In Vitro Reconstitution of Light-harvesting Complexes of Plants and Green Algae
Summary
This protocol details the reconstitution of light-harvesting complexes in vitro. These integral membrane proteins coordinate chlorophylls and carotenoids and are responsible for harvesting light in higher plants and green algae.
Abstract
In plants and green algae, light is captured by the light-harvesting complexes (LHCs), a family of integral membrane proteins that coordinate chlorophylls and carotenoids. In vivo, these proteins are folded with pigments to form complexes which are inserted in the thylakoid membrane of the chloroplast. The high similarity in the chemical and physical properties of the members of the family, together with the fact that they can easily lose pigments during isolation, makes their purification in a native state challenging. An alternative approach to obtain homogeneous preparations of LHCs was developed by Plumley and Schmidt in 19871, who showed that it was possible to reconstitute these complexes in vitro starting from purified pigments and unfolded apoproteins, resulting in complexes with properties very similar to that of native complexes. This opened the way to the use of bacterial expressed recombinant proteins for in vitro reconstitution. The reconstitution method is powerful for various reasons: (1) pure preparations of individual complexes can be obtained, (2) pigment composition can be controlled to assess their contribution to structure and function, (3) recombinant proteins can be mutated to study the functional role of the individual residues (e.g., pigment binding sites) or protein domain (e.g., protein-protein interaction, folding). This method has been optimized in several laboratories and applied to most of the light-harvesting complexes. The protocol described here details the method of reconstituting light-harvesting complexes in vitro currently used in our laboratory, and examples describing applications of the method are provided.
Introduction
The photosynthetic apparatus of plants and algae include integral membrane proteins that bind chlorophyll a (chl a), b (chl b) and carotenoids (car). These pigment-protein complexes are active in harvesting light energy and transferring that excitation energy to the reaction centers, where it is used to promote charge separation2. They are also involved in regulatory feedback mechanisms that protect the photosynthetic apparatus from high light damage3,4. The light harvesting complexes (LHCs) are comprised of a large family of related proteins in plants and algae5.
The homogeneous purification of each member of the family has been complicated by the highly similar chemical and physical properties of the complexes. In addition, purification procedures often result in loss of pigments or other potential cofactors such as lipids. In vitro reconstitution represents a powerful method to overcome these problems. The LHC associated with Photosystem II (LHC-II) was first reconstituted in vitro by Plumley and Schmidt in 19871. The researchers extracted delipidated protein and pigments separately from plant chloroplasts, and then combined the heat denatured protein with pigments in the presence of Lithium Dodecyl Sulfate (LDS), followed by three cycles of freezing and thawing1. They showed that the spectral properties of the reconstituted LHC complexes were very similar to complexes purified from plants. The ease of reconstituting LHC pigment-protein complexes, likely due to some inherent self-assembly feature, along with the difficulty in isolating purified complexes from organisms, led to the quick adoption of the method by other researchers. The reconstitution of photosynthetic proteins overexpressed in Escherichia coli (E. coli) was achieved by Paulsen and colleagues in 19906. In E. coli, overexpressed membrane proteins are typically contained in inclusion bodies, which facilities their purification. Reconstitution is achieved through heat denaturation of the inclusion bodies containing recombinant protein in the presence of LDS, followed by the addition of pigments which initiates the protein folding. Folding of the LHCII complex is a two-step process: first, chlorophyll a is bound in less than 1 min; second, chlorophyll b is bound and stabilized over several minutes7.
In addition to providing insight into the folding dynamics, in vitro reconstitution combined with site-directed mutagenesis has allowed the identification of specific amino acids important for stability (e.g., 8,9) or pigment coordination (e.g., 10). Manipulation of refolding conditions by adjusting parameters such as pigment composition or detergents have also identified elements critical for proper folding, such as the requirement of Xanthophylls for the LHCII complex (e.g., 1,11). In addition, investigation of the properties of individual pigments bound to the complexes has been possible using complexes reconstituted in vivo (e.g., 10).
The method described here begins with isolation of pigments (chlorophylls and carotenoid) from spinach and the green alga Chlamydomonas reinhardtii. The expression and purification of a LHC protein from E. coli in the form of inclusion bodies is then detailed, followed by the reconstitution of LHC and subsequent purification by Ni affinity column. In the final step, the reconstituted complexes are further purified by sucrose gradient centrifugation to remove free pigments and unfolded apoprotein. This protocol represents an optimized procedure incorporating several modifications that have been introduced by different laboratories over time1,6,10,12–14.
Protocol
1. Total Pigment Extraction from Spinach Leaves
- Homogenize one handful of spinach leaves (~20 g) in 100 ml of cold Grinding Buffer (see Table 1) using a blender for 20 sec.
- Filter the solution through a two layers of nylon cloth with a pore diameter of 20 μm and centrifuge the filtrate at 1,500 x g for 10 min at 4 °C.
- Resuspend the pellet containing the chloroplasts with a soft artists paint brush in 1 ml of cold Wash Buffer (see Table 1). Once the pellet is resuspended, add 50 ml of Wash Buffer and centrifuge the solution at 10,000 x g for 10 min at 4 °C.
- Remove the supernatant and gently resuspend the pellet (thylakoids) in 50 ml of Wash Buffer (see Table 1).
- Centrifuge the solution at 10,000 x g for 10 min at 4 °C and remove the supernatant completely. At this point, carry out the following steps in the dark, to avoid pigment oxidation.
- Add ~20 ml of 80% acetone buffered with Na2CO3 (see Table 1) to extract the pigments. Leave the solution on ice for 10 min, vortexing occasionally.
- Pellet the cellular components by centrifugation at 12,000 x g for 15 min at 4 °C.
NOTE: If the pigments are not totally extracted, the pellet will have a green color and step 1.6 should be repeated. - Collect the supernatant into a separatory funnel. Add 0.4 volumes of diethylether, shake vigorously and open the valve to vent the gas.
- Add 0.8 volumes of 0.33 M NaCl and mix vigorously. Allow ~10 min for the layers to separate. The ether phase on top contains the extracted pigments. Remove the clear lower phase.
NOTE: If the separation is not clear, freeze and thaw the solution to improve the phase separation. - Remove the ether by pouring it from top of the separatory funnel into a suitable glass container. Dry by adding a spoonful of granular anhydrous sodium sulfate. Swirl the solution and allow ~5 min for the desiccant to absorb water from the ether.
NOTE: Repeat this step if the sodium sulfate appears completely clumped together; there should be some free-floating crystals when the ether is sufficiently dried. If a water layer forms, remove this with a Pasteur pipet before adding additional anhydrous sodium sulfate. - Decant the ether to a new glass container, leaving the sodium sulfate solid behind.
- Evaporate the ether in a rotary speedvac or under a stream of N2.
- Dissolve the pigments completely in 10 ml of 100% acetone.
- Dilute a small amount (~3 µl) into 1 ml of 80% acetone and measure the absorption spectra and determine the Chl a/b ratio and the Chl concentration with the method described by Porra et al. (1989)15.
- Aliquot and dry the pigments in a rotary speedvac or under N2 stream until the acetone is completely evaporated. Store the dried pigments at -80 °C.
2. Extraction of Carotenoids from Spinach
- Follow steps 1.1 to 1.5. At this point, carry out the following steps in the dark, to avoid pigment oxidation.
- Resuspend the thylakoid pellet in ~50 ml 96% ethanol buffered with Na2CO3 (see Table 1) to extract the pigments. Leave the solution on ice for 5 min.
- Pellet the cellular components by centrifugation at 12,000 x g for 15 min at 4 °C.
NOTE: If the pigments are not totally extracted, the pellet will have a green color and step 2.2 should be repeated. - Collect the supernatant and add 0.1 volume of 80% KOH (w/v) to initiate saponification.
- Leave the solution at 4 °C O/N, tightly capped and protected from the light.
- Collect the solution into a separatory funnel. Add 1 volume of diethyl ether and mix gently.
- Add 0.8 volumes of 0.33 M NaCl and mix gently. Allow ~10 min for the layers to separate. The orange ether phase on top contains the saponified carotenoids. Remove the green lower phase by draining through the stopcock of the funnel.
- Add 3 volumes of water and mix gently to remove the potassium hydroxide. Allow the layers to separate. NOTE: If the upper phase appears cloudy, add a small amount of NaCl (e.g., 3 g of NaCl in 200 ml of solution) and swirl gently to dissolve.
- Remove the lower phase by draining through the stopcock of the funnel.
- Follow steps 1.10 to 1.13.
- Dilute a small amount (~3 µl) into 1 ml of 80% acetone and measure the absorption spectra at 440 nm in 80% acetone. To determine the concentration, use the average coefficient extinction for the carotenoids (ε440 = 255)16 in the following formula: Car [mg/ml] = (Abs440 nm/225) x 11 (optical path) = 1 cm.
- Aliquot and dry the carotenoids in a speedvac or under N2 stream until all diethylether has been evaporated. Store the dried pigments at -80 °C.
3. Total Pigment and Carotenoid Extraction from Chlamydomonas reinhardtii
- Grow C. reinhardtii on solid TAP medium17 in a petri dish by spreading a small amount of liquid culture onto the surface. Grow under continuous illumination flux of 20 μmol photos PSA m-2 sec-1 until a green layer of cells is visible.
- Using a sterile inoculating loop, harvest a small amount of C. reinhardtii from the solid TAP medium and put the cells into 500 ml of TAP medium17 in a 1 L flask. Grow the culture at 25 °C with 170 rpm agitation under a continuous illumination flux of 20 μmol photos PSA m-2 sec-1.
- After 5-6 days, the culture should reach the end of the logarithmic phase (6 x 106 cell/ml or 2-2.5 optical density at 750 nm). Centrifuge the culture at 4,000 x g for 15 min at 4 °C.
- For total pigment extraction, follow steps 1.6 to 1.15.
- The yield of total pigment extract starting from 500 ml of full growth culture of C. reinhardtii is around 5 ml of solution with a concentration of 0.5 mg chl a+b / ml.
- For carotenoids extraction, follow steps 2.2 to 2.12.
4. Purification of Inclusion Bodies
- Clone the coding sequence of the LHC protein of interest into an expression vector that results in a fused C-terminal His tag using standard molecular biology procedures. Transform this construct into E. coli host strain such as BL21 (DE3).
- Prepare Lysis buffer, Detergent buffer, Triton buffer, TE (Table 1), 1 M Isopropyl β-D-1-thiogalactopyranoside (IPTG) and LB medium18 with the appropriate antibiotics.
- Pick a single E. coli colony containing the expression clone from a freshly streaked plate into ~5 ml of LB medium with the appropriate antibiotics using standard procedures6. Grow at 37 °C with 220 rpm agitation for at least 16 hr.
- Add 2.5 ml of the O/N culture into a 1 L Erlenmeyer flask with 250 ml of LB supplemented with the appropriate antibiotic.
- Grow the cells for 2-3 hr (or until the OD600 is ~0.6) at 37 °C at 220 rpm.
- Add IPTG to a final concentration of 1 mM. Continue to grow the cells at 37 °C with 220 rpm 3-4 hr.
- Centrifuge the culture for 10 min at 5,000 x g at 4 °C in a pre-weighed centrifuge tube. Discard the supernatant thoroughly and determine the weight the pellet by weighing again and subtracting the centrifuge tube weight.
- Resuspend the E. coli cell pellet in 0.8 ml/g of Lysis buffer by vigorous vortexing.
NOTE: Alternatively, the cell pellet can be frozen at -80C for later use. If starting with a frozen pellet, allow to thaw completely before adding the lysis buffer. - Add 2 mg of lysozyme per gram of cells, and incubate on wet ice with occasional vortexing for 30 min.
- Add 20 μg/ml DNAse, 10 mM MgCl2, 1 mM NaCl, 20 µg/ml RNAse. Vortex and put on ice for 30 min.
- Add 2 ml of cold Detergent buffer per gram of cells. Mix well and keep RT for 5 min.
- Transfer to 2 ml centrifuge tubes (split into two tubes if necessary). Centrifuge for 10 min at 12,000 x g at 4 °C to pellet the inclusion bodies.
- Add 1 ml of cold Triton buffer and completely resuspend the pellet by sonification (3 pulses x 5 sec x 50% power with 20 sec intervals). NOTE: Have the tube in a small beaker surrounded by ice water to keep it cold during the sonification. In the case of multiple tubes, combine the resuspended inclusion bodies into one tube after resuspension.
- Centrifuge for 10 min at 12,000 x g at 4 °C to pellet the inclusion bodies.
- Repeat step 4.13 and 4.14 two times.
- Resuspend the inclusion bodies in 1 ml of cold TE with sonification for a final wash to remove the Triton buffer. Centrifuge for 10 min at 12,000 x g at 4 °C to pellet the inclusion bodies.
- Resuspend the pellet in 1 ml of cold TE by sonification.
- Assess the protein concentration by standard methods such as the Bradford assay19. Store aliquots of the inclusion bodies at -20 °C.
5. Reconstitution
This protocol typically yields 1-2 ml of reconstituted protein with an OD of 4 when absorbance is measured in the Qy region (600-750 nm). Quantity can be adjusted as desired, although care should be taken to maintain the proper ratios during the procedure.
- Prepare the following solutions as described in Table 1: 2x Reconstitution Buffer, 20% OG, 2 M KCl, TE. Perform the following steps in dim light.
- Resuspend 800 μg of LHC Inclusion Bodies in a total of 400 μl TE in a 2 ml microfuge tube. Add 400 μl of the 2x Reconstitution Buffer and vortex briefly.
- Add 0.6 μl of β-Mercaptoethanol (stock 14.8 M) to have a final concentration of 10 mM. Heat the protein for 1 min at 98 °C. Vortex briefly and place at RT for 3 min.
- Resuspend 500 μg of total dried chlorophyll pigments plus 80 μg carotenoid pigments in 30 μl 100% EtOH by vigorously vortexing for 1 min or place in a bath sonicator for 1-2 min.
- Spin the pigment mix ~30 sec at 15,800 x g at 4 °C and confirm that there is no pellet. If there is a pellet, repeat vortexing and/or sonification. IMPORTANT: After the resuspension and spin, immediately add pigment to the protein, or it can aggregate and will need to be resuspended again.
- Slowly add the pigment mix to the cooled protein while vortexing. Continue to vortex 5-10 sec and place tube on wet ice. Be careful not to vortex too vigorously as the protein can overflow the top of the tube.
- Add 94 μl of 20% Octyl β-D-glucopyranoside (OG) (final concentration 2%), vortex briefly and keep on ice 10 min.
- Add 90 μl of KCl 2 M (final concentration 150-200 mM), vortex briefly and keep on ice 20 min. NOTE: column preparation (Section 6) can be initiated at this time.
- Spin for 10 min at 15,800 x g at 4 °C. Remove the supernatant without disturbing the pellet (precipitated LDS) to a 10 ml tube. Keep cold and protected from light.
6. Nickel Column Purification
- Prepare the following solutions as described in Table 1: OG buffer, OG rinse buffer, Elution buffer.
- Connect a Ni-Sepharose column (1 ml) or equivalent to a peristaltic pump ensuring that no air gets inside the column during this step and the following steps.
- Set the speed of the pump to 1 ml/min and rinse the column with 5-10 ml of water to remove the storage solution.
- Equilibrate the column with 3-4 ml of OG buffer.
- Add 3-4 ml of OG buffer to the protein sample and load to the column. NOTE: if the protein has been sitting on ice for longer than 10 min after removal of LDS, spin again at 15,800 x g at 4 °C for 1 min to remove any additional LDS precipitation.
- Rinse the column with 5 ml of OG buffer.
- Rinse the column with 2 ml of OG rinse buffer.
- Elute the bound protein with 3 ml elution buffer. Collect the green elute which contains the reconstituted protein. NOTE: This is usually about 1 ml in total.
7. Sucrose Gradient Centrifugation
- Prepare the following solutions as described in Table 1: Sucrose solution, 0.06% β-DM, 0.01 M HEPES, pH 7.6.
- Fill ultracentrifuge tubes with the sucrose solution and freeze at -20 °C O/N or -80 °C for at least 1 hr.
- Remove the tube from freezer and allow to thaw undisturbed at 4 °C. NOTE: The freeze/thaw process creates a gradient from 0.1 to 1 M sucrose. A 15 ml tube typically thaws in about 3 hr.
- Carefully remove from the top the same volume as the green fraction eluted from the nickel Sepharose column in step 6.8. Then load the reconstituted sample on top slowly to avoid disturbing the gradient.
- Balance tubes and spin at 200,000 x g at 4 °C in an ultracentrifuge using a SW-41 or SW-60 swinging bucket rotor for 18 hr, set to slow acceleration and stopping without brakes.
- Carefully take out the gradient from the tube holder with forceps. Use a syringe with a long needle that has a blunt opening to collect the fraction from the top. NOTE: Alternately, collect fractions from the bottom by piercing the tube with a needle and collecting drops.
Representative Results
This protocol details a method to reconstitute chorophyll a/b binding proteins in vitro. This technique permits the folding of these pigment-protein complexes in vitro starting from the apoprotein, which can be obtained by overexpression in a heterologous system, and pigments extracted from plant or algae. After reconstitution, the refolded pigment-protein complex is purified from the excess of pigments and the unfolded apoprotein in two steps. The first step (Figure 1 A-B) is based on the presence of His-tag at the C-terminal of the protein, which permits the removal of large part of the unbound pigments. The second purification step utilizes sucrose density gradient centrifugation, (Figure 2) where the unfolded protein usually migrates slower than the green band containing the reconstituted protein. The goal of the reconstitution in vitro is to obtain complexes with the same properties as the native ones. To illustrate this outcome, the spectroscopic properties of an in vivo light-harvesting complex is compared with the same LHC complex reconstituted in vitro13,20,21. The absorption spectrum of the LHCs in the visible range (350 nm and 750 nm) depends on the pigment composition of the complex, as well as on the pigment’s environment (which includes the protein) and it is thus a sensitive tool to check the quality of the reconstitution. In Figure 3, the absorption spectrum of CP24, a chlorophyll a/b binding protein from Arabidopsis thaliana, reconstituted in vitro, is compared with the spectrum of the same complex purified from Arabidopsis thylakoids21. In the spectra, it is possible to recognize the Qy and the Soret transition of Chl a (peaks at 671/439 nm) and Chl b (peaks at 649/466 nm). The native and reconstituted complexes show identical absorption spectra, indicating a virtually identical pigment composition and organization. Fluorescence spectroscopy can be used to assess the quality of the reconstituted complex. The fluorescence emission spectra is measured upon excitation at different wavelengths, which excite preferentially different pigments: Chl a at 440 nm, Chl b at 475 nm, and Xanthophylls at 500 nm. In a properly folded protein-pigment complex, Chl b and Xanthophylls transfer their excitation energy primarily to Chl a within a few picoseconds, and the fluorescence originates from a thermally equilibrated system resulting in a single peak with the same shape and maxima at all three excitation wavelengths (Figure 4A–B). The presence of Chl b not coordinated to the protein can be recognized by an additional peak or shoulder around 650 nm upon 475 nm excitation (Figure 4C). The presence of free Chl a instead leads to additional emission around 675 nm, which is mainly present upon 440 nm excitation. The fluorescence emission spectra upon 475 nm excitation of both reconstituted and the native CP24 complexes (Figure 4D) show a single peak at 681 nm, indicating that reconstituted complex is correctly folded. An additional confirmation that the pigment-protein complex is correctly reconstituted comes from circular dichroism (CD) measurements. The CD signal in the visible region depends on the excitonic interactions between pigments and it is thus very sensitive to even small changes in the organization of the chromophores22. Figure 5 shows the CD spectra of reconstituted and native CP24, with the typical fingerprint peaks at 681 nm, 650 nm and 481 nm. In conclusion, the high similarity between the spectroscopic properties of native and the reconstituted CP24 confirms that the reconstitution procedure yields native-like complexes suitable for in vitro study of light-harvesting proteins.
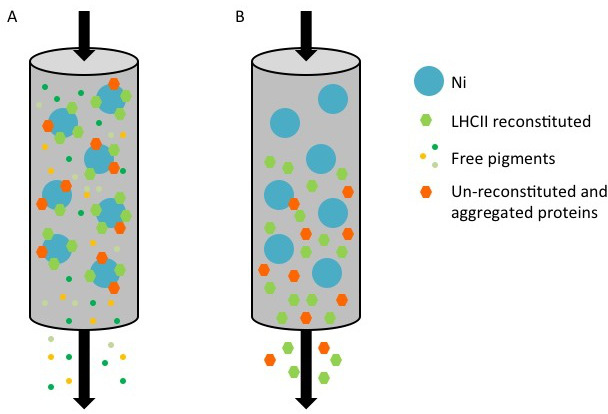
Figure 1. Representation of the purification of recombinant LHC proteins with a His tag using a nickel column. (A) During the purification, His-tagged protein, comprised of both reconstituted complexes (green hexagon) and un-reconstituted/aggregated protein (orange hexagon) are bound to the surface of the Ni-Sepharose (blue spot), while unbound pigments (small colored spots) flow through. (B) When the column is washed with the elution buffer containing imidazole, the reconstituted and un-reconstituted proteins are collected in the flow through.
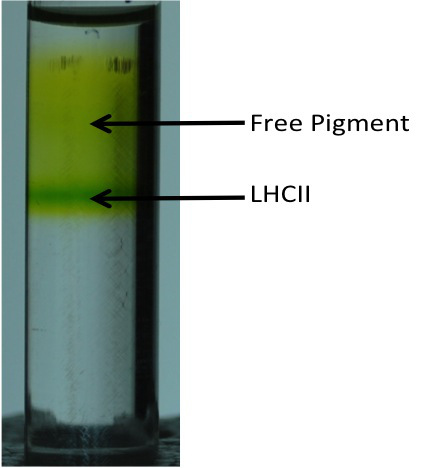
Figure 2. Sucrose gradient of reconstituted LHCII after purification by nickel column. The reconstituted complexes are separated from the free pigment by the density gradient. The dark green band represents reconstituted LHCII and the pale green background is composed of free pigments.
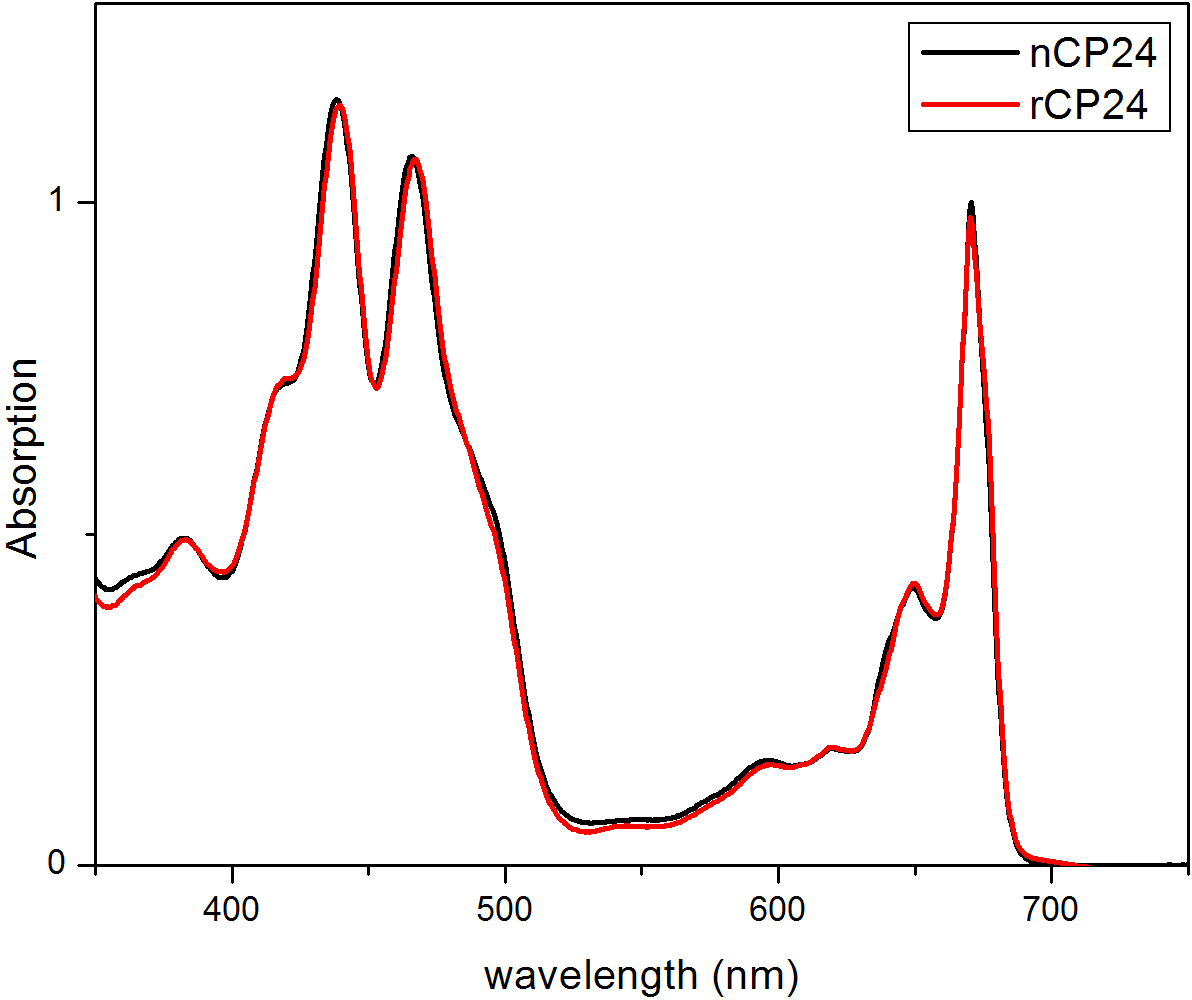
Figure 3. Absorption spectra of reconstituted protein CP24 (rCP24, red line) and the native one (nCP24, black line) isolated from Arabidopsis thaliana. In both spectra, it is possible to recognize the Qy and the Soret transition of Chl a (peaks at 671/439 nm) and Chl b (peaks at 649/466 nm). This figure has been modified from Passarini et al. 201421.

Figure 4. Fluorescence emission spectra. The fluorescence emission spectra of reconstituted CP24 wildtype complex (A) and normalized to the maximum (B) showing efficient energy transfer from Chl b and Xanthophyls to Chl a. (C) Fluorescence emission spectra of reconstituted CP24 (rCP24) and the native complex (nCP24) isolated from Arabidopsis thaliana. The spectra are normalized to the maximum of the peak (D). Please click here to view a larger version of this figure.
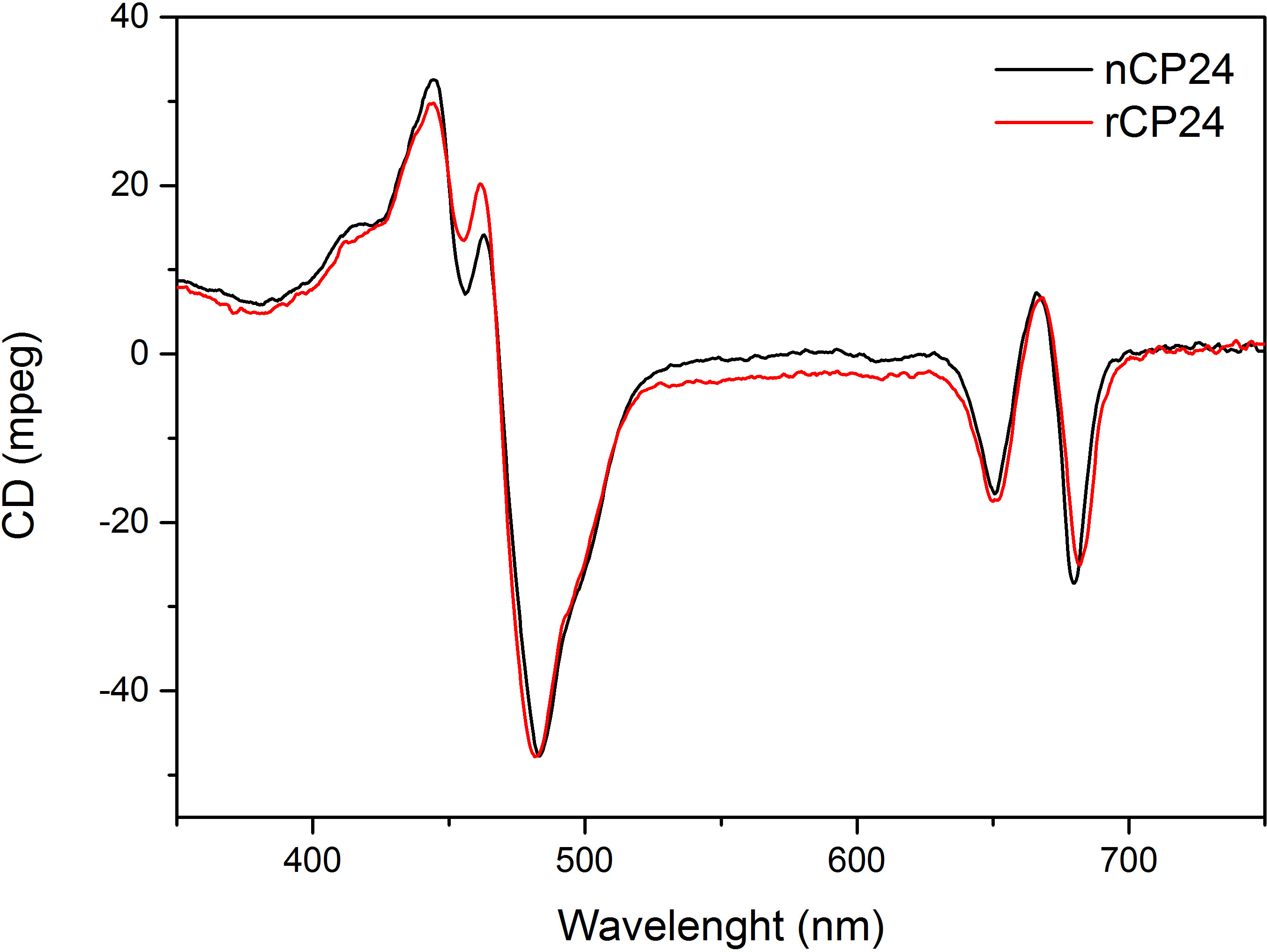
Figure 5. Circular Dichroism Spectra. Reconstituted CP24 (rCP24, red line) and the native complex (nCP24, black line) isolated from Arabidopsis thaliana shows very similar spectra.
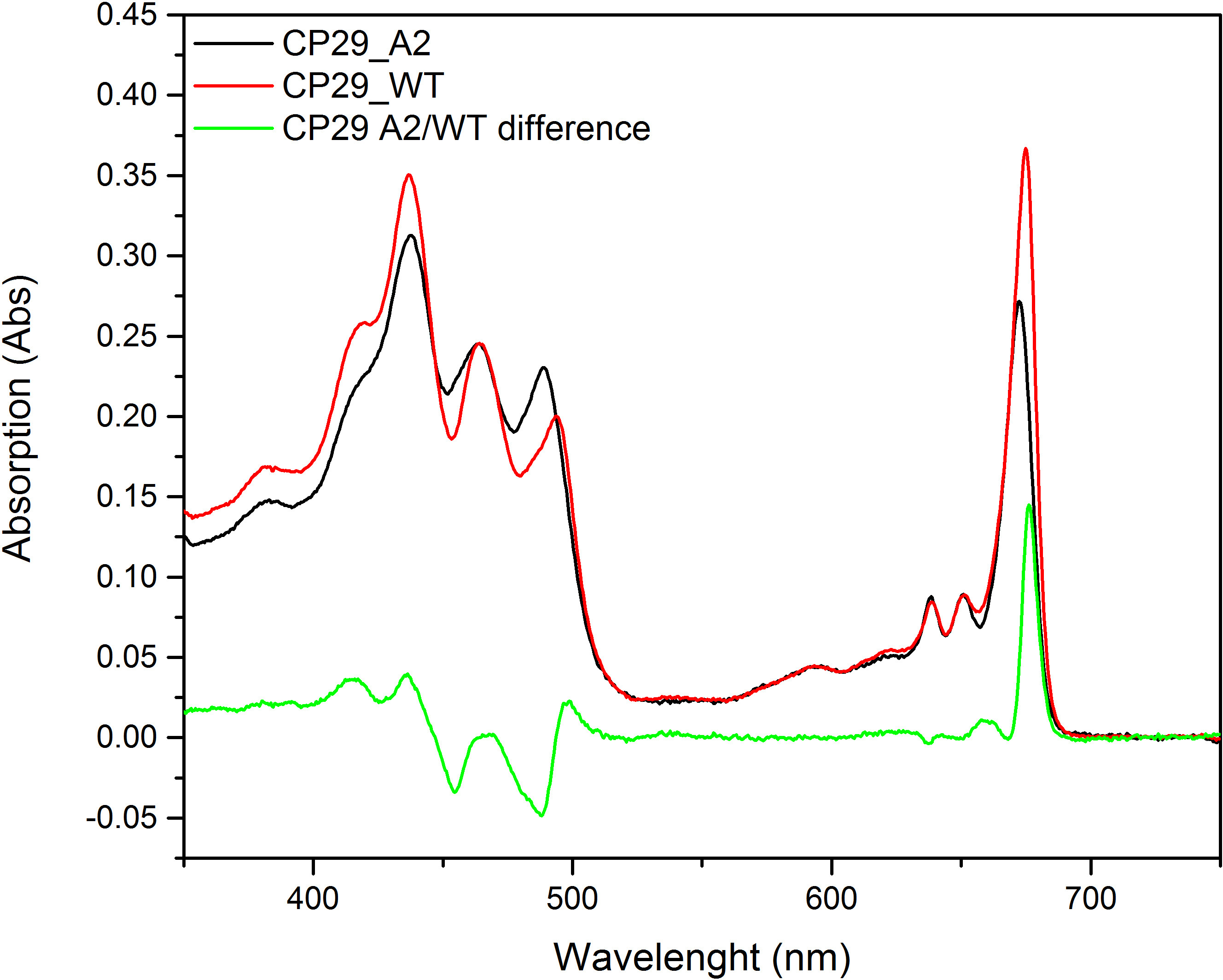
Figure 6. Absorption spectra of CP29 wild type (CP29_WT) and mutated CP29 (CP29_A2). The green line shows the differences between the two plots.
| All the buffers can be stored at 4 °C. | |||
| Components | Final Concentration | Additional notes | |
| Grinding Buffer | Sorbitol | 0.4 M | |
| Tricine | 0.1 M | pH 7.8 | |
| NaCl | 10 mM | ||
| MgCl2 | 5 mM | ||
| Milk Powder | 0.5% w/v | ||
| Wash Buffer | Sorbitol | 50 mM | |
| Tricine | 5 mM | pH 7.8 | |
| EDTA | 10 mM | pH 8 | |
| Lysis Buffer | Tris | 50 mM | pH 8 |
| Sucrose | 2.5% w/v | ||
| EDTA | 1 mM | pH 8 | |
| Detergent buffer | NaCl | 200 mM NaCl | |
| Deoxycholic acid | 1% w/v | ||
| NONIDET P-40 | 1% w/v | ||
| Tris | 20 mM | pH 7.5 | |
| EDTA | 2 mM | pH 8 | |
| beta-mercaptoethanol | 10 mM | ||
| Triton Buffer | Triton X-100 | 0.5% w/v | |
| Tris | 20 mM | pH 7.5 | |
| beta-mercaptoethanol | 1 mM | ||
| Buffer TE | Tris | 50 mM | pH 8 |
| EDTA | 1 mM | pH 8 | |
| Reconstitution Buffer | HEPES | 200 mM | |
| Sucrose | 5% w/v | ||
| Lithiumdodecylsulfate (LDS) | 4% w/v | ||
| Benzamidine | 2 mM | ||
| Aminocaproic Acid | 10 mM | ||
| OG Buffer | Octylglucoside | 1% w/v | |
| Sucrose | 12.5% w/v | ||
| NaCl | 0.2 M | ||
| HEPES | 20 mM | ||
| Imidazole | 10 mM | ||
| OG Rinse Buffer | n-Dodecyl-beta-D-Maltoside (β-DM) | 0.06% w/v | |
| HEPES | 40 mM | pH 7.5-9 | |
| NaCl | 0.2 M | ||
| Elution Buffer | Imidazole | 0.5 M | |
| n-Dodecyl-beta-D-Maltoside (β-DM) | 0.06% w/v | ||
| HEPES | 40 mM | pH 8 | |
| NaCl | 0.2 M | ||
| Sucrose Solution | Sucrose | 20% w/v | |
| n-Dodecyl-beta-D-Maltoside (β-DM) | 0.06% w/v | ||
| HEPES | 0.01 M | pH 7.6 | |
| Acetone 80% buffered with Sodium Carbonate | Acetone | 80% v/v | |
| Sodium Carbonate | 1 M | ||
| Ethanol 96% buffered with Sodium Carbonate | Ethanol | 96% v/v | |
| Sodium Carbonate | 1 M | ||
Table 1. List of buffers and solutions used in this protocol.
| Chla a/b mix | Chla a/b | Chl a | Chl b | Neo | Viola | Lute | Chl tot | Chl/Car | |
| nCP26 | – | 2.2±0.05 | 6.2 | 2.8 | 0.61 | 0.38 | 1.02 | 9 | 4.5±0.1 |
| rCP26 | 8 | 2.71±0.05 | 6.57 | 2.43 | 0.72 | 0.32 | 0.97 | 9 | 3.9±0.04 |
| rCP26 | 5.5 | 2.25±0.05 | 6.23 | 2.77 | 0.77 | 0.3 | 0.96 | 9 | 4.0±0.1 |
| rCP26 | 3 | 2.08±0.04 | 6.08 | 2.92 | 0.76 | 0.3 | 1.04 | 9 | 4.1±0.1 |
| rCP26 | 1 | 1.7±0.05 | 5.7 | 3.3 | 0.7 | 0.3 | 0.9 | 9 | 4.3±0.05 |
| rCP26 | 0.3 | 1.11±0.04 | 4.7 | 4.28 | 0.7 | 0.3 | 0.9 | 9 | 4.2±0.2 |
| rCP26 | 0.05 | 0.23±0.01 | 1.4 | 5.6 | 0.58 | 0.24 | 1.11 | 7 | 3.1±0.06 |
| rCP26 | <0.01 | 0.11±0.01 | 0.7 | 6.3 | 0.64 | 0.3 | 1.08 | 7 | 3.06±0.06 |
Table 2. Pigment content of CP26 native complex compared to reconstituted protein-pigment complexes with different Chl a/b Ratios39.
Discussion
Membrane proteins are not so easy to study. Isolation of native membrane proteins is complicated by the need to solubilize the lipid bilayer with detergents, which can damage the protein and remove essential cofactors. These proteins might also be present at low levels in biological membranes, or be mixed with closely related proteins, as in the case of the light harvesting complexes, that makes purification of single complexes difficult. Heterologous protein expression in E. coli and in vitro reconstitution offers the possibility to avoid these problems. In vitro reconstitution and purification of folded proteins results in complexes that possess characteristics very similar to those of the native complexes20,21,23 and thus can be used to study complexes that cannot be purified to homogeneity 24–27.
This method uses spinach, which is easily attainable year-round, as a source for the total pigment and carotenoid preparations. For some reconstitutions of proteins native to algae, use of pigments purified from algae is preferred due to different pigment compositions. The Chl a/b ratio and Chl/car ratio remains the same regardless of pigment source.
It is important to realize that the efficiency of the reconstitution is usually around 35%28. Thus it is necessary to remove the non-bound pigments and the unfolded apoprotein from the solution after the reconstitution. A two-step purification protocol is presented in this protocol (see also results). However, it should be noted that the sucrose gradient step does not allow the complete separation of apo- and holo-protein. For most analyses this is not a problem, as the apoprotein does not contain pigments and thus does not interfere with the functional measurements. However, in case it is necessary to fully remove the apoprotein from the fraction containing the reconstituted complex (for example, to calculate the pigment to protein stoichiometry), an anionic exchange column can be used (see Passarini et al. 200929 for details).
The capacity to refold recombinant light harvesting proteins with isolated pigments in vitro provides an opportunity to “manipulate” the complexes by modifying the reconstitution “environment” in various ways, thereby changing the characteristics of the resulting complex. For example, changing the pigment composition during reconstitution can result in a complex with altered pigment composition. This feature can be utilized to study the influence various pigments have on the structure and stability of the complex. Usually the pigment preparation obtained from spinach has a Chl a/b ratio of 3:1 and a Chl/car ratio of 2.9:1. This ratio typically produces a reconstituted protein with the same properties as the native one. However, adjustment of the Chl a/b ratio by the addition of purified Chl a or b can influence the binding of different pigments due to varying selectivity of the binding sites30–33. This is possible because most of the pigment binding sites are not completely selective for Chl a or Chl b, but can accommodate both, although with different affinity10,30,34. In a similar way, the carotenoid binding sites were also shown to be able to accommodate more than one Xanthophyll species8,35–38. Different reconstitutions of CP26, another pigment-protein complex of higher plants, using various pigment compositions are shown in Table 2 39. These reconstitutions were used to assess the affinity of binding sites for particular pigments39. It is interesting to note that in order to obtain a complex with the same pigment composition as the native one, the Chl a/b ratio of pigment mix must be 3:1. This seems to be the case for all LHC complexes of higher plants20,40.
The combination of molecular biology with the reconstitution technique allows the properties of a Chl-binding complex to be studied in more detail. The importance of different protein domains on the stability and folding of the complexes, or their involvement in the protein-protein interactions, have been determined by truncating the apoprotein or performing random mutagenesis8,41–44. Single amino acid residues important for the coordination of different pigments can be altered through site-directed mutagenesis in order to analyze the properties of individual pigments or assess their contribution to the function and stability of the complex10,28,29,45–52. Figure 6 shows reconstituted Lhcb4 (CP29) with a mutation of the histidine at position 21653. A comparison of the pigment composition of wildtype and mutant complexes shows that the mutation induces the loss of one Chl a molecule, indicating that the targeted site accommodates a Chl a in the WT complex. The differences of the absorption spectra of WT and mutant, upon normalization to the pigment content, also shows the absorption properties of the lost pigment. In this case, the difference can be seen in the main peak at 680 nm, indicating that the Chl a coordinated by His216 absorbs at this wavelength (for more details about this mutant and the spectroscopic properties see Mozzo et al. 200853). Mutation analysis can also be used to determine the effect of the environment on the spectroscopic properties of the pigments54.
In conclusion, light harvesting proteins can readily be reconstituted in vitro resulting in pigment-protein complexes with very similar properties to native complexes. In this way, the difficulties of isolating native proteins are eliminated, while also delivering protein preparation with high yield and purity for further study. The importance of a 3:1 Chl a/b ratio in producing an authentic complex is emphasized, and examples of reconstituted wildtype and mutant LHCs are provided to illustrate applications of the technique.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by the European research council by a ERC starting/consolidator grant to RC and by the Dutch Foundation for research on matter (FOM) via a FOM program (10TM01).
Materials
| Name of Material/ Equipment | Company | Catalog Number | Comments/Description |
| HisTrap HP | GE Healthcare | 17-5247-01 | |
| Nylon cloth | 20 μm pores | ||
| Soft artists paint brush | |||
| NONIDET P-40 | Sigma | 74385 | |
| Beta-DM | Sigma | D4641 | |
| DNAase | ThermoScientific | EN0525 | |
| Milk Powders | |||
| RNAase | ThermoScientific | EN0531 | |
| Sonicator | |||
| Octyl β-D-glucopyranoside | Sigma | O8001 | |
| Ultracentrifuge XL | Beckman-Coulter | ||
| TAP medium | see reference 17 | ||
| LB medium | see reference 19 |
References
- Plumley, F. G., Schmidt, G. W. Reconstitution of chlorophyll a/b light-harvesting complexes: Xanthophyll-dependent assembly and energy transfer. Proceedings of the National Academy of Sciences of the United States of America. 84, 146-150 (1987).
- Croce, R., van Amerongen, H. Light-harvesting and structural organization of Photosystem II from individual complexes to thylakoid membrane. Journal of photochemistry and photobiology B Biology. 104 (1-2), 142-153 (2011).
- Li, Z., Wakao, S., Fischer, B. B., Niyogi, K. K. Sensing and responding to excess light. Annual review of plant biology. 60, 239-260 (2009).
- De Bianchi, S., Ballottari, M., Dall’osto, L., Bassi, R. Regulation of plant light harvesting by thermal dissipation of excess energy. Biochemical Society transactions. 38 (2), 651-660 (2010).
- Neilson, J. A. D., Durnford, D. G. Structural and functional diversification of the light-harvesting complexes in photosynthetic eukaryotes. Photosynthesis research. 106 (1-2), 57-71 (2010).
- Paulsen, H., Rümler, U., Rüdiger, W. Reconstitution of pigment-containing complexes from light-harvesting chlorophyll a/b-binding protein overexpressed in Escherichia coli. Planta. 181 (2), 204-211 (1990).
- Horn, R., Grundmann, G., Paulsen, H. Consecutive binding of chlorophylls a and b during the assembly in vitro of light-harvesting chlorophyll-a/b protein (LHCIIb). Journal of molecular biology. 366 (3), 1045-1054 (2007).
- Cammarata, K. V., Schmidt, G. W. In vitro reconstitution of a light-harvesting gene product: deletion mutagenesis and analyses of pigment binding. Biochemistry. 31 (10), 2779-2789 (1992).
- Paulsen, H., Hobe, S. Pigment-binding properties of mutant light-harvesting chlorophyll-a/b-binding protein. European journal of biochemistry / FEBS. 205 (1), 71-76 (1992).
- Bassi, R., Croce, R., Cugini, D., Sandonà, D. Mutational analysis of a higher plant antenna protein provides identification of chromophores bound into multiple sites. Proceedings of the National Academy of Sciences of the United States of America. 96 (18), 10056-10061 (1999).
- Paulsen, H., Finkenzeller, B., Kühlein, N. Pigments induce folding of light-harvesting chlorophyll a/b-binding protein. European journal of biochemistry / FEBS. 215 (3), 809-816 (1993).
- Caffarri, S., Croce, R., Cattivelli, L., Bassi, R. A look within LHCII differential analysis of the Lhcb1-3 complexes building the major trimeric antenna complex of higher-plant photosynthesis. Biochemistry. 43 (29), 9467-9476 (2004).
- Giuffra, E., Cugini, D., Croce, R., Bassi, R. Reconstitution and pigment-binding properties of recombinant CP29. European journal of biochemistry / FEBS. 238 (1), 112-120 (1996).
- Rogl, H., Kosemund, K., Kühlbrandt, W., Collinson, I. Refolding of Escherichia coli produced membrane protein inclusion bodies immobilised by nickel chelating chromatography. FEBS letters. (1-2), 21-26 (1998).
- Porra, R. J., Thompson, W. A., Kriedemann, P. E. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica et Biophysica Acta (BBA) – Bioenergetics. 975 (3), 384-394 (1989).
- Davies, B. H. Identification of carotenoids by their absorption characteristics. Biochem J. 103 (2), (1967).
- Gorman, D. S., Levine, R. P. Cytochrome f and plastocyanin their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proceedings of the National Academy of Sciences of the United States of America. 54 (6), 1665-1669 (1965).
- Bertani, G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. Journal of bacteriology. 62 (3), 293-300 (1951).
- Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical biochemistry. 72, 248-254 (1976).
- Wientjes, E., Croce, R. The light-harvesting complexes of higher-plant Photosystem I Lhca1/4 and Lhca2/3 form two red-emitting heterodimers. The Biochemical journal. 433 (3), 477-485 (2011).
- Passarini, F., Xu, P., Caffarri, S., Hille, J., Croce, R. Towards in vivo mutation analysis knock-out of specific chlorophylls bound to the light-harvesting complexes of Arabidopsis thaliana – the case of CP24 (Lhcb6). Biochimica et biophysica acta. , (2014).
- Georgakopoulou, S., van der Zwan, G., Bassi, R., van Grondelle, R., van Amerongen, H., Croce, R. Understanding the changes in the circular dichroism of light harvesting complex II upon varying its pigment composition and organization. Biochemistry. 46 (16), 4745-4754 (2007).
- Croce, R., Müller, M. G., Caffarri, S., Bassi, R., Holzwarth, A. R. Energy transfer pathways in the minor antenna complex CP29 of photosystem II a femtosecond study of carotenoid to chlorophyll transfer on mutant and WT complexes. Biophysical journal. 84 (4), 2517-2532 (2003).
- Schmid, V. H., Cammarata, K. V., Bruns, B. U., Schmidt, G. W. In vitro reconstitution of the photosystem I light-harvesting complex LHCI-730 heterodimerization is required for antenna pigment organization. Proceedings of the National Academy of Sciences of the United States of America. 94 (14), 7667-7672 (1997).
- Castelletti, S., Morosinotto, T., Robert, B., Caffarri, S., Bassi, R., Croce, R. Recombinant Lhca2 and Lhca3 subunits of the photosystem I antenna system. Biochemistry. 42 (14), 4226-4234 (2003).
- Storf, S., Jansson, S., Schmid, V. H. R. Pigment binding, fluorescence properties, and oligomerization behavior of Lhca5, a novel light-harvesting protein. The Journal of biological chemistry. 280 (7), 5163-5168 (2005).
- Mozzo, M., Mantelli, M., Passarini, F., Caffarri, S., Croce, R., Bassi, R. Functional analysis of photosystem I light-harvesting complexes (Lhca) gene products of Chlamydomonas reinhardtii. Biochimica et biophysica acta. 1797 (2), 212-221 (2010).
- Remelli, R., Varotto, C., Sandonà, D., Croce, R., Bassi, R. Chlorophyll binding to monomeric light-harvesting complex. A mutation analysis of chromophore-binding residues. The Journal of biological chemistry. 274 (47), 33510-33521 (1999).
- Passarini, F., Wientjes, E., Hienerwadel, R., Croce, R. Molecular basis of light harvesting and photoprotection in CP24 unique features of the most recent antenna complex. The Journal of biological chemistry. 284 (43), 29536-29546 (2009).
- Giuffra, E., et al. Analysis of some optical properties of a native and reconstituted photosystem II antenna complex, CP29 pigment binding sites can be occupied by chlorophyll a or chlorophyll b and determine spectral forms. Biochemistry. 36 (42), 12984-12993 (1997).
- Pagano, A., Cinque, G., Bassi, R. In vitro reconstitution of the recombinant photosystem II light-harvesting complex CP24 and its spectroscopic characterization. The Journal of biological chemistry. 273 (27), 17154-17165 (1998).
- Kleima, F. J., et al. Decreasing the chlorophyll a/b ratio in reconstituted LHCII structural and functional consequences. Biochemistry. 38 (20), 6587-6596 (1999).
- Croce, R., Morosinotto, T., Castelletti, S., Breton, J., Bassi, R. The Lhca antenna complexes of higher plants photosystem I. Biochimica et biophysica acta. 1556 (1), 29-40 (2002).
- Hobe, S., Trostmann, I., Raunser, S., Paulsen, H. Assembly of the major light-harvesting chlorophyll-a/b complex Thermodynamics and kinetics of neoxanthin binding. The Journal of biological chemistry. 281 (35), 25156-25166 (2006).
- Croce, R., Weiss, S., Bassi, R. Carotenoid-binding sites of the major light-harvesting complex II of higher plants. The Journal of biological chemistry. 274 (42), 29613-29623 (1999).
- Hobe, S., Niemeier, H., Bender, A., Paulsen, H. Carotenoid binding sites in LHCIIb. Relative affinities towards major xanthophylls of higher plants. European journal of biochemistry / FEBS. 267 (2), 616-624 (2000).
- Jahns, P., Depka, B., Trebst, A. Xanthophyll cycle mutants from Chlamydomonas reinhardtii indicate a role for zeaxanthin in the D1 protein turnover. Plant Physiology and Biochemistry. 38 (5), 371-376 (2000).
- Wehner, A., Grasses, T., Jahns, P. De-epoxidation of violaxanthin in the minor antenna proteins of photosystemII, LHCB4, LHCB5, and LHCB6. The Journal of biological chemistry. 281 (31), 21924-21933 (2006).
- Croce, R., Canino, G., Ros, F., Bassi, R. Chromophore organization in the higher-plant photosystem II antenna protein CP26. Biochemistry. 41 (23), 7334-7343 (2002).
- Caffarri, S., Passarini, F., Bassi, R., Croce, R. A specific binding site for neoxanthin in the monomeric antenna proteins CP26 and CP29 of Photosystem II. FEBS letters. 581 (24), 4704-4710 (2007).
- Hobe, S., Förster, R., Klingler, J., Paulsen, H. N-proximal sequence motif in light-harvesting chlorophyll a/b-binding protein is essential for the trimerization of light-harvesting chlorophyll a/b complex. Biochemistry. 34 (32), 10224-10228 (1995).
- Kuttkat, A., Hartmann, A., Hobe, S., Paulsen, H. The C-terminal domain of light-harvesting chlorophyll-a/b-binding protein is involved in the stabilisation of trimeric light-harvesting complex. European journal of biochemistry / FEBS. 242 (2), 288-292 (1996).
- Rupprecht, J., Paulsen, H., Schmid, V. H. Protein domains required for formation of stable monomeric Lhca1- and Lhca4-complexes. Photosynthesis research. 63 (3), 217-224 (2000).
- Yang, C., et al. The negatively charged amino acids in the lumenal loop influence the pigment binding and conformation of the major light-harvesting chlorophyll a/b complex of photosystem II. Biochimica et biophysica acta. 1777 (11), 1463-1470 (2008).
- Rogl, H., Kühlbrandt, W. Mutant trimers of light-harvesting complex II exhibit altered pigment content and spectroscopic features. Biochemistry. 38 (49), 16214-16222 (1999).
- Yang, C., Kosemund, K., Cornet, C., Paulsen, H. Exchange of pigment-binding amino acids in light-harvesting chlorophyll a/b protein. Biochemistry. 38 (49), 16205-16213 (1999).
- Morosinotto, T., Castelletti, S., Breton, J., Bassi, R., Croce, R. Mutation analysis of Lhca1 antenna complex. Low energy absorption forms originate from pigment-pigment interactions. The Journal of biological chemistry. 277 (39), 36253-36261 (2002).
- Morosinotto, T., Breton, J., Bassi, R., Croce, R. The nature of a chlorophyll ligand in Lhca proteins determines the far red fluorescence emission typical of photosystem I. The Journal of biological chemistry. 278 (49), 49223-49229 (2003).
- Ballottari, M., Mozzo, M., Croce, R., Morosinotto, T., Bassi, R. Occupancy and functional architecture of the pigment binding sites of photosystem II antenna complex Lhcb5. The Journal of biological chemistry. 284 (12), 8103-8113 (2009).
- Croce, R., et al. Origin of the 701-nm fluorescence emission of the Lhca2 subunit of higher plant photosystem I. The Journal of biological chemistry. 279 (47), 48543-48549 (2004).
- Morosinotto, T., Mozzo, M., Bassi, R., Croce, R. Pigment-pigment interactions in Lhca4 antenna complex of higher plants photosystem I. The Journal of biological chemistry. 280 (21), 20612-20619 (2005).
- Mozzo, M., Morosinotto, T., Bassi, R., Croce, R. Probing the structure of Lhca3 by mutation analysis. Biochimica et biophysica acta. 1757 (12), 1607-1613 (2006).
- Mozzo, M., Passarini, F., Bassi, R., van Amerongen, H., Croce, R. Photoprotection in higher plants the putative quenching site is conserved in all outer light-harvesting complexes of photosystem II. Biochimica et biophysica acta. 1777 (10), 1263-1267 (2008).
- Wientjes, E., Roest, G., Croce, R. From red to blue to far-red in Lhca4 how does the protein modulate the spectral properties of the pigments. Biochimica et biophysica acta. 1817 (5), 711-717 (2012).

