Expression of Recombinant Cellulase Cel5A from Trichoderma reesei in Tobacco Plants
Summary
Tobacco plants were used to produce a fungal cellulase, TrCel5A, via a transient expression system. The expression could be monitored using a fluorescent fusion protein, and the protein activity was characterized post-expression.
Abstract
Cellulose degrading enzymes, cellulases, are targets of both research and industrial interests. The preponderance of these enzymes in difficult-to-culture organisms, such as hyphae-building fungi and anaerobic bacteria, has hastened the use of recombinant technologies in this field. Plant expression methods are a desirable system for large-scale production of enzymes and other industrially useful proteins. Herein, methods for the transient expression of a fungal endoglucanase, Trichoderma reesei Cel5A, in Nicotiana tabacum are demonstrated. Successful protein expression is shown, monitored by fluorescence using an mCherry-enzyme fusion protein. Additionally, a set of basic tests are used to examine the activity of transiently expressed T. reesei Cel5A, including SDS-PAGE, Western blotting, zymography, as well as fluorescence and dye-based substrate degradation assays. The system described here can be used to produce an active cellulase in a short time period, so as to assess the potential for further production in plants through constitutive or inducible expression systems.
Introduction
Degradation of lignocellulosic biomass and its conversion into liquid fuel has been envisioned as a method to reduce reliance on fossil fuels. One significant hurdle in establishing economically feasible biomass processing systems is in the enzymatic degradation of cellulose and hemicellulose1. Plant expression systems show great potential for the production of enzymes on an industrial scale. Agricultural systems to harvest plants economically and in volume are already established, as they have been for thousands of years. The production of cellulases in plants is desirable due to factors like the ease of autohydrolysis2, and the potential for maximizing the use of lower enzyme levels by increasing the digestibility of plant cell walls1. Finally, plant systems allow the targeting of recombinant proteins to specific areas of plant cells and are able to posttranslationally modify enzymes where required3.
Nicotiana tabacum (tobacco) is very commonly used as a model organism for heterologous protein expression studies in plants, due to its rapid growth and biomass accumulation features4. Transient expression of recombinant proteins is a technique which enables protein production in short time periods5, while maintaining flexibility as to the localization and thus posttranslational modifications of the selected proteins, i.e. cellulases. This enables the production of the cellulase(s) for basic analysis, while also laying the groundwork for further expression strategies in plants. Using such a transient expression strategy, an endoglucanase from glycosyl hydrolase family 5, Cel5A, derived from the fungal host Trichoderma reesei (Hypocrea jecorina)6 is produced (hereafter referred to as TrCel5A). TrCel5A is a 42 kDa protein which is natively glycosylated and is highly active in hydroylzing cellulose chains7.
The transient expression technique described here is based on a relatively commonly used system, infiltrating the plant leaves with Agrobacteria carrying the gene of interest in an appropriate expression vector. To allow for rapid analysis of successful in planta expression, TrCel5A can also be expressed as a fusion protein with mCherry, a monomeric fluorescent protein originally derived from Discosoma sp. protein DsRed8, with an additional six Histidine residues (His-tag) fused to the C-terminus (TrCel5A-mCherry). Expression of the heterologous fusion protein, TrCel5A-mCherry, can thus be monitored within the growing plant by using green light to examine mCherry dispersal. If desired, thin sections of the plant material can be examined under green light microscopically to establish the specific localization of the protein. For this work, signal and transit peptides were incorporated in the construct, to localize the heterologous protein export to the endoplasmic reticulum9.
To analyze the activity of plant expressed cellulases, including TrCel5A, a number of cellulase activity tests can be run. After the extraction of the total soluble plant proteins, TrCel5A can be partially purified using a thermal incubation technique. Protein size is established using SDS-PAGE followed by Western blotting. Zymography can be used to analyze activity against substrates, e.g. cellulose which has been carboxy-methylated (making carboxymethyl cellulose: CMC) for solubility10. Cellulase and glucosidase activity can be monitored using the fluorophore 4-methylumbelliferyl (4-MU) associated with β-D-cellobioside (combined: 4-MUC). Another method to assess endoglucanase activity involves the spectrometric analysis of CMC which has been associated with an azo-dye (Remazolbrilliant Blue R)11. In addition, protein activity of both endoglucanases and a range of cellulases can be monitored by the use of a sugar analysis test, such as the p-hydroxy benzoic acid hydrazide (PAHBAH) assay12. These techniques can be used to elucidate and quantify the activity of the expressed cellulase of interest.
Protocol
1. Growth of Wild Type N. tabacum Plants
- To grow N. tabacum L. cv. Petit Havana SR1 plants on soil, place tobacco seeds on appropriate pots filled with normal potting soil for germination. After 2 weeks, transfer seedlings to individual pots. Incubate plants in a greenhouse with constant 22 °C (dark period) or 25 °C (light period) and 70% relative air humidity. Provide illumination in a 16/8 hr light/dark cycle (e.g. Philips IP65 400 W lamps; 180 μmol·sec-1·m-2; λ= 400-700 nm) and water daily.
2. Transient Expression of TrCel5A and TrCel5A-mCherry Proteins
- Under sterile conditions, pick single colonies of Agarobacterium tumefaciens (GV3101::pMP90RK GmR, KmR, RifR) containing either pTRAkc-ER-TrCel5A or pTRAkc-ER -TrCel5A-mCherry and transfer to an Erlenmeyer flask containing 10 ml yeast extract broth (YEB, prepared as per manufacturers specifications) medium and kanamycin (50 mg/ml), rifampicin (50 mg/ml), and carbenicillin (100 mg/ml) for selection. Incubate for 36-40 hr at 26 °C and 180 rpm. Note: The design of appropriate constructs for protein expression is outside the scope of this paper, but for guidance, the methods of Maclean et al.13 and Munro and Pellham14 can be followed for construct design.
- Take 200 µl of each culture to inoculate 20 ml of YEB with the antibiotic concentrations mentioned above and incubate under the same conditions as before.
- After 36-40 hr, preinduce the cultures by adding 2 µl acetosyringon (200 µM final concentration), 100 µl 40% (w/v) glucose solution and 200 µl of 1 M MES buffer pH 5.6. Incubate overnight.
- Prepare 100 ml of a 2x infiltration buffer (100 g/L sucrose, 3.6 g/L glucose, 8.6 g/L Murashige and Skoog basal salts, adjust pH to 5.6 using potassium hydroxide).
- Measure OD600 of the cultures with 1:5 dilutions until it reaches an OD600 of 5-6. Calculate culture volume needed for 30 ml infiltration medium at OD600. Add 15 ml of 2x infiltration buffer, then add deionized H2O to make the buffer up to 30 ml.
- Take plants from greenhouse and use a pipette tip to nick the abaxial side of the 3rd to 5th leaf from the top to ease infiltration.
- Use a pipette tip to nick the abaxial side of the leaves to ease infiltration.
- Fill a syringe with infiltration medium and put it (without needle) on one of the previously made nicks. Support syringe with a finger on the adaxial leaf side. Press the medium carefully into the tissue of interveinal leaf zones. Note: After infiltration, cultivate plants in a greenhouse under the same conditions as described above. Also, maintain an equal proportion of untreated, non-transient expression plants (NTEPs) as controls, under the same conditions as infiltrated plants.
3. Assessment of TrCel5A-mCherry Expression in Plant Leaves
- After 4-6 days, take plants infiltrated with A. tumefaciens containing pTRAkc-ER-TrCel5A-mCherry from the greenhouse and place them in a dark room for fluorescence analysis.
- To visualize the fluorescence in leaf sections expressing TrCel5A-mCherry, light plants using a portable light source emitting green light at 515 nm with a red filter (620-750 nm). Note: In depth characterization of protein expression in the plant leaves can be achieved by thin-sectioning leaves with a vibratome and examination under light and fluorescence microscopy. For this follow the steps below:
- Cut small sections of approximately 5 x 5 mm size from an infiltrated leaf and embed them in 4% agarose dissolved in 50 mM Phosphate buffer (pH 5.7).
- Cut section of 80-90 µm size from embedded leaves using a vibratome. Place them on a microscope slide, cover with buffer or a cover slip and examine them using a microscope at 10X magnification with a fluorescence detector, using the same wavelength and filter stated above.
4. TrCel5A Extraction from Tobacco Leaves
- Cut pieces from tobacco leaves to an approximate weight of 1 g from plants infected with A. tumefaciens containing pTRAkc-ER-TrCel5A and place in a mortar precooled with liquid nitrogen. Add an appropriate amount of liquid nitrogen to the samples. Grind leaves to powder using a precooled pestle.
- Add 2 ml of phosphate buffered saline (PBS) containing 1 mM phenylmethylsulfonyl fluoride to the ground leaf matter and mix until the majority of leaf matter is in suspension. Centrifuge extract at 15,000 x g for 20 min at 4 °C and take protein-containing supernatant.
- Centrifuge the extract at 15,000 x g for 20 min at 4 °C and take protein-containing supernatant being careful not to disrupt the pellet. Discard the pellet.
- Partially purify TrCel5A by incubating the protein-containing supernatant at 55 °C for 10 min. Allow to rest at room temperature for a further 10 min, then centrifuge at 15,000 x g for 10 min at RT. Repeat partial purification process for non-infiltrated plants to obtain control samples. Use these samples for all following experiments. Note: Protein solutions can be stored at 4 °C for up to 1 week or at -20 °C for up to 3 months.
- Repeat the partial purification process for non-infiltrated plants to obtain control samples. Use these samples for all following experiments. Note: Protein solutions can be stored at 4 °C for up to 1 week or at -20 °C for up to 3 months.
5. SDS-PAGE, Western Blotting, and Azo-CMC Zymography of TrCel5A-mCherry
- Purchase or prepare 12% (w/v) SDS-PAGE gels as per established methods15.
- Dissolve CMC in water to obtain a 1.5% (w/v) solution. To make CMC-SDS-PAGE gels, substitute 1 ml of 1.5% CMC for 1 ml of water in a 10 ml SDS-PAGE gel mix solution, resulting in a 0.1% (w/v) CMC + 12% (w/v) SDS-PAGE gel mix. Pour gel normally. Note: Make certain that the CMC in the original solution is completely dissolved by a combination of stirring and heating to 40 °C (repeat directly prior to preparing the gel solution).
- Denature samples by boiling in the presence of SDS, as per established methods12. As a positive control, use a 1:500 dilution of a commercially available cellulase mixture from T. reesei, such as T. reesei ATCC 26921. As a negative control use a similarly prepared protein solution from non-infiltrated leaves (NTEPs).
- Carry out the electrophoresis as per normal protocols15, e.g. use 200 V for ~50 min with the electrophoresis halted once the dye front reached the bottom of the gel.
- Stain one SDS-PAGE gel using 0.1% (w/v) Coomassie brilliant blue and destain using a methanol/glacial acetic acid mixture. Note: Rapid staining and destaining can be performed by gently heating the staining and destaining solutions, i.e. by using a microwave for ~15 sec to warm (but not boil) first the staining and then the destaining solutions, changing the destaining solution after 10 min, or when cooled, and repeat with new destaining solution.
- Perform Western blotting of one SDS-PAGE gel using established protocols16,17. Use the following antibodies: a polyclonal rabbit antibody against a His-tag (primary) and a polyclonal goat anti-rabbit antibody Fc region which has been conjugated to an alkaline phosphatase (secondary). Note: secondary antibodies should be selected to enable visualization using nitroblue tetrazolium chloride/5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt (NBT/BCIP), as described18.
- Perform in-gel protein refolding on the 0.15% (w/v) CMC SDS-PAGE gel by incubating the gel in 1.5% (w/v) β-cyclodextrin, 20 mM phosphate-citrate buffer, pH 6.5, at room temperature with gentle shaking for 15 min19.
- Discard the β-cyclodextrin solution and briefly wash the gel with H2O. Incubate at RT in 50mM sodium acetate buffer (pH 4.8) for a further 15 min. Perform CMC zymography using the refolded 0.15% CMC SDS-PAGE gel by incubating the gel in 30 mM sodium acetate buffer pH 4.8 for 20 min at 50 °C.
- Perform CMC zymography using the refolded 0.15% (w/v) CMC SDS-PAGE gel by incubating the gel in 30 mM sodium acetate buffer (pH 4.8) for 20 min at 50 °C.
- Stain the gel for 10 min using a 0.1% w/v Congo red solution and destain for 30 min or until clear bands are observed using 1 M NaCl. Wash the gel with 0.3% (v/v) acetic acid for clearer imaging.
6. 4-MUC Activity Assay of TrCel5A
- Prepare a stock solution of 10 mM 4-MUC in 50% (v/v) methanol, and store it in the dark at RT
- Immediately prior to the assay, prepare a 4-MUC 2x working solution, composed of 1 mM 4-MUC and 60 mM sodium acetate, pH 4.8.
- Perform this test on samples containing 100 μl of leaf protein (with or without TrCel5A inducibly expressed), 100 μl a commercially available cellulase mixture from T. reesei 1:1,000, and pure H2O, respectively. Run each sample in triplicate.
- Pipette 100 μl of the 4-MUC working solution into separate wells of a 96-well plate, then add 100 μl of each sample.
- Determine 0 min timepoint of 4-MU fluorescence through fluorescent spectroscopy, using an excitation wavelength of 360 nm and an emission wavelength of 465 nm.
- Incubate plates for 20 min at 50 °C, covered with adhesive lids to prevent evaporation. Stop the reaction by freezing (or by combining 100 μl 0.15 M glycine, pH 10.0, and 100 μl of sample in a separate plate).
- Determine the endpoint of 4-MU fluorescence through fluorescent spectroscopy, using an excitation wavelength of 360 nm and an emission wavelength of 465 nm. Subtract 0 min from endpoint (20 min) timepoint to obtain the change in fluorescence values.
7. Azo-carboxymethyl Cellulose Activity Assay of TrCel5A
- Prepare a stock of 1.5% (w/v) Azo-CMC which is stored at RT, and a precipitation solution containing 18 mM zinc acetate, 0.5 M sodium acetate, pH 5 and 80% ethanol (v/v), store at room temperature.
- Immediately prior to the assay, prepare an Azo-CMC 2x working solution, composed of 1% (w/v) Azo-CMC (in H2O) and 60 mM sodium acetate, pH 4.8.
- Combine 50 μl of sample and 50 μl of the working solution in 1.5 ml tubes.
- Test samples containing 50 μl of leaf protein (with or without TrCel5A transiently expressed); 100 μl of a commercially available cellulase mixture from T. reesei diluted 1:1,000; and pure H2O. Run samples and standards in triplicate.
- Incubate samples for 20 min at 50 °C. Stop the reaction by adding 250 μl of precipitation solution. Vortex each sample vigorously for 10 sec to ensure solutions were completely combined.
- Incubate samples at RT for 10 min and then vortex again. Centrifuge samples at 1,000 x g for 10 min. Remove 200 μl of each sample supernatant to a 96-well plate, taking care not to disrupt the pellet.
- Assay using absorbance spectroscopy at a wavelength of 590 nm.
8. PAHBAH Assay of TrCel5A
- Use a hole-punch to cut a circle (~5.5 mm diameter) of filter paper. Add 100 μl 60 mM NaAc, pH 4.8, and 100 μl of samples, either containing leaf protein (with or without TrCel5A inducibly expressed) or a commercial T. reesei cellulase mixture, diluted 1:1,000, and incubate with filter paper circles for 20 hr at 50 °C with 300 rpm shaking. For each sample incubated with filter paper, also incubate a duplicate solution without filter paper as individual sample-controls.
- Prepare a 330 mM p-hydroxy benzoic acid hydrazide (PAHBAH) solution A by dissolving PAHBAH in H2O with the addition of 5 ml concentrated HCl for a total of 100 ml solution, and store at RT. Note: Best results are achieved by adding the HCl to a smaller volume of PAHBAH slurry, i.e. 30 ml, which is stirred under temperature to help dissolution before making the volume to 100 ml with H2O.
- Prepare PAHBAH solution B, containing 50 mM sodium citrate, 10 mM calcium chloride, and 500 mM NaOH and store at RT.
- Immediately prior to the assay, prepare a 1.5x working solution of 1 ml PAHBAH reagent to 9 ml buffer solution, store on ice until use (to be used within 4 hr).
- Prepare samples in thermostable 0.2 or 0.5 ml tubes. Combine 5 μl of each solution incubated with filter paper (step 8.1) 45 μl of H2O and 100 μl of PAHBAH working solution. Sample controls (samples incubated without filter paper) should be run under the same concentrations (5 μl sample, 45 μl H2O, 100 μl PAHBAH working solution). Also test 5 standards (5 μl) containing between 0 and 0.2 g/L of glucose, and pure H2O. Run samples and standards in triplicate.
- Incubate samples for exactly 10 min at 100 °C, then cool at RT for exactly 10 min. Pipette solutions into a 96 well plate and assay spectroscopically at a wavelength of 410 nm. Subtract individual sample-controls (incubated without filter paper) from samples to obtain final values.
Representative Results
TrCel5A and TrCel5A-mCherry were expressed successfully in tobacco plants using the transient expression system (Figures 1A and 1B). Examination of the expression pattern of the TrCel5A-mCherry fusion protein revealed healthy leaves (Figure 1C), which showed widespread protein expression under green light (Figure 1D).
Extraction of the total soluble proteins was performed on tobacco plants which had leaves that contained the transiently expressed TrCel5A protein and SDS-PAGE showed a large variety of proteins were present (Figure 2A, lane 1). The total soluble protein sample from TrCel5A expressing plants and a total soluble protein sample derived from NTEPs, were incubated for 10 min at 55 °C. TrCel5A remains stable and active at 55 °C while many other (native tobacco) proteins are not stable at this temperature. Thus, many non-essential proteins were precipitated by this treatment, leaving a partially purified sample of TrCel5A. After this thermal-precipitation step, a strong protein band was observed at ~40 kDa (Figure 2A, lane 1). This is likely to be the catalytic domain (CD) of the TrCel5A, which has been shown previously to run at this size when expressed in plants4. The negative and positive control samples (Figure 2A, lanes 3 and 4, respectively) showed low concentrations of remaining thermo-tolerant tobacco proteins from NTEPs (negative control, Figure 2A, lane 3) and multiple bands indicating a mixture of T. reesei proteins (positive control, Figure 2A, lane 4).
The size of the TrCel5A was confirmed by antibody staining directed at the His-6tag incorporated into the TrCel5A C-terminus, which was seen to be absent for both controls (Figure 2B). The TrCel5A was demonstrated to retain endoglucanase activity by CMC zymography (Figure 2C, lanes 1 and 2), where the refolded protein created a strong cleared area, indicating CMC degradation, at the same height as in the Coomassie stained SDS-PAGE gel and the Western blot. No activity was observed for the non-cellulase protein mixtures from NTEPs (Figure 2C, lane 3), and the mixture of T. reesei proteins run as a positive control showed endoglucanase activity from multiple components (Figure 2C, lane 4).
To quantify the cellulolytic activity of TrCel5A three assays were used. Firstly, the degradation of 4-MUC was analyzed by observing the increased fluorescence emission at 465 nm (excited at 360 nm) of the released 4-MU. TrCel5A was shown to degrade 4MUC, as did a 1:1,000 dilution of a commercially available mixture of T. reesei cellulases (Figure 3A).
A quantifiable analysis of the activity of TrCel5A against CMC was done by measuring the release of an Azo-dye at 590 nm from Azo-CMC. TrCel5A activity under these conditions, was equivalent in activity to a 1:1,000 dilution of the cellulase mixture mentioned above (Figure 3B).
The ability of TrCel5A to degrade filter paper was also analyzed. For this assay, an initial saccharification assay was performed (i.e. with the TrCel5A solution degrading the filter paper) and then the supernatant was analyzed by the PAHBAH assay to ascertain the quantity of reducing sugars released (Figure 3B).
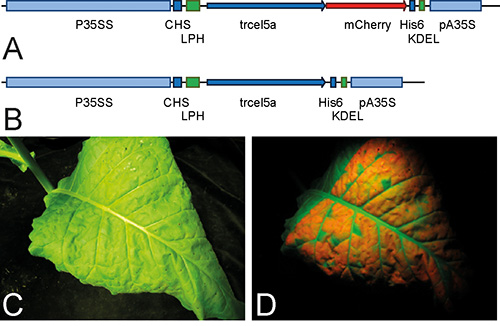
Figure 1. Plant expression cassettes for TrCel5A constructs and heterologous expression of TrCel5A-mCherry in tobacco leaves visualized by fluorescence under green light. Schematic representations of the plant expression cassette used for TrCel5A (A) and TrCel5A-mCherry (B) are shown. After transient TrCel5A-mC expression, tobacco leaves were examined under normal (C) or green light (D). For panels (A) and (B) the CaMV promoter (P35SS) and terminator signal (pA35S) are indicated in light blue. 5'-UTR of chalcone synthase (CHS), and the His6 coding sequence (His6) are indicated in blue. The plant codon-optimized leader peptide (LPH) derived from the heavy chain of the murine mAb24 is depicted in green. LPH achieves the secretion of the recombinant protein to the apoplast, after which a C-terminal KDEL signal, indicated in red, retards the protein to the ER. Arrows label the binding site for primer to amplify the CaMV 35SS expression cassette. For panels (C) and (D) the green light had an excitation 515 nm. Images were taken without magnification.
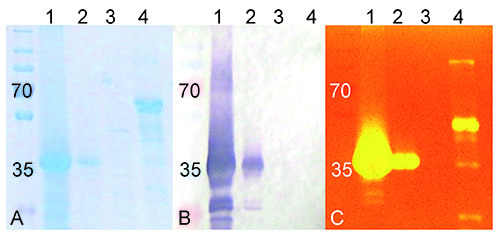
Figure 2. Size and activity of TrCel5A shown by SDS-PAGE, Western blotting, and CMC zymography. Separation of the total soluble proteins: from tobacco plants where TrCel5A was transiently expressed before (1) and after (2) thermal precipitation purification; from tobacco plants where TrCel5A was not present, also after thermal precipitation (3); or of a commercially available mixture of T. reesei cellulases (4). SDS-PAGE separated proteins are visualized with Coomassie Blue staining (A), Western blotting against the His-tag region of TrCel5A (B), and the cellulase activity shown by zymography using CMC visualized using Congo Red (C). PageRuler Plus (Fermentas) was used as the molecular weight marker (M).
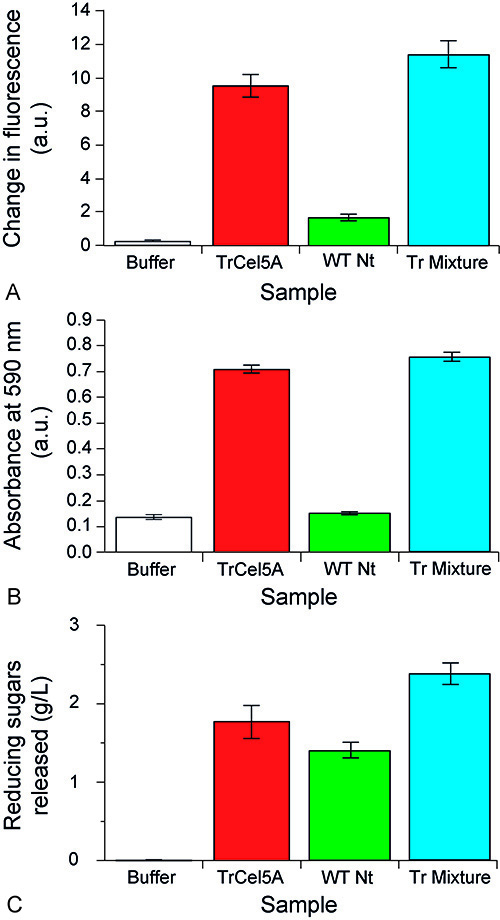
Figure 3. Enzyme activity quantification of TrCel5A. TrCel5A was incubated with 4-MUC (A), Azo-CMC (B), and filter paper (C) and the substrate degradation measured by release of the fluorophore 4-MU (A), an Azo-dye (B), or by quantifying reducing sugars by monitoring the color change of PAHBAH (C). Each assay shows the results for three replicates. Error bars indicate standard deviation. In all three assays the samples analyzed were buffer alone, TrCel5A after thermal precipitation purification, NTEP (WT Nt) after thermal precipitation purification, and a commercially available mixture of T. reesei cellulases.
Discussion
Recombinant expression of cellulases is a field of great interest, due to the desire for more efficient biofuel production systems1. Plants offer many possibilities for successful cellulase production, with features such as economic harvesting techniques and autohydrolysis methods being very desirable properties for large-scale biofuel production. Further, the use of different localizations for producing proteins allow, if required, posttranslational modifications20. Altering plants to enable constitutive expression of cellulases, while offering obvious benefits, is a time consuming method which may or may not be successful. Techniques such as sequestration of the expressed proteins and inducible expression mechanisms increase the likelihood of successful cellulase production in plants, however these still take many months to establish. Here, transient expression methods are demonstrated, to illustrate the potential for a specific cellulase to be produced through plant systems within a matter of days.
A fusion with the fluorescent protein mCherry was used here to demonstrate the success of the transient protein expression methods. TrCel5A-mCherry was clearly distributed across the majority of leaves which had been infiltrated with the vector-containing Agrobacteria. Localization to the endoplasmic reticulum enables posttranslational modifications of the expressed protein. The ability of an expression system to enable e.g. glycosylation is particularly important for those cellulases which are originally derived from fungal organisms, like T. reesei 21. Yeast expression systems, which are increasingly popular for heterologous cellulase expression, frequently result in hyper-glycosylation of the enzymes, which is debated to be a potential hindrance to full cellulase activity22. Plant glycosylation mechanisms are without the drawback of hyper-glycosylation. Additionally, expression in plant systems allows for localization to areas which allow or do not allow posttranslational modifications, thus enabling tailoring to the source of the protein. The system demonstrated here can also be used, by the application of different signal sequences14, for localization to the apoplast, chloroplast, cytosol or golgi vesicles. The use of fluorescent protein fusions, such as TrCel5A-mCherry, can demonstrate the success of protein expression as well as the localization of the protein. However, such conjugates are not required for further activity tests. Thus it is recommended that a non-mCherry TrCel5A construct be used for all further tests, as was shown here, to obtain more accurate activity data of the expressed cellulase. Regarding controls for further experiments, samples from non-transiently infiltrated plants are commonly viewed to be adequate controls.
Characterization of the expressed protein, TrCel5A, by SDS-PAGE and Western blotting revealed a band at approximately 40kDa, indicating that the protein linker region had been cleaved and that the major product derived was a peptide containing the catalytic domain. This is in line with previous findings of TrCel5A protein expression in plants5. Zymography analysis of activity against CMC indicated that the remaining peptide was active as an endoglucanase. While SDS-PAGE and Western blotting are extremely common assays, CMC zymography is a much more specific test for cellulase (and in particular endoglucanase) activity. Some important factors to keep in mind for successful zymography results are a) that the substrate (in this case CMC) must be completely solubilized and spread evenly throughout the gel; b) that the protein needs to be successfully refolded, preferably in a short period to limit the amount of protein diffusion through the gel; and c) that the activity assay should be at the optimal temperature for the enzyme being assayed (for TrCel5A between 50 and 60 °C) and should be brief to enable sharper bands of degradation.
Quantitative analysis of the cellulase activity of expressed TrCel5A demonstrated that the enzyme was active against a range of substrates, including 4-MUC, CMC and filter paper. Each of the substrates was examined using a different spectroscopic test, either via a fluorophore (4-MU) or by light scatter analysis of a color reaction (Azo-dye or PAHBAH). All three substrates and assays are widely used for the assessment of cellulase activity, and are relatively straight forward assays. 4-MUC is used as a basic substrate to determine glycosyl hydrolase activity, as a wide variety of these enzymes (endoglucanases, exoglucanases and β-glucosidases) are active against it. The speed of the assay, limited number of requirements, high sensitivity and ability of many cellulases to cleave the substrate, makes this a useful assay for general screening for cellulases, or for probing the success of an expression run. One point to keep in mind when using 4-MUC is that this substrate is particularly suited to the activity of β-glucosidases, making the activity of endoglucanases (such as TrCel5A) or exoglucanases appear weak in comparison to β-glucosidases or enzyme mixtures which contain β-glucosidases. When performing a 4-MUC assay with low protein concentrations or otherwise low activity levels it is recommended to utilize the addition of glycine (pH 10) as a stop solution, as the pH alteration enhances the fluorescence of 4-MU. CMC is a more specific substrate for the enzyme expressed here, as it is only degraded by endoglucanases. The Azo-CMC assay is more complicated than the one for 4-MUC, due to the need for precipitation of the cellulose (and methanol) by zinc. The major benefit of this substrate, and thus assay, is that it is specific for endoglucanase activity, making it very useful for establishing the nature of the cellulase being produced. Filter paper degradation is another common test for establishing cellulase activity. The amount of degradation observed varies significantly dependent upon the properties of the cellulase being assessed, and it is a preferred substrate for examining the activity of enzyme mixtures. The PAHBAH assay, used here to examine filter paper degradation, recognizes the amount of sugars released after saccharification and so is also applicable to a wide range of model substrates not demonstrated here, including CMC, lichenan, β-glucan, α-cellulose, AVICEL, xylan and xyloglucan. For this assay, care should be taken to include standards on the same plate (or in the same run) as the samples being assessed, as variation in incubation times can lead to differences in the results. It is thus also important to repeat the assay to ensure accuracy. Additionally, a sample control should always be run, where the sample is incubated without the cellulose substrate, to control for any reducing sugars naturally present in the samples which might be identified by the PAHBAH, and would otherwise give false positive results.
In conclusion, methods for the transient expression and characterization of a recombinant cellulase are clearly demonstrated. These techniques provide a straightforward way to generate sufficient amounts of protein for further examination of one or more cellulases, using activity tests like those demonstrated here. In particular, these methods provide a swift way to examine the potential of designated cellulases for in planta production.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by the BMBF Forschungsinitiative “BioEnergie 2021 – Forschung für die Nutzung von Biomasse” and the Cluster of Excellence “Tailor-made Fuels from Biomass”, which is funded through the Excellence Initiative by the German federal and state governments to promote science and research at German universities.
Materials
| 4-methylumbelliferyl β-D-cellobioside | SIGMA-ALDRICH | M6018 | |
| Acetic acid | ROTH | 3738 | |
| Acetosyringon (concentrated) | ROTH | 6003 | |
| Antibiotic – kanamycin | ROTH | T832 | |
| Antibiotic – rifampicin | ROTH | 4163 | |
| Antibiotic – carbenicillin | ROTH | 6344 | |
| Antibody – rabbit anti His(6) pAb | ROCKLAND | 600-401-382 | |
| Antibody – goat anti rabbit AP, FC pAb | DIANOVA | 111-055-008 | |
| Azo-carboxymethyl cellulose | MEGAZYME | S-ACMC | |
| β-cyclodextrin | SIGMA-ALDRICH | C4805 | |
| Calcium chloride dihydrate | SIGMA-ALDRICH | C5080 | |
| Carboxymethyl cellulose | ROTH | 3333.1 | |
| Cellulase from Trichoderma reesei ATCC 26921 | SIGMA-ALDRICH | C2730 | |
| Congo red | SIGMA-ALDRICH | 75768 | |
| Coomasie brilliant blue G 250 | ROTH | 9598.2 | |
| D(+)-glucose | ROTH | 8337 | |
| Ethanol | ROTH | K928 | |
| Filter paper – Rotilabo blotting paper | ROTH | CL67 | |
| NBT/BCIP | SIGMA-ALDRICH | 72091 | |
| MES buffer | ROTH | 4256 | |
| Methanol | ROTH | 8388 | |
| Sodium citrate | ROTH | 3580 | |
| Sodium dodecylsulfate | SERVA | 20765 | |
| Sodium hydroxide | ROTH | 6771 | |
| Soil, Einheitserde classic | PATZER GMBH | ||
| Spectrometer – Infinite M200 | TECAN | ||
| Sucrose | SIGMA-ALDRICH | S-5390 | |
| 4-Hydroxy benzhydrazide | SIGMA-ALDRICH | H9882 | |
| Phosphate buffered saline | ROTH | 1058 | |
| UV light source – BLAK RAY | UVP | ||
| Vibratome – VT1000S | Leica |
References
- Garvey, M., Klose, H., Fischer, R., Lambertz, C., Commandeur, U. Cellulases for biomass degradation: comparing recombinant cellulase expression platforms. Trends in Biotechnology. 31, 581-593 (2013).
- Klose, H., Günl, M., Usadel, B., Fischer, R., Commandeur, U. Ethanol inducible expression of a mesophilic cellulase avoids adverse effects on plant development. Biotechnology for Biofuels. 6, 53 (2013).
- Klose, H., Röder, J., Girfoglio, M., Fischer, R., Commandeur, U. Hyperthermophilic endoglucanase for in planta lignocellulose conversion. Biotechnology for Biofuels. 5, 63 (2012).
- Sharma, A. K., Sharma, M. K. Plants as bioreactors: recent developments and emerging opportunities. Biotechnology Advances. 27 (6), 811-832 (2009).
- Tremblay, R., Wang, D., Jevnikar, A. M., Ma, S. Tobacco, a highly efficient green bioreactor for production of therapeutic proteins. Biotechnology Advances. 28 (2), 214-221 (2010).
- Lee, T. M., Farrow, M. F., Arnold, F. H., Mayo, S. L. A structural study of Hypocrea jecorina Cel5A. Protein Science. 20, 1935-1940 (2011).
- Saloheimo, M., et al. EGIII, a new endoglucanase from Trichoderma reesei: the characterization of both gene and enzyme. Gene. 63, 11-21 (1988).
- Davidson, M. W., Campbell, R. E. Engineered fluorescent proteins: innovations and applications. Nature Methods. 6, 713-717 (2009).
- Pelham, H. R. The retention signal for soluble proteins of the endoplasmic reticulum. Trends in Biochemical Sciences. 15, 483-486 (1990).
- Béguin, P. Detection of cellulase activity in polyacrylamide gels using Congo red-stained agar replicas. Analytical Biochemistry. 131, 333-336 (1983).
- Jørgensen, H., Mørkeberg, A., Krogh, K. B. R., Olsson, L. Growth and enzyme production by three Penicillium species on monosaccharides. Journal of Biotechnology. 109, 295-299 (2004).
- Lever, M. A new reaction for colorimetric determination of carbohydrates. Analytical Biochemistry. 47, 273-279 (1972).
- Maclean, J., et al. Optimization of human papillomavirus type 16 (HPV-16) L1 expression in plants: comparison of the suitability of different HPV-16 L1 gene variants and different cell-compartment localization. Journal of General Virology. 88, 1460-1469 (2007).
- Munro, S., Pelham, H. R. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 48, 899-907 (1987).
- Laemmli, U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227, 680-685 (1970).
- Burnette, W. N. “Western Blotting”: Electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Analytical Biochemistry. 112, 195-203 (1981).
- Towbin, H., Staehelin, T., Gordon, J. Elelctrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets – procedure and some applications. Proceedings of the National Academy of Sciences of the United States of America. 76, 4350-4354 (1979).
- Blake, M. S., Johnston, K. H., Russell-Jones, G. J., Gotschlich, E. C. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Analytical Biochemistry. 136, 175-179 (1984).
- Yamamoto, E., Yamaguchi, S., Nagamune, T. Effect of β-cyclodextrin on the renaturation of enzymes after sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Analytical Biochemistry. 381, 273-275 (2008).
- Fischer, R., Vaquero-Martin, C., Sack, M., Drossard, J., Emans, N., Commandeur, U. Towards molecular farming in the future: transient protein expression in plants. Biotechnology and Applied Biochemistry. 30 (2), 113-116 (1999).
- Sammond, D. W., et al. Cellulase linkers are optimized based on domain type and function: Insights from sequence analysis, biophysical measurements, and molecular simulation. PLoS One. 7, (2012).
- Boonvitthya, N., Bozonnet, S., Burapatana, V., O’Donohue, M. J., Chulalaksananukul, W. Comparison of the heterologous expression of Trichoderma reesei endoglucanase II and cellobiohydrolase II in the yeasts Pichia pastoris and Yarrowia lipolytica. Molecular Biotechnology. 54, 158-169 (2013).

