Transient Gene Expression in Tobacco using Gibson Assembly and the Gene Gun
Summary
This work describes a novel method for selectively targeting subcellular organelles in plants, assayed using the BioRad Gene Gun.
Abstract
In order to target a single protein to multiple subcellular organelles, plants typically duplicate the relevant genes, and express each gene separately using complex regulatory strategies including differential promoters and/or signal sequences. Metabolic engineers and synthetic biologists interested in targeting enzymes to a particular organelle are faced with a challenge: For a protein that is to be localized to more than one organelle, the engineer must clone the same gene multiple times. This work presents a solution to this strategy: harnessing alternative splicing of mRNA. This technology takes advantage of established chloroplast and peroxisome targeting sequences and combines them into a single mRNA that is alternatively spliced. Some splice variants are sent to the chloroplast, some to the peroxisome, and some to the cytosol. Here the system is designed for multiple-organelle targeting with alternative splicing. In this work, GFP was expected to be expressed in the chloroplast, cytosol, and peroxisome by a series of rationally designed 5’ mRNA tags. These tags have the potential to reduce the amount of cloning required when heterologous genes need to be expressed in multiple subcellular organelles. The constructs were designed in previous work11, and were cloned using Gibson assembly, a ligation independent cloning method that does not require restriction enzymes. The resultant plasmids were introduced into Nicotiana benthamiana epidermal leaf cells with a modified Gene Gun protocol. Finally, transformed leaves were observed with confocal microscopy.
Introduction
This work is a metabolic engineering / synthetic biology project wherein plant cells are engineered to express a reporter protein in multiple organelles but with only a single DNA construct.
One approach to target proteins to more than one location involves cloning multiple genetic copies, each containing a different localization peptide. Each copy must be introduced by successive retransformation, or alternatively, by backcrossing single transforms1. This involves additional cloning, and is limited by one localization tag per terminus.
Another way to localize a protein to multiple locations is through alternative splicing2–5. RNA is transcribed from a single gene, but different copies of the transcript are processed differently, often in more than one way per cell. This can result in more than one messenger RNA in the cell transcribed from a single gene. These different messenger RNAs can encode for different isoforms of the same protein, or in the case of a frameshift, a different protein altogether. Although alternative splicing has been described in the literature for many years, the mechanisms of action and conserved donor and receptor splice sites are only being elucidated more recently6. As these sites are being better described, they open up opportunities for engineering.
Plant metabolic engineers are faced with a challenge when expressing a protein in multiple organelles. For a protein that is to be localized to more than one organelle, the engineer must clone the same gene multiple times, each with a separate signal sequence directing it to the organelle of interest. For a single gene in three organelles, this is simply three genes. But for a six-gene metabolic pathway, this expands to 18 genes, a significant cloning effort. Combining multiple localization sequences into a single, alternatively-spliced gene significantly reduces this effort. For example, re-engineering photorespiration7,8 and isoprenoid synthesis9,10 involves both the chloroplasts and peroxisomes. In our case we took advantage of splice sites as observed in a natural Arabidopsis thaliana system described previously6. We rationally redesigned the mRNA sequence leaving the natural splice sites alone, but placed sequences that would encode chloroplast- or peroxisome- targeting tags within the alternatively-spliced introns (Figure 1). The expressed protein may or may not have a tag, depending on whether the pre-mRNA that encoded it was excised as an intron (Figures 1g and 1h). For more information on the design of the constructs presented in this work, please see the companion article11.
Because this is still a significant cloning effort, Gibson assembly, a new method for cloning DNA constructs, is used in construction. The Gibson method may be used for any sequence, regardless of restriction sites (Figure 2)12–14. The specific mix of enzymes allows for a one-step, isothermal assembly. In this method, several double-stranded linear DNA parts are designed such that they have overlapping sequences of ~50 bp. The Gibson assembly enzyme mix partially digests the linear DNA parts, exposing single strands of homologous sequences. These partial single-stranded sequences are re-annealed in the reaction mixture, resulting in a rapid, one-step, sequence-independent, ligation-free subcloning reaction.
This work describes 1) rational design alternatively spliced constructs for expression in plant chloroplasts, peroxisomes, and cytosols, 2) their cloning using the new ligation-free method of Gibson assembly, 3) their delivery into tobacco leaf cells with the Gene Gun, and 4) results showing organelle targeting, as observed with GFP and confocal microscopy.
Protocol
1. Design of Alternatively-spliced Sequences for Multiple Organelle Targeting
- Determine the sites for final protein expression. For this work, the interest is in targeting the chloroplast, peroxisome, and cytosol.
- Use the literature to identify protein and DNA sequences known to target proteins to organelles of interest. In this case a chloroplast targeting sequence from Arabidopsis thaliana L-isoaspartate methyltransferase6,15, and a peroxisome targeting sequence from Arabidopsis thaliana transthyretin-like S-allantoin synthase16,17 were determined.
- Identify an alternative-splicing regime that should target the proteins of interest to the correct organelles. For this work, splice sites as observed in a natural Arabidopsis system6. The mRNA sequence was redesigned leaving the natural splice sites, but sequences were placed encoding chloroplast- or peroxisome- targeting tags within the alternatively-spliced introns (Figure 1).
2. Gibson Assembly of DNA Parts
See also Figure 2.
The Gibson assembly method depends on the action of several enzymes in a pre-made mix to 1) partially digest the ends of the double stranded DNA’s to make single strands and 2) anneal complimentary base pairs of neighboring DNA parts. This allows for cloning of DNA fragments without the limitations of restriction sites.
- Choose an order for the elements in the DNA plasmid construct. As an example TriTag-1 has elements of: a) a plasmid vector (pORE, containing a GFP construct known to work in plants, a kanamycin resistance marker, and origins of replication for E. coli and Agrobacterium tumefaciens hosts), b) a promoter sequence, (PENTCUP2, shown to be constitutive in plants), c) a chloroplast targeting sequence (CTS), c) a peroxisome targeting sequence (PTS), and d) a series of splice sites as described previously6.
- Design the construct base-for-base with in silico design software. ApE is an open source freeware package useful for this work.
Note: TriTag-1, has the order: plasmid vector, promoter, ATG, splice donor site, chloroplast target sequence, splice donor site, neutral site, splice acceptor site, peroxisome target sequence, splice acceptor site, GFP as contained in plasmid vector.- Design the in silico DNA construct such that there is a 50-bp overlap between each piece when ordering primers for PCR. It is possible to use the 50 bp overlap as the primers: 25 bp each direction.
- Be sure to keep track of the origin of each of the sequence pieces. Is it as: a plasmid to be grown and miniprepped; a linear sequence that can be used as a PCR template; or a sequence that will need to be synthesized commercially? Several companies offer low-cost DNA synthesis services for fragments <500 bp. In this case, the TriTag itself is 437 bp so it was advantageous to order it commercially.
- The best results will come if all the fragments are PCR-amplified and purified from their template DNA. The resistance marker is a good spot to divide this large DNA into two smaller templates that are more easily PCR-amplified.
- Restriction Restriction enzyme DpnI cuts methylated DNA. If the PCR template was a miniprep from E. coli, DpnI treatment will remove the contaminating template DNA.
- Design the in silico DNA construct such that there is a 50-bp overlap between each piece when ordering primers for PCR. It is possible to use the 50 bp overlap as the primers: 25 bp each direction.
- Purify all the DNA sequences separately and elute in water, TE or Buffer EB.
- pORE vector part 1, ~3,000 bp (green arrows). PCR amplified and DpnI treated.
- pORE vector part 2, ~3,000 bp (black arrows). PCR amplified and DpnI treated.
- TriTag insert, 437 bp (blue arrows). Ordered commercially. Resuspend in ddH2O.
- PENTCUP promoter, ~750 bp (orange arrows). PCR-amplified and DpnI treated.
- Measure the concentrations of each of the DNA pieces. Aim for 150 ng/μl of the vector pieces.
- Take an aliquot of the Gibson assembly mix out of the freezer and thaw on ice. This is commercially available, but also simple to make in lab12–14.
- There are two steps to this recipe: a viscous 5x master mix and 15 μl reaction-size 1.33x aliquots. The 5x master mix contains: 3 ml 1 M Tris-HCl pH 7.5, 300 μl 1 M MgCl2, 600 μl 10 mM of each dNTP, 300 μl 1 M DTT, 1.5 g PEG-8000, 20 mg NAD, and ddH2O to 6 ml. Prepare 320 μl aliquots (18) and freeze all but one at -20 °C. The solution will be quite viscous. Label these “5X isotherm buffer”
- To the one remaining (320 μl), add 1.2 μl T5 Exonuclease, 20 μl Phusion High-Fidelity DNA Polymerase, 160 μl Taq DNA Ligase and 700 μl ddH2O. Prepare 15 μl aliquots (~80) on ice in PCR tubes and store at -20 °C.
- Determine volumes for equimolar amounts of each of the fragments. There are a few online calculators, including Gibthon.org. There should be a total of 5 μl of all the DNA pieces. Add them to the 15 μl Gibson mix aliquot. If this requires volumes too small to pipette, dilute the DNA fragments and/or use more than one aliquot.
- Do not forget to prepare a control of the vector DNA alone (e.g. the DNA part containing the antibiotic resistance marker) and make up the volume with water.
- Incubate the tubes in a thermal cycler at 50-55 °C for 30-60 minutes. Replace on ice and transform into E. coli via heat-shock chemically competent cells. Do not use electrocompetent cells. The high salt concentration and the relatively low DNA concentration may cause the cells to arc.
- Apply all 20 μl to a commercial preparation of 200 μl calcium-chloride competent E. coli.
Incubate on ice 30 min and heat shock 60 sec at 42 °C - Return to ice 2 min, apply 750 μl SOC or LB medium and recover shaking in a microcentrifuge tube 30 min at 37 °C. Plate the mixture and appropriate dilutions on LB+Kan (or other appropriate antibiotic). This may result in higher background (and a higher number of non-target recombination events) than traditional ligation-based cloning but it is worthwhile to screen about 10 colonies.
- Apply all 20 μl to a commercial preparation of 200 μl calcium-chloride competent E. coli.
- Confirm clone via sequence and store an aliquot at -80 °C
3. Biolistic Transfection of Tobacco with the Gene Gun
This is a technique that is established in JoVE18,19. Key steps and differences are described below.
- Grow 50-100 ml E. coli or 200 ml Agrobacterium. Maxi-prep the culture. A concentration of about 1,500 ng/μl is required to continue.
- Prepare gene gun bullets as previously described. Load them into the Gene Gun.
- For transfection of Nicotiana benthamiana tobacco leaf epidermal tissue, choose a large leaf close to the base of a 3 month old plant. Carefully cut it and place it bottom side up on wet paper towels in a 15 cm Petri dish.
- Set the He pressure for the Gene Gun to 200-250 psi.
- Carefully aim the Gene Gun between ribs on the bottom side of a leaf, about 3-5 cm above it. Release the safety and deliver the particles to the leaf. Each bullet may be used twice in the same location on the leaf. If the leaf explodes, discard and start a new leaf—there is some variance in leaf toughness due to growth conditions such as age, moisture, etc. For a 12-cm leaf, expect to fit about 6 shots.
- Store the leaf, covered with the top of a Petri dish for 2-3 days in low light on the bench.
- Examine the leaf in a dissecting microscope equipped with UV fluorescence, and search for individual cells expressing GFP. Dissect 5-10 mm sections of leaf.
- Submerge the leaf in a deep well microscope slide under ~200 μl 0.1% Triton X-100. The detergent helps prevent air bubbles from forming on the surface, and the deep well microscope slide allows for a larger tissue imaging area.
4. Confocal Microscopy of Transfected Tissue
These instructions vary for every instrument, so it is essential to get properly trained.
- For fluorescence detection experiments, set the excitation laser to 489 nm, and set the photomultiplier detectors to 500-569 nm for GFP fluorescence and 630-700 nm for chlorophyll auto-fluorescence. Image cells using a 40x water-immersion objective.
Representative Results
The design effort was a result of significant planning. Novel to this project is the use of alternative splicing to create a pre-mRNA that is translated into differentially expressed proteins. These proteins are expressed in different organelles, in this case the chloroplast, peroxisome and/or cytosol. We adapted natural Arabidopsis gene that is alternatively spliced6, and placed known chloroplast6 and peroxisome17 targeting sequences in alternate exons (TriTag-1 and TriTag-2). TriTag-3 consists of a peroxisome signal embedded within a low-complexity region of the chloroplast sequence. These were placed in frame with GFP (Figure 1).
The plasmids used in this study thus contained several elements: the native alternative splicing regimen from the PIMT2 gene6, chloroplast and peroxisome targeting sequences, GFP, and a plasmid backbone. Since each has very specific required sequence, a sequence-independent method is necessary. The Gibson assembly protocol12,13 can be used on any sequences so long as the constituent parts are designed with sequence similarity to each other, and do not contain repeating sequences. In brief, linear DNA fragments are constructed (either via PCR amplification, commercial synthesis, or digestion of a pre-made plasmid) such that they have ~50 bp of overlapping homologous sequence (Figure 2). Equimolar quantities of each of the DNA fragments are combined in a single tube containing the Gibson assembly mix, incubated at 50 °C for one hour, and transformed into E. coli. The plasmids resulting from the Gibson assembly protocol were confirmed by Sanger sequencing.
In this study the three different GFP organelle-targeting constructs were compared to untagged GFP and a Rubisco-tagged GFP. Confocal microscopy was used to detect GFP fluorescence in the single cells transiently transformed. In control experiments, the transient expression observed was correlated to the chloroplasts (easily identifiable by the red autofluorescence of chlorophyll), the nuclei (large organelle that is not a chloroplast and has only one per cell), and/or small organelles believed to be peroxisomes. Green (GFP) and red (chlorophyll autofluorescence) channels were overlaid to study intracellular localization (Figure 3). For a more complete discussion of these results, please see our companion article11.
As predicted, in all three dual targeting constructs, GFP was observed in the chloroplasts (Figure 4) in overlay confocal micrographs. TriTag-1 and TriTag-2 target the GFP also to the cytosol and the small organelle believed to be the peroxisome (Figures 4a and 4b, respectively). TriTag-3 targets the GFP to only the chloroplast and the peroxisome, but not the cytosol (Figure 4c). A summary of the results is diagrammed (Figure 5).
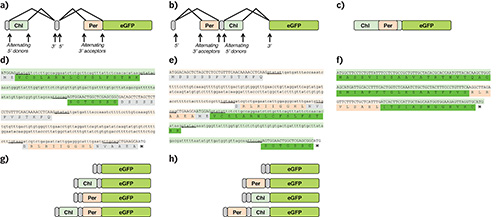
Figure 1. Design of alternative splicing constructs for differential organelle expression in Nicotiana benthamiana. Splice diagrams of (a) TriTag-1, (b) TriTag-2, and (c) TriTag-3 showing non-targeting sequences (gray), chloroplast targeting sequences (light green), peroxisome targeting sequences (orange), and the GFP coding sequence used in transient expression experiments. TriTag-1 and TriTag-2 consist of alternative splicing constructs; TriTag-3 consists of a peroxisome signal embedded within a low-complexity region of the chloroplast sequence. Sequences of (d) TriTag-1, (e) TriTag-2, and (f) TriTag-3. The ATG codon at the end of these sequences corresponds to the GFP start codon. Alternatively spliced targeting regions are underlined. Donor and acceptor dimers are underlined. The light green DNA sequences derive from the PIMT2 5’ coding region6 and include sequences encoding the chloroplast targeting sequence (green). The light orange DNA sequences derive from the TTL 5’ coding region17 and include sequences encoding the peroxisome targeting sequence (orange). TriTag-3 consists of a peroxisome signal embedded within a low-complexity region of the chloroplast sequence. Schematics of final mRNAs believed to be expressed for (g) TriTag-1 and (h) TriTag-2. Reprinted with permission11. Click here to view larger image.
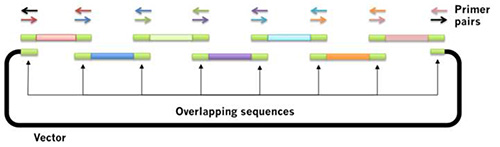
Figure 2. Schematic for Gibson assembly: a novel method for assembly of overlapping DNA fragments. The Gibson assembly method for plasmid construction12,13 is rapidly replacing traditional restriction-ligation cloning in many laboratories. Essentially, one can design a construct in silico that contains the DNA of interest exactly, composed of PCR products amplified from various sources. Design primers that are complimentary to each other, and PCR-amplify each fragment with the colored primers. Add all the pieces into a single tube with the enzyme mix, and transform E. coli competent cells. Click here to view larger image.
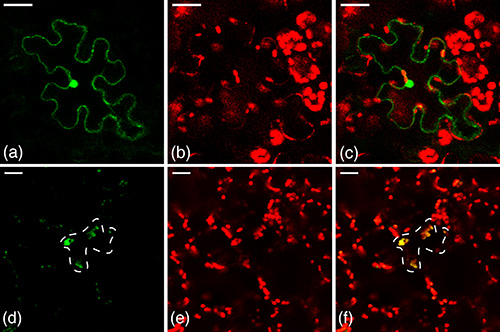
Figure 3. Control treatments bombarded into N. benthamiana leaf epidermal cells. (a-c) Untagged GFP. (a) Green channel. Untagged GFP is expressed in the nucleus and around the cell periphery, corresponding to the cytosol, but not the vacuole. (b) Red chloroplasts indicate autofluorescence of chlorophyll. (c) Overlay. In this construct fluorescence is observed in the nucleus and cytosol only, but not the chloroplasts. (d-f) Chloroplast-tagged GFP control. (d) Green channel. GFP is expressed in the large organelles, with a cell as outlined. (e) Red chloroplasts indicate autofluorescence of chlorophyll. (f) Overlay. In this case yellow chloroplasts are observed, indicating that the GFP (green) is targeted to the chloroplast. Reprinted with permission11. Click here to view larger image.
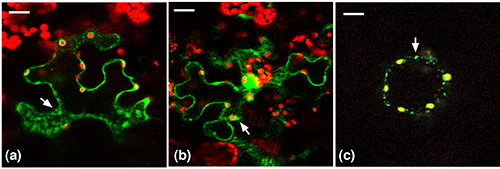
Figure 4. Tri-Tag-1, 2, 3 tagged GFP merged images. (a) Tri-Tag1. In this case yellow chloroplast peripheries are observed, indicating that the GFP is targeted to the chloroplast. GFP is also observed in the cytosol as well as small punctate organelles that may be peroxisomes. (b) Tri-Tag2. In this case yellow chloroplast peripheries are observed, indicating that the GFP is targeted to the chloroplast. GFP is also observed in the cytosol as well as small punctate organelles that may be peroxisomes. (c) Tri-Tag3. In this case yellow chloroplasts are observed, as well as small punctate organelles that may be peroxisomes. But GFP is not observed in the cytosol. Reprinted with permission11. Click here to view larger image.
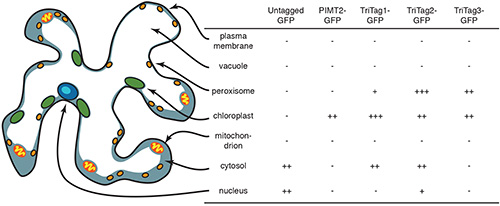
Figure 5. Compartments of a typical tobacco leaf epidermal cell. Here their relative sizes and locations within the cell are shown, and the relative expression levels observed via confocal microscopy. Reprinted with permission11. Click here to view larger image.
Discussion
In this study, simple strategies are described for localizing a single transgenic protein to multiple cellular compartments in plants. The goal was to design construct that would express a single gene in more than one organelle in Nicotiana benthamiana. Strategies include rational design of GFP-based DNA constructs, Gibson assembly, delivery of the plasmids to leaf cells with the Gene Gun, and observation of the results with confocal microscopy.
Three different short, N-terminal tags were designed for simultaneous chloroplast, peroxisome and cytosol targeting11. These “TriTags” were designed by combining naturally occurring DNAs encoding alternatively spliced mRNAs that direct the encoded proteins to either the chloroplast plus cytoplasm6 or the peroxisome plus cytoplasm17. TriTag-3 does not rely on alternative splicing and consists of a chloroplast targeting sequence in which a naturally unstructured portion has been replaced with a peroxisomal targeting sequence15,17. The TriTags function in vivo to target GFP in Nicotiana benthamiana leaf epidermal cells (Figure 5).
Confocal micrographs showed that TriTag-1 and TriTag-2 target GFP to the chloroplast, peroxisome, and cytosol effectively, but with preference for the chloroplast and peroxisome, respectively (Figures 4a and 4b). TriTag-3 targets GFP only to the chloroplast and the peroxisome, but not the cytosol (Figure 4c). In all, these tags may reduce time and resources spent on cloning for plant metabolic engineering, specifically for those needing specific expression in the chloroplasts and peroxisomes.
Modifications and Troubleshooting
The TriTags as currently constructed are fairly static. However, the underlying technology, alternative splicing to allow translation of different location-specific tags, is flexible. The existing chloroplast and/or peroxisome specific tags could be changed for any other organelle, be it the secretion pathway, the mitochondrion, an ER residence tag, the vacuole. These could also be combined with tissue-specific promoters, e.g. for the seeds, the flowers, the roots, the leaves, stems, etc.
For a project such as this one, the Gibson assembly method12–14 for cloning constructs is ideal. It is a simple, powerful tool to subclone any sequence without the limitations of restriction sites. In one-step reactions, Gibson assembly rapidly created the constructs and controls with minimal effort in cloning. The Gibson assembly technique is nearly infinitely modifiable for any desired sequence so long as the template sequences can be PCR-amplified. Although in this example Gibson assembly was shown with a digested plasmid, PCR-amplifying the vector tends to work better than using a digested miniprep.
The Gene Gun is a rapid, effective method for transient transfection of cells, as has been established by this journal18,19. Nicotiana benthamiana has large, lobate leaf epidermal cells, which are both ideal for transfection with the Gene Gun, but also for observing with confocal microscopy. The protocol for making bullets is well established. Gold has been used as a microcarrier in most experiments in the literature thus far, but tungsten microcarriers are recently available at low cost.
Limitations of the Techniques
One major limitation of the Gibson method is the presence of repeated sequences: the longer the overlapping region is the fewer side products result. Digestion of the PCR products with DpnI should remove contaminating plasmid templates; which are usually the source of side products. The enzymes digest the DNA fragments from double-stranded DNA into single-stranded. The presence of repeated sequences near the ends (~1 kb) of the DNA fragments significantly increases the possibility of improper recombination. As well, repeats within the sequences also pose a limitation: it is not well known, and difficult to control how far the T5 exonuclease chews back single-stranded DNA. The rapid activity of the exonuclease is tempered by high reaction temperature (50-55 °C) eventually denaturing the enzyme, and the viscosity decreasing the overall motion in the solution. For more fine-tuning of the reaction steps, sequence and ligation independent cloning (SLIC)20 is preferred.
A major limitation of the gene gun technique is that it only produces on the order of one to ten transiently transformed cells per shooting event.
Significance with Respect to Existing Methods
The Gibson assembly technique is enjoying rapid adoption for its ease, flexibility, and adaptability. It is a faster protocol than traditional restriction/ligation cloning, and does not require specific sequences such as those required for restriction digestion. It is not limited to two or three fragments to be recombined; the original researchers have demonstrated it successfully for up to 10 parts, albeit with limited efficiency. For many researchers Gibson assembly makes for a larger number of positive clones.
For more transformants with Biolistic techniques, or for the creation of transformed callus tissue, the use of a cabinet-style biolistic device (e.g. the BioRad PDS-1000) should be used. An alternative method is Agrobacterium infiltration.
Future Applications
Gibson assembly allows larger and larger clusters of DNA to be assembled, to the point where a maximum limit has not been observed. The original authors have used it to assemble several hundred kilobases DNA13.
The ability to selectively target multiple organelles with a single genetic construct has the potential to decrease the number of genes needing to be transformed. The delivery of larger and larger DNA fragments (and thus larger metabolic pathways) is a near-term goal. Isoprenoid pathways have been delivered to the chloroplast by biolistic methods; other gene clusters such as alternative carbon fixation pathways are on the horizon.
Critical Steps Within the Protocol.
In Gibson assembly, be sure the template sequences do not have homology to each other except at the target recombination sites.
In biolistic transformation of the Nicotiana plants, be careful to keep the gene gun about 10-15 cm above the leaves and keep the helium pressure at 200-250 psi. Closer than that, the fragile leaf will explode, and farther than that they will not be well transformed. Two days after transfection a bruise should appear on the leaf if it is properly transformed.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank Jen Sheen of Massachusetts General Hospital for the generous donation of Nicotiana benthamiana seedlings. Jen Bush helped us greatly in advice in growing plants and setting up a growth chamber area. Tom Ferrante of the Wyss Institute offered crucial help with confocal microscopy. The authors would especially like to thank Don Ingber of Children’s Hospital Boston and the Wyss Institute for the generous donation of a Gene Gun and associated supplies. Funding for this project was provided through a cooperative agreement with the Department of Energy Advanced Research Projects Agency (ARPA- E Award # DE-000079) for PAS, JCW, and MdM, and through Chimerion Biotechnology, Inc. for MJV.
Materials
| Oligonucleotide primers | IDT | (custom) | Design specifically for construct |
| GeneBlocks | IDT | (custom) | 500 bp oligonucleotides |
| ApE software | U Utah | (download) | http://biologylabs.utah.edu/jorgensen/wayned/ape/ |
| MinElute kit | Qiagen | 28004 | Used to purify PCR products |
| QIAprep spin miniprep kit | Qiagen | 27104 | Used to prepare cloning-appropriate amounts of plasmid |
| Phusion Master Mix with GC Buffer | NEB | M0532S | Used to PCR-amplify gene of interest |
| Gibson assembly reaction mix | NEB | E2611L | Master mix of the following 9 ingredients |
| 1 M Tris-HCl pH7.5 | Teknova | T1075 | Gibson assembly mix |
| 1 M MgCl2 | G Biosiences | 82023-086 | Gibson assembly mix |
| dNTP mix | Fermentas | R0192 | Gibson assembly mix |
| 1 M DTT | Fermentas | R0861 | Gibson assembly mix |
| PEG-8000 | Affymetrix | 19966 | Gibson assembly mix |
| NAD | Applichem | A1124,0005 | Gibson assembly mix |
| T5 exonuclease | Epicentre | T5E4111K | Gibson assembly mix |
| Phusion polymerase | NEB | F530S | Gibson assembly mix |
| Taq DNA ligase | NEB | M0208L | Gibson assembly mix |
| Gibthon.org | Website to simplify calculations | ||
| Plasmid PLUS Maxi kit | Qiagen | 12963 | Used to prepare DNA for gene gun bullets |
| Gene Gun system | BioRad | 165-2451 | Includes all parts necessary |
| Nicotania benthamiana | (n/a) | (n/a) | Gift of Jen Sheen, MGH |
| Confocal microscope | Leica | SP5 X MP | Imaging of resultant cells |
| Deep well slides | Electron Microscopy Sciences | 71561-01 | Used for confocal imaging |
| A Plasmid Editor (ApE) | University of Utah | http://biologylabs.utah.edu/jorgensen/wayned/ape | |
| Gibthon:Ligation calculator | http://django.gibthon.org/tools/ligcalc/ |
References
- Que, Q., et al. Trait stacking in transgenic crops: challenges and opportunities. GM crops. 1 (4), 220-229 (2010).
- Reddy, A. S. N., Rogers, M. F., Richardson, D. N., Hamilton, M., Ben-Hur, A. Deciphering the plant splicing code: experimental and computational approaches for predicting alternative splicing and splicing regulatory elements. Frontiers in Plant Science. 3 (18), (2012).
- Hickey, S. F., et al. Transgene regulation in plants by alternative splicing of a suicide exon. Nucleic Acids Research. 40 (10), 4701-4710 (2012).
- Syed, N. H., Kalyna, M., Marquez, Y., Barta, A., Brown, J. W. S. Alternative splicing in plants – coming of age. Trends in Plant Science. 17 (10), 616-623 (2012).
- Black, D. L. Mechanisms of alternative pre-messenger RNA splicing. Annual Review of Biochemistry. 72, 291-336 (2003).
- Dinkins, R. D., et al. Changing transcriptional initiation sites and alternative 5′- and 3′-splice site selection of the first intron deploys Arabidopsis protein isoaspartyl methyltransferase2 variants to different subcellular compartments. The Plant Journal: for Cell and Molecular Biology. 55 (1), 1-13 (2008).
- Kebeish, R., et al. Chloroplastic photorespiratory bypass increases photosynthesis and biomass production in Arabidopsis thaliana. Nature Biotechnology. 25 (5), 593-599 (2007).
- Maier, A., et al. Transgenic Introduction of a Glycolate Oxidative Cycle into A. thaliana Chloroplasts Leads to Growth Improvement. Frontiers in Plant Science. 3 (38), 1-12 (2012).
- Kumar, S., et al. Remodeling the isoprenoid pathway in tobacco by expressing the cytoplasmic mevalonate pathway in chloroplasts. Metabolic Engineering. 14 (1), (2012).
- Sapir-Mir, M., et al. Peroxisomal localization of Arabidopsis isopentenyl diphosphate isomerases suggests that part of the plant isoprenoid mevalonic acid pathway is compartmentalized to peroxisomes. Plant Physiology. 148 (3), 1219-1228 (2008).
- Voges, M. J., Silver, P. A., Way, J. C., Mattozzi, M. D. Targeting a heterologous protein to multiple plant organelles via rationally designed 5′ mRNA tags. Journal of Biological Engineering. 7 (20), (2013).
- Gibson, D. G. Enzymatic assembly of overlapping DNA fragments. Methods in Enzymology. 498, 349-361 (2011).
- Gibson, D. G., et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods. 6 (5), 12-16 (2009).
- Gibson, D. G., Smith, H. O., Hutchison, C. A., Venter, J. C., Merryman, C. Chemical synthesis of the mouse mitochondrial genome. Nature Methods. 7 (11), 901-903 (2010).
- Lowenson, J. D., Clarke, S. Recognition of D-aspartyl residues in polypeptides by the erythrocyte L-isoaspartyl/D-aspartyl protein methyltransferase. Implications for the repair hypothesis. The Journal of Biological Chemistry. 267 (9), 5985-5995 (1992).
- Lanyon-Hogg, T., Warriner, S. L., Baker, A. Getting a camel through the eye of a needle: the import of folded proteins by peroxisomes. Biology of the Cell / under the auspices of the European Cell Biology Organization. 102 (4), 245-263 (2010).
- Reumann, S., et al. Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. The Plant Cell. 19 (10), 3170-3193 (2007).
- Woods, G., Zito, K. Preparation of gene gun bullets and biolistic transfection of neurons in slice culture. J. Vis. Exp. (12), (2008).
- Hollender, C. A., Liu, Z. Bimolecular Fluorescence Complementation (BiFC) Assay for Protein-Protein Interaction in Onion Cells Using the Helios Gene. (40), (2010).
- Li, M. Z., Elledge, S. J. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nature Methods. 4 (3), 251-256 (2007).

