens networks can be superimposed to the host interactome to provide an appraisal of the global impact of pathogen proteins in the host cellular network.
- Extract the GO (Gene Ontology) terms associated with the targeted cellular factors with the DAVID bioinformatics database 12 to assess the functional enrichment of the dataset.
Representative Results
A major strength of HT-GPCA lies in its high sensitivity, as illustrated by the assessment of false positive and false negative rates for the HPV E2 protein in Figure 2 (adapted from reference 13). To determine the false negative rate, known interactions of E2 from HPV16 were assessed by HT-GPCA (Figure 2A). Four out of 18 interactions were not recovered (corresponding to a 22% false negative rate). The false positive interactions were measured to be 5.8% using 12 HPV E2 proteins against a random set of cellular proteins (Figure 2B). Furthermore, the strong specificity of the method is highlighted in Figure 2C showing that a single point mutation known to interfere with E2 binding to its cell partner BRD4 annihilates the NLR ratio (Figure 2C).
This large-scale comparative interactomic approach has been recently successfully applied to three early proteins of the Human Papillomaviruses (HPV): E2, E6 and E7 originating from different genotypes representative of the HPV natural diversity in tropisms and pathologies 13,14. For each of the early proteins, hierarchical clustering of the interaction profiles mostly recapitulates HPV phylogeny, proving the robustness of the approach and the pertinence of interaction datasets (Figure 3 adapted from references 13 and 14). It can be used to correlate specific virus-host interaction profiles with pathological traits, thereby giving clues of the involvement of viral proteins in pathogenesis. Genotype-specific interactions can be extracted, which potentially correspond to tropism or pathogenic biomarkers as shown in Figure 4 for the E6 proteins of HPV (Figure 4 adapted from reference 14).
Furthermore, functional insights can emerge from such large-scale analyses. For example, in the interactomic study of the HPV E2 proteins, the functional enrichment analysis led to the identification of a prominent family consisting in cellular transcription factors, in line with the primary role of E2 as a viral transcriptional regulator. It also revealed an unexpected functional targeting of the intracellular trafficking machinery, opening new perspectives for the role of E2 in HPV pathogenesis 13.
Overall, the comparative interatomic studies performed with the HPV early proteins greatly improve the comprehension of HPV pathogenesis by correlating specific interactions profiles to phenotypic differences.
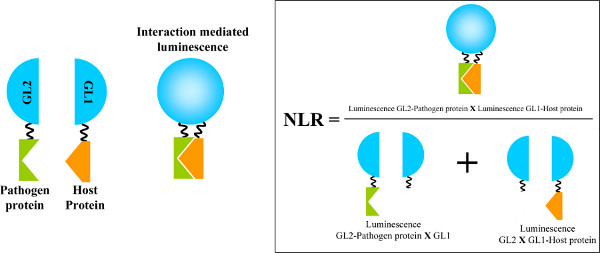
Figure 1. Identification of protein-protein interactions using HT-GPCA. Both pathogen and host proteins are fused to an inactive half of the Gaussia princeps luciferase (GL2 and GL1 parts, respectively). When the proteins are interacting, a luciferase activity is induced by the proximity of both halves of the enzyme. The restored Gaussia luciferase activity is calculated from a Normalized Luminescence Ratio (NLR) as shown on the right. Figure adapted from reference 8. Click here to view larger figure.
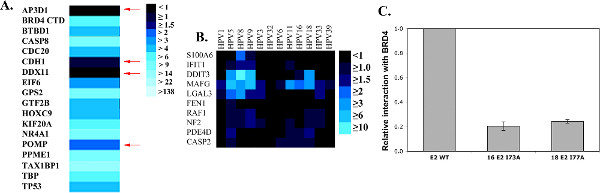
Figure 2. HT-GPCA characteristics. (A) 18 interactions extracted from the literature involving the E2 protein from HPV16 and cellular proteins were tested by HT-GPCA in order to estimate the false-negative rate associated with this technique. The NLR ratio are represented as a color gradient (heat map) with a scale from black (no interaction) to light blue (strong interaction). Four known interactions were not recovered (red arrows) giving a rough estimate of the false-negative rate around 22%. (B) 12 HPV E2 proteins were tested with 10 randomly picked cellular proteins, corresponding to a priori negative interactions, in order to determine the false-positive rate of the HT-GPCA, which scored around 5.8%. (C) The interaction between the BRD4 cellular protein and HPV16 or HPV18 E2 proteins is known to depend on the amino acids I73 and I77 respectively. When these amino acids are mutated, the HT-GPCA NLR drops with a 5-time decrease demonstrating a high specificity of detecting interactions. Figure adapted from reference 13. Click here to view larger figure.
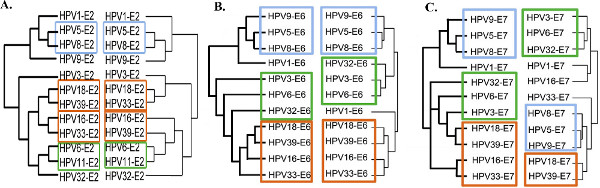
Figure 3. Comparison between experimental interaction-based dendrograms and HPV phylogenetic trees. The phylogeny of HPV based on the early proteins E2 (A), E6 (B) or E7 (C) sequences are represented on the left of each graph. Interaction dendrograms were obtained by hierarchical clustering of the interaction profiles of each protein and are represented on the right. A significant congruence was observed between both tree types, with similar clustering of the pathological types (colored rectangles), proving the pertinence of the interaction datasets. Figure adapted from references 13 and 14. Click here to view larger figure.
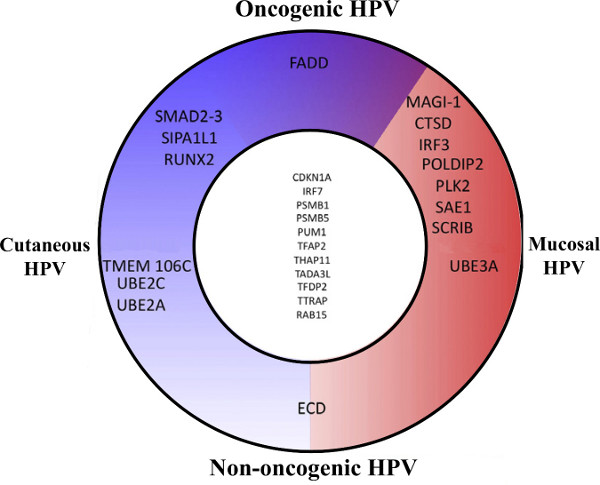
Figure 4. Schematic representation of the E6 main cellular targets. Cellular targets were sorted based on the interaction intensity and grouped according to interacting HPV type. This representation allows the identification of specific biomarkers such as FADD, which interacts only with the E6 proteins of oncogenic HPV. Such targeting must be crucial for the carcinogenic trait of all oncogenic HPV and thus constitutes a good candidate to be used either as a surrogate marker for HPV infection or as a therapeutic target. Figure adapted from reference 14.
Discussion
Independently, yeast-two hybrid and mammalian interaction assays, such as GST pull-down, LUMIER or MAPPIT, have proven to be effective tools to detect protein-protein interactions, but are limited by the high rate of false-positive and false-negative interactions associated with these techniques 15. Moreover, evidence is growing that combining orthogonal methods increases the reliability of the obtained interaction dataset 7. The recent development of the HT-GPCA technique described here has not only improved the sensitivity of detecting binary protein-protein interaction in mammalian cells, but has also offered the possibility to handle a large amount of interactions at once.
The strategy described here improves the coverage of individual Y2H screenings by probing interactions of multiple strain variants. Moreover, retesting interactions with an orthogonal detection method provides stringent comparative interaction datasets, by overcoming limitations and escaping artifacts inherent to the two-hybrid methodology. Indeed, since HT-GPCA takes place in mammalian cells, the post-translational modifications and natural subcellular localization of the proteins are unaffected. Our approach can thus produce in a single step interaction maps that are improved when compared to those primarily based on yeast two-hybrid for interaction sensing 4.
At this point although, we would recommend being careful when studying trans-membrane proteins, since the detection of interactions may depend on the localization of the Gaussia fragments on either side of the membrane. In these cases, it could be necessary to fuse the Gaussia fragments in the C-terminal end of one or both partners. Additionally, the luciferase expression could be influenced by numerous parameters such as transfection efficiency and cell number. Therefore, we would strongly recommend following the normalization procedure described in the protocol section in order to take into account experimental variations.
By using ordered arrays of ORF taken from the increasing collection of the Human ORFeome, we foresee a point in the near future where HT-GPCA could be used directly as a screening method. This should first drastically improve the screening coverage since each protein pairs would then be tested, and second, it should facilitate the automation. However, a more drastic high throughput expansion of this assay should be brought by the development of an in vitro HT-GPCA assay using in vitro expression systems based on human cell extracts. This should also enable to monitor protein-protein interactions in a controlled biochemical environment.
Lastly, upcoming developments are expected to improve the visualization of complex-mediated luminescence in living cells. In that case, the HT-GPCA could be used to detect dynamic interactions in the context of the addition of drugs or complex inhibitors thus allowing live monitoring of the disruption of an interaction.
In all, this approach constitutes an efficient outline for any studies of pathogen-host interplay and we feel that comparative interactome analyses could provide a solid framework to understand microbe hijacking of host cells.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported in part by funding from the Institut Pasteur and by grants from the Ligue nationale contre le Cancer (grants R05/75-129 and RS07/75-75), the Association pour la Recherche sur le Cancer (grants ARC A09/1/5031 and 4867), and the Agence Nationale de la Recherche (ANR07 MIME 009 02 and ANR09 MIEN 026 01). M.M was a recipient of a M.E.N.R.T fellowship.
Materials
| Name of Reagent/Material | Company | Catalog Number | Comments |
| Yeast strains | Clontech | ||
| Minimal SD base | US biological | D9500 | |
| Amino acids | Sigma | ||
| 3-amino-1,2,4-Triazole (3-AT) | Acros organics | 264571000 | |
| Zymolase | Seikagaku | 120491 | |
| DMEM | Gibco-Life Technologies | 31966 | |
| Fetal bovine serum | BioWest | S1810 | |
| Phosphate buffer Saline (PBS) | Gibco-Life Technologies | 14190 | |
| Penicillin-Streptomycin | Gibco-Life Technologies | 15140 | |
| Trypsin-EDTA | Gibco-Life Technologies | 25300 | |
| Renilla luciferase assay | Promega | E2820 | |
| White culture plate | Greiner Bio-One | 655083 | |
| 96-wellPCR plates | 4titude | 4t-i0730/C | |
| Incubator (30 °C) | Memmert | ||
| Incubator (37 °C) | Heraeus | ||
| Luminometer | Berthold | Centro XS-LB 960 |
References
- Davey, N. E., Trave, G., Gibson, T. J. How viruses hijack cell regulation. Trends in Biochemical Sciences. 36, 159-169 (2011).
- Calderwood, M. A. Epstein-Barr virus and virus human protein interaction maps. Proc. Natl. Acad. Sci. U.S.A. 104, 7606-7611 (2007).
- de Chassey, B., et al. Hepatitis C virus infection protein network. Mol. Syst. Biol. 4, 230 (2008).
- Simonis, N., et al. Host-pathogen interactome mapping for HTLV-1 and -2 retroviruses. Retrovirology. 9, 26 (2012).
- Dyer, M. D., Murali, T. M., Sobral, B. W. The landscape of human proteins interacting with viruses and other pathogens. PLoS Pathog. 4, e32 (2008).
- Rual, J. F., et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 437, 1173-1178 (2005).
- Venkatesan, K., et al. An empirical framework for binary interactome mapping. Nat. Methods. 6, 83-90 (2009).
- Cassonnet, P., et al. Benchmarking a luciferase complementation assay for detecting protein complexes. Nat. Methods. 8, 990-992 (2011).
- Ito, T., et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. U.S.A. 98, 4569-4574 (2001).
- Saldanha, A. J. Java Treeview–extensible visualization of microarray data. Bioinformatics. 20, 3246-3248 (2004).
- Smoot, M. E., Ono, K., Ruscheinski, J., Wang, P. L., Ideker, T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 27, 431-432 (2011).
- Huang da, W., Sherman, B. T., Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44-57 (2009).
- Muller, M., et al. Large scale genotype comparison of human papillomavirus E2-host interaction networks provides new insights for e2 molecular functions. PLoS Pathog. 8, e1002761 (2012).
- Neveu, G., et al. Comparative analysis of virus-host interactomes with a mammalian high-throughput protein complementation assay based on Gaussia princeps luciferase. Methods. , (2012).
- Braun, P., et al. An experimentally derived confidence score for binary protein-protein interactions. Nat. Methods. 6, 91-97 (2009).

