A Colorimetric Assay that Specifically Measures Granzyme B Proteolytic Activity: Hydrolysis of Boc-Ala-Ala-Asp-S-Bzl
Summary
We describe a simple, quantitative colorimetric assay that specifically measures the proteolytic activity of human, mouse or rat Granzyme B (GzmB). This protocol can be easily adapted for determining protease activity of other granule serine proteases by the hydrolysis of other synthetic peptide substrates with an appropriate recognition sequence.
Abstract
The serine protease Granzyme B (GzmB) mediates target cell apoptosis when released by cytotoxic T lymphocytes (CTL) or natural killer (NK) cells. GzmB is the most studied granzyme in humans and mice and therefore, researchers need specific and reliable tools to study its function and role in pathophysiology. This especially necessitates assays that do not recognize proteases such as caspases or other granzymes that are structurally or functionally related. Here, we apply GzmB’s preference for cleavage after aspartic acid residues in a colorimetric assay using the peptide thioester Boc-Ala-Ala-Asp-S-Bzl. GzmB is the only mammalian serine protease capable of cleaving this substrate. The substrate is cleaved with similar efficiency by human, mouse and rat GzmB, a property not shared by other commercially available peptide substrates, even some that are advertised as being suitable for this purpose. This protocol is demonstrated using unfractionated lysates from activated NK cells or CTL and is also suitable for recombinant proteases generated in a variety of prokaryotic and eukaryotic systems, provided the correct controls are used. This assay is a highly specific method to ascertain the potential pro-apoptotic activity of cytotoxic molecules in mammalian lymphocytes, and of their recombinant counterparts expressed by a variety of methodologies.
Introduction
Granzymes are a family of serine proteases found in the secretory lysosomes of natural killer (NK) cells and cytotoxic T lymphocytes (CTL) 1. Five different granzymes exist in humans (A, B, H, K and M), and ten in mice (A – G, K, M and N) 2,3. Granzyme A and Granzyme B (GzmA, GzmB) are the most abundant and have been extensively investigated in the human and rodent setting.
The classic function of GzmB is the induction of apoptosis in target cells executed in conjunction with the pore-forming protein perforin, which permits the granzyme to access the target cell cytosol 4. Although GzmB expression is unequivocally found in cytotoxic lymphocytes, recent studies have been addressing a variety of other GzmB-expressing cell types, including but not limited to keratinocytes 5, basophils 6, mast cells 7, plasmacytoid dendritic cells 8, and B cells 9,10. In this context, non-apoptotic GzmB functions were revealed ranging from participation in inflammatory processes, tissue remodelling and other immunoregulatory properties 11-14.
Given that a broader biological role has been proposed for GzmB than previously suspected, researchers require reliable and specific tools for its detection. Of advantage is GzmB’s specific requirement to cleave on the carboxyl side of aspartic acid residues, a property unique among eukaryotic serine proteases 15. Mouse, human and rat GzmB are structurally very similar, however the extended substrate specificity of mouse GzmB differs subtly from that of both human and rat 16, which means that certain generic substrates with Asp at the terminal (P1) can be cleaved efficiently by GzmB from all three species, whereas other substrates with more complicated sequences upstream of P1 may give widely variable results. In both the past and more recent literature, this fact has caused considerable confusion and misinterpretation of the biological significance of some experimental findings, even though carefully controlled, kinetic studies have sought to correct the situation 17.
In this paper we have sought to illustrate these points using two commercially available substrates, namely Boc-Ala-Ala-Asp-SBzl and N-acetyl-Ile-Glu-Pro-Asp-p-nitroanilide. The two reagents do generate different reactive groups following cleavage (a free sulphydryl versus a fluorescent free paranitroanilide), but this has no effect whatever on proteolytic cleavage. The described protocol is a modern adaption of a very old protocol 18, but should help investigators to use the different GzmB substrates appropriately, while also providing a methodological framework for detecting the activity of other granzymes, such as GzmA and GzmH.
Protocol
NOTE: Spleens were derived from mice (6-10 weeks of age) and all animal experiments were performed according to the animal ethics guidelines of the Peter MacCallum Cancer Centre (E486).
1. Preparation of Samples.
- Preparation of activated mouse NK cells.
- Isolate primary naive NK cells from single-cell suspensions of the spleens of C57BL/6 or B6.GrzmB-/- (GzmB gene null) mice by negative selection using commercially available kits.
- Follow the manufacturers’ protocol. Briefly, magnetically label “non-NK” cells such as T and B cells, dendritic cells, granulocytes, macrophages, and red blood cells using biotin-conjugated antibodies against their specific surface markers. Use magnetic separation with anti-Biotin beads to deplete “non-NK” cells.
- Culture isolated NK cells for 5-7 days in media containing IL-2 (1,000 U/ml) at 700,000 cells/ml in 24-well plates at 37 °C.
- Alternatively, use activated T cells 19, primary antigen-restricted CTL from OT1 T cell receptor transgenic mice 20, CTL generated in mixed lymphocyte cultures 21, or supernatants from activated NK cells to determine the activity of secreted GzmB 19.
- Maintenance of human NK cell line and isolation of primary NK cells.
- Maintain human NK leukemia (YT cells) in Roswell Park Memorial Institute (RPMI) medium supplemented with 10% fetal calf serum. Alternatively, purify primary NK cells from the peripheral blood of healthy subjects and stimulate for 2-3 days in medium containing IL-2 at 100 U/ml as previously described 22.
- Generation of cell lysates.
- Wash cells three times in PBS (pH 7.4), then suspend in Nonidet P-40 lysis buffer (0.1% NP-40, 250 mM NaCl, 25 mM Hepes, 2.5 mM EDTA). Ideally, lyse a minimum of 5 x 106 NK cells, or 1 x 107 T cells in 100 µl buffer. Do not add protease inhibitors, as many are irreversible inhibitors of serine proteases, and in particular tryptases such as GzmA. Incubate on ice for 20 min, and then pellet the nuclei in a microcentrifuge at 15,000 x g for 10 min. Discard the nuclear pellet and transfer the supernatant to a fresh tube.
- Determine the protein concentration of the lysates.
- Use a commercially available protocol of choice such as, Bradford or bicinchoninic acid (BCA) and determine the protein concentration. Ensure that the concentration is ~2 mg/ml minimum. Store lysates at -20 °C until required (granzyme activity is stable for many months at -20 °C).
2. Granzyme B Activity Assay
- Preparation of Reagents
- Prepare a 10 mM stock of the substrates (Boc-AAD-S-Bzl or Ac-IEPD-pNA) in DMSO (~5 mg/ml) and store in aliquots at -20 °C. Prepare a working dilution freshly and discard after each day, as Boc-AAD-S-Bzl partially hydrolyzes on storage in DMSO.
- Prepare a 250 mM stock of the colorimetric reagent (5,5’-dithio-bis(2-nitrobenzoic acid), DTNB) in DMSO (0.09 g/ml) and store at -20 °C. Prepare a fresh working dilution each day. This reagent is only required for substrates with SBzl indicator groups, not pNA.
- Make up fresh buffer (0.1M HEPES, 0.05 M MgCl2, pH 7.3) every 2-3 weeks.
- Bring DTNB and synthetic substrates to room temperature. Dilute substrates (1:16, e.g. 100 µl in 1.6 ml of buffer) and DTNB (1:250, e.g. 10 µl in 2.5 ml buffer) and mix well.
- Dilute a minimum of 100 µg protein lysate of each sample in buffer to a final volume of 160 µl. Using a multichannel pipette add 50 µl per well in triplicate (~33.3 µg/well) in a ‘‘U’’ bottom vinyl 96-well plate. Include a “buffer-only” control (also in triplicate).
- Add 100 µl of diluted DTNB to the samples/buffer control (omit this step when working with Ac-IEPD-pNA.
NOTE: Cleavage of Ac-IEPD-pNA by GzmB directly releases fluorogenic pNA (Figure 4B). - Use a multichannel pipette to quickly add 50 µl of diluted substrates to each set of triplicates, starting with the buffer then any negative controls and finishing with expected positive samples.
- Place the plate in the plate reader. Measure Asp-ase activity by hydrolysis of Boc-Ala-Ala-Asp-S-Bzl/Ac-IEPD-pNA in a microplate reader at OD 405 nm. Use a kinetic assay protocol, take 25 readings, every 15 sec (ca. 6 min total reading time).
- Use the absorbance readings generated at each time point to manually calculate the rate of substrate cleavage if the plate-reader’s software cannot generate a value for Vmax.
3. Analysis of Data
NOTE: The data generated is presented as maximum velocity, defined as change of OD over time (mOD/min).
- To analyze the data, determine the maximum rate of substrate cleavage, which is calculated from the rate of change in absorbance. Do this automatically using the plate reader software, which typically has this option.
- Alternatively after viewing the kinetic plots of change in absorbance, which are generated during the kinetic assay on the plate reader, manually select the data points which give the steepest straight line. For most assays the maximum rate is achieved within the first 2 min of the assay.
- Subtract the initial OD reading from the highest OD reading on this straight line and divide by the time interval. Transfer raw data to Microsoft Excel/Graph pad for statistical analysis and graphic display.
Representative Results
The Boc-AAD-S-Bzl substrate is specific for GzmB
The serine protease GzmB is a major constituent of cytotoxic lymphocytes (CTL and NK cells) and is predominantly responsible for inducing rapid apoptotic death in target cells, such as virus-infected or transformed cells. This is largely due to its substrate preference for cleavage after certain specific aspartate residues in selected proteins, an attribute shared with the caspases, which also cleave after aspartate residues but are from a different class of proteases, the cysteine protease family 15. This difference in the proteolytic mechanism means that AAD-SBzl cannot be cleaved by any caspase. GzmB expression is induced following activation of cytotoxic lymphocytes and enzymatic activity can be monitored in cell lysates prepared from these cells (Figure 1). The availability of granzyme gene knockout mice has allowed us to establish that the synthetic peptide substrate Boc-Ala-Ala-Asp (AAD)- thiobenzyl ester (S-Bzl) is specifically hydrolysed by GzmB. Robust cleavage of the synthetic substrate can be measured in a cell lysate from B6 primary NK cells, but not in B6.GrzmB-/- NK cells. As a control, the proteolytic activity of GzmA, which has trypsin-like (tryptase) substrate specificity and is present in both lysates was equivalent, as measured by the hydrolysis of N-α-CBZ-L-lysine (BLT)-S-Bzl.
Species-specific GzmB substrate recognition sequence
Early studies describing potential target cell molecules cleaved by GzmB caused some confusion until it emerged that mouse and human GzmB have a slightly altered substrate recognition preference, which is dictated by as many as 8-10 residues mapping surrounding the P1 aspartate 23. This was particularly evident in studies that examined cleavage of the pro-apoptotic BH3–only molecule Bid, which can be cleaved efficiently by human, but far less efficiently by mouse GzmB. In intact cells, this difference results in the activation of different cell death pathways, with human GzmB operating preferentially through the mitochondrial pathway, and mouse GzmB via direct caspase activation. Analysis of tetrapeptide sequences revealed that human but not mouse GzmB could cleave after the sequence IEPD, which matches the recognition sequence in both human and mouse Bid 16. In both the literature and in commercial product catalogues the tetrapeptide substrate, Ac-IEPD-pNA has been used generically to test for GzmB activity (Table 1). It is now clear that whilst both human and mouse can cleave Boc–AAD-S-Bzl, with very similar efficiency, mouse GzmB in NK cell lysates fails to efficiently hydrolyse Ac-IEPD-pNA (Figure 2). In contrast, decreasing the amount of human NK YT lysate from 30 µg to 10 µg still resulted in sufficient GzmB to hydrolyse the Ac-IEPD-pNA substrate (Figure 3).
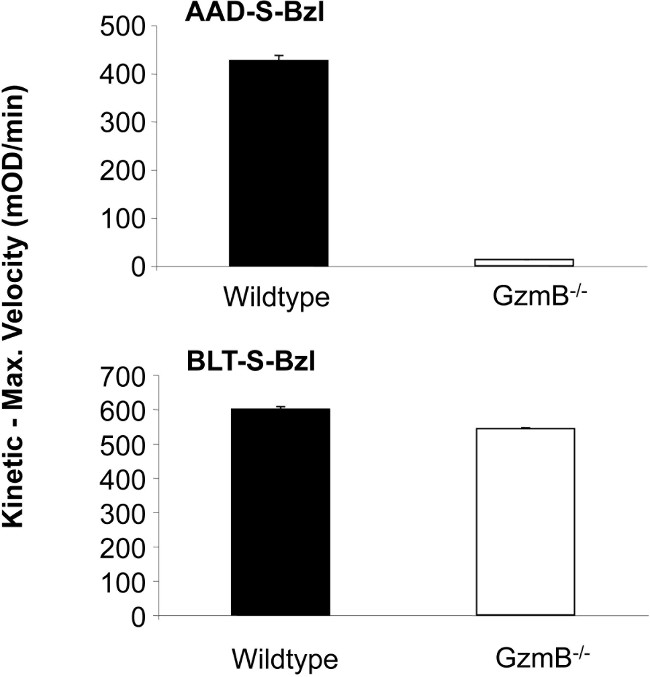
Figure 1: Granule enzyme activity in murine NK cells. Granzyme B (upper panel) and granzyme A (lower panel)activity in NK cell lysates from B6 wildtype and B6.GrzmB knockout mice was determined by the maximum rate of hydrolysis of the Boc-Ala-Ala Asp-S-Bzl and N-α-CBZ-L-lysine-S-Bzl peptide substrates. Data depict one representative experiment performed in triplicate (n >3).
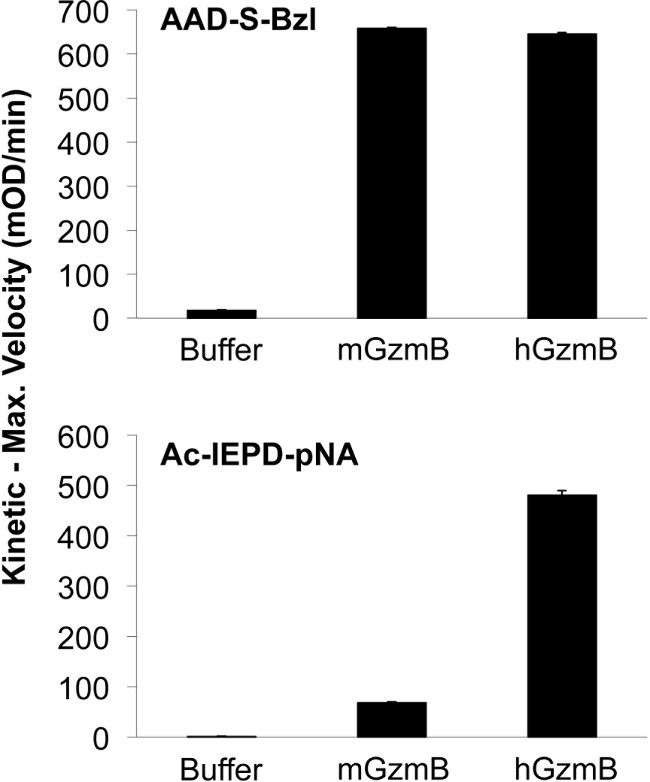
Figure 2: Species-specific hydrolysis of Ac-IEPD- pNA substrate. Mouse and human GzmB hydrolysed Boc-Ala-Ala-Asp-S-Bzl equally but only human GzmB hydrolysed Ac-IEPD-pNA. The human NK cell line YT and primary IL-2 activated mouse NK were used as a source of GzmB. Bar graph shows one representative experiment performed in triplicate (n >3).
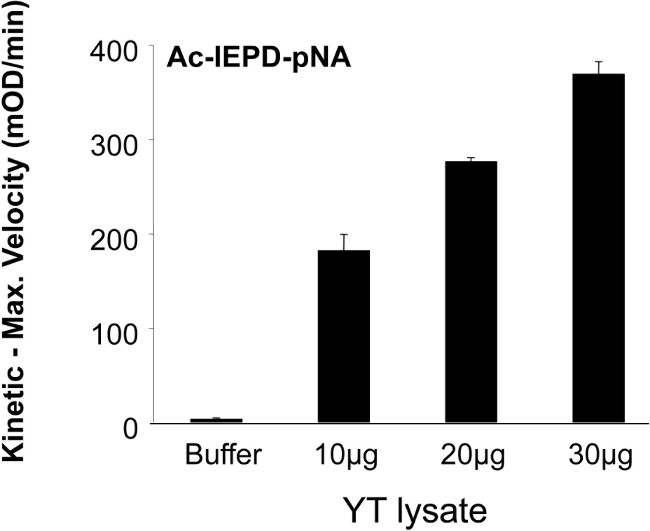
Figure 3: Reduction in protein concentration did not abrogate GzmB activity. The rate of hydrolysis of Ac-IEPD-pNA induced by human GzmB was compared using decreasing protein concentration of YT NK cell line lysate. Experiment representative for n >3.
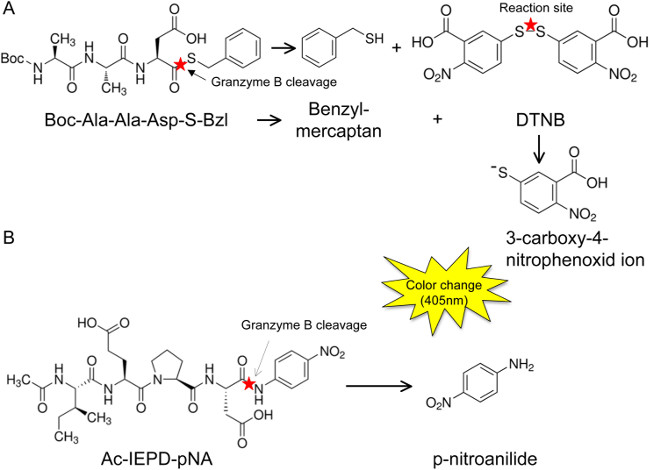
Figure 4: Graphical depiction of the assay principle. GzmB cleaves the substrates Boc-AAD-S-Bzl (A) and Ac-IEPD-pNA (B) after the P1 aspartate, thereby releasing either a benzyl mercaptan, which in turn interacts with DTNB to form a fluorogenic 3-carboxy-4-nitrophenoxide ion (A), or by directly releasing p-nitroanilide, which is a chromophore itself (B). Fluorescence emissions of both these molecules can be detected at 405 nm.
| Serine protease | Substrate | Activity |
| Granzyme A | N-α-CBZ-L-lysine (BLT) thiobenzyl ester (S-Bzl) | Tryptase22 |
| Granzyme B | 1. Boc-Ala-Ala-Asp (AAD)-S-Bzl | ASPase14 |
| 2. N-acetyl-Ile-Glu-Pro-Asp (IEPD)-p-nitroanilide (pNA) | ||
| Granzyme H | Suc-Phe-Leu-Phe-S-Bzl | Chymase23 |
Table 1: List of substrates available for detection of granzyme activity. Tryptase activity of GzmA can be detected by hydrolysis of the specific N-α-CBZ-L-lysine (BLT) thiobenzyl ester (S-Bzl). GzmB specifically cleaves after aspartate residues (Asp-ase), which can be assessed by hydrolysis of either Boc-Ala-Ala-Asp (AAD)-S-Bzl (detects mouse and human GzmB activity), or N-acetyl-Ile-Glu-Pro-Asp (IEPD)-p-nitroanilide (pNA) (detects only human GzmB activity). Chymase activity of GzmH is assessed by cleavage of Suc-Phe-Leu-Phe-S-Bzl.
Discussion
Historically the granzymes were identified as key effector molecules of cytotoxic lymphocytes (CTL and NK cells) capable of inducing a rapid apoptotic death in target cells. This was principally due to the action of GzmB, which cleaved target substrate molecules at aspartate (D) residues and thus was able to activate the caspase cascade by both cleaving pro-caspases, as well as several of their downstream targets. However, it is now appreciated that GzmB expression is not confined to lymphoid cytotoxic cells and its function may be extended well beyond target cell recognition and cell death.
In this paper we describe a protocol that enables the detection of active GzmB in cell lysates derived from both mouse and human CTL/NK cells using one simple colorimetric assay. The simplicity and specificity of this assay as ascertained by these findings allow the application of this protocol to examine GzmB in cellular samples from other sources. However, it is important that the cellular lysates have a protein concentration of at least 2mg/ml, and appropriate positive and negative controls are included in the experimental protocol. The specificity of the Boc-AAD-SBzl reagent for GzmB is evident from the lack of substrate–cleavage activity in the lysate derived from GzmB null animals, as activity is completely lost (Figure 1). In any future studies it is recommended that such a negative control (s. protocol section 1.1) is used to verify that any activity detected is specific for GrzB. (Note: Although like GrB, caspases have an absolute specificity for cleavage at aspartate residues in the P1 position, preferred substrates are tetrapepetides where the P4 amino acid is also a critical determinant of specificity 24.). If examining recombinant material the active site serine to alanine mutant protease (inactive protease prepared in exactly the same way as the active enzyme) should be included, and this is particularly important if assaying non-mammalian cell lysates, particularly those from yeast or bacterial cells.
Using the above protocol for assays of mammalian cells, GzmB activity is not diminished by the presence of endogenous cognate serine protease inhibitors (serpins), which can be greatly up-regulated in activated cytotoxic lymphocytes (CL) 25, as well as in cells derived from non-immune tissues, including transformed cells 26. The cytosolic serpinB9, (formerly PI-9) is highly specific for human GzmB; however in the mouse, there are several close but les specific orthologues of which serpin b9, or SPI-6 has significant inhibitory activity for mouse GzmB 27. Although lysis in NP-40 buffer can be permissive for generating an irreversible interaction between GzmB and serpin, incubation at 37 °C is required for optimal complex formation 28. We do not detect complex formation in the lysates by western blot, which would result in loss of protease activity. This protocol is carried out at typical ambient laboratory temperature, up to ~22 °C.
From more than 20 years of experience performing these assays we have found that the commercial source of the Boc-AAD-SBzl reagent is of upmost importance for obtaining consistent, sensitive and reliable results. We would only recommend the supplier we have referenced, although other commercial sources are also suitable for the substrates required to detect the activity of other granzymes. The protocol described in this paper can be easily adapted for determining protease activity of other granule serine proteases by the hydrolysis of synthetic peptide substrates with an appropriate recognition sequence as listed in Table 1. The N-α-CBZ-L-lysine-S-Bzl substrate can be used to detect the tryptase activity of both GzmA in mouse and human CL and theoretically GzmK in mouse CL. Although GzmK mRNA expression has been demonstrated by RT-PCR in killer cells generated from GzmAB gene knockout mice in response to influenza peptides 29, we are not aware of GzmK protease activity having been reported in this or any other physiologically relevant context. The activity of either recombinant human GzmH, or the protease purified from NK cells, can be assessed with Suc-Phe-Leu-Phe-S-Bzl 30, however it is not possible to ascribe the hydrolysis of this substrate specifically to this granzyme in cell lysates. Lymphocytes contain a number of other endolysosomal proteases such as the cathepsins, which will also have chymotrypsin-like (chymase) activity. In the mouse, the GzmH gene is replaced by a cluster of genes (Gzms C-F) all predicted to have similar (chymase) activity.
Although human GzmB was found to effectively cleave the BH3-only pro-apoptotic molecule Bid, and thus directly engage the mitochondrial death pathway 31, apparently conflicting results were initially obtained with mouse GzmB. This confusion was resolved by elegant studies by a number of investigators, which determined that the preferred substrate recognition sequence differed between mouse and human (and rat) GzmB, despite the high degree (~80%) of overall sequence homology of the proteases 16,17,23. The optimal recognition sequence for human GzmB is I/V, E/M/Q, P/Xaa (S/T) and D in the P1 position whereas for mouse GzmB it is L/I/V, E, F/Y/Q, D 16,17. The major difference was at the P2 position, as P in particular is not tolerated by mouse GzmB, hence the mouse Bid (IEPD 75) cleavage site would not be accommodated by the GzmB substrate binding cleft. Furthermore, the synthetic peptide substrates IEPD-pNA, routinely described as specific for GzmB, and IETD-pNA, are poorly hydrolyzed by mouse GzmB. We have further consolidated the finding made by others 23 using recombinant granzymes that human GzmB, but not GzmB present in mouse NK and CTL hydrolyse IETD-pNA and IEPD-pNA. Kaiserman et al. 17 compared the cleavage activity of recombinant human and mouse GrB produced in Pichia pastoris, using Boc-AAD-SBzl with the Ac-IETD–pNA substrates. Similarly, the authors found that whilst both enzymes cleaved Boc-AAD-SBzl with comparable efficiency, hydrolysis of Ac-IETD–pNA by mouse GrB was 30-fold less efficient. Whilst T and P (as well as S) in the P2 position are preferred by human GzmB, they are not tolerated mouse GzmB. By contrast, we clearly demonstrated comparable GzmB activity in both human and mouse NK cell lysates using the more generic Boc-AAD-SBzl reagent. However, only human GzmB activity could be detected with the IEPD-pNA reagent.
Together these findings highlight the fact that differences of the fine specificity of GzmB of different species can have a major impact on how to choose an appropriate substrate for in vitro analysis. In summary, if detection of mouse, human or rat GzmB activity is investigated, Boc-AAD-SBzl is the substrate of choice.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work received support through grant HA 6136/1-1 from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) to MH. JAT is supported by Program and Project Grants from the National Health and Medical Research Council of Australia.
Materials
| Product | Company | Catalogue number | Comment/description |
| Boc-Ala-Ala-Asp-S-Bzl | SM Biochemicals LLC, CA | SMSB05 | Granzyme B substrate (mouse and human) |
| Ac-IEPD-pNA | SM Biochemicals LLC, CA | SMPNA009 | Granzyme B substrate (only human) |
| N-a-CBZ-L-lysine-S-Bzl | Sigma-Aldrich | C3647 | Granzyme A substrate |
| Suc-Phe-Leu-Phe-S-Bzl | SM Biochemicals LLC, CA | SB025 | Granzyme H substrate |
| 5,5’-dithio-bis(2-nitrobenzoic acid) | Sigma-Aldrich | D8130 | DTNB, Ellman’s Reagent |
| NK cell isolation kit II mouse | Miltenyi Biotec GmbH | 130-096-892 | negative selection kit |
| NK cell isolation kit human | Miltenyi Biotec GmbH | 130-092-657 | negative selection kit |
| Plate reader | Biorad iMark | Biorad Microplate Manager Software Version MPM6.3 | |
| Serocluster U-bottom vinyl 96-well plate | Corning, MA, USA | 2797 |
References
- Trapani, J. A., Browne, K. A., Dawson, M., Smyth, M. J. Immunopurification of functional Asp-ase (natural killer cell granzyme B) using a monoclonal antibody). Biochem Biophys Res Commun. 195 (2), 910-920 (1993).
- Ewen, C. L., Kane, K. P., Bleackley, R. C. A quarter century of granzymes. Cell Death Differ. 19 (1), 28-35 (2012).
- Hoves, S., Trapani, J. A., Voskoboinik, I. The battlefield of perforin/granzyme cell death pathways. J Leukoc Biol. , (2009).
- Peters, P. J., et al. Cytotoxic T lymphocyte granules are secretory lysosomes, containing both perforin and granzymes. J Exp Med. 173 (5), 1099-1109 (1991).
- Berthou, C., et al. Acquisition of granzyme B and Fas ligand proteins by human keratinocytes contributes to epidermal cell defense. J Immunol. 159 (11), 5293-5300 (1997).
- Tschopp, C. M., et al. Granzyme B, a novel mediator of allergic inflammation: its induction and release in blood basophils and human asthma. Blood. 108 (7), 2290-2299 (2006).
- Strik, M. C., et al. Human mast cells produce and release the cytotoxic lymphocyte associated protease granzyme B upon activation. Mol Immunol. 44 (14), 3462-3472 (2007).
- Jahrsdorfer, B., et al. Granzyme B produced by human plasmacytoid dendritic cells suppresses T cell expansion. Blood. , (2009).
- Hagn, M., et al. Human B cells secrete granzyme B when recognizing viral antigens in the context of the acute phase cytokine IL-21. J Immunol. 183 (3), 1838-1845 (2009).
- Hagn, M., et al. Human B cells differentiate into granzyme B-secreting cytotoxic B lymphocytes upon incomplete T-cell help. Immunol Cell Biol. 90 (4), 457-467 (2012).
- Froelich, C. J., Pardo, J., Simon, M. M. Granule-associated serine proteases: granzymes might not just be killer proteases. Trends Immunol. 30 (3), 117-123 (2009).
- Hagn, M., Jahrsdorfer, B. Why do human B cells secrete granzyme B? Insights into a novel B-cell differentiation pathway. Oncoimmunology. 1 (8), 1368-1375 (2012).
- Susanto, O., Trapani, J. A., Brasacchio, D. Controversies in granzyme biology. Tissue Antigens. 80 (6), 477-487 (2012).
- Walch, M., et al. Cytotoxic cells kill intracellular bacteria through granulysin-mediated delivery of granzymes. Cell. 157 (6), 1309-1323 (2014).
- Odake, S., et al. Human and murine cytotoxic T lymphocyte serine proteases: subsite mapping with peptide thioester substrates and inhibition of enzyme activity and cytolysis by isocoumarins. Biochemistry. 30 (8), 2217-2227 (1991).
- Casciola-Rosen, L., et al. Mouse and human granzyme B have distinct tetrapeptide specificities and abilities to recruit the bid pathway. J Biol Chem. 282 (7), 4545-4552 (2007).
- Kaiserman, D., et al. The major human and mouse granzymes are structurally and functionally divergent. J Cell Biol. 175 (4), 619-630 (2006).
- Powers, J. C., Kam, C. M. Peptide thioester substrates for serine peptidases and metalloendopeptidases. Methods Enzymol. 248, 3-18 (1995).
- Hagn, M., et al. Activated mouse B cells lack expression of granzyme. B. J Immunol. 188 (2), 3886-3892 (2012).
- Konjar, S., et al. Human and mouse perforin are processed in part through cleavage by the lysosomal cysteine proteinase cathepsin L. Immunology. 131 (2), 257-267 (2010).
- Sutton, V. R., et al. Residual active granzyme B in cathepsin C-null lymphocytes is sufficient for perforin-dependent target cell apoptosis. J Cell Biol. 176 (4), 425-433 (2007).
- Trapani, J. A., Smyth, M. J., Apostolidis, V. A., Dawson, M., Browne, K. A. Granule serine proteases are normal nuclear constituents of natural killer cells. J Biol Chem. 269 (28), 18359-18365 (1994).
- Cullen, S. P., Adrain, C., Luthi, A. U., Duriez, P. J., Martin, S. J. Human and murine granzyme B exhibit divergent substrate preferences. J Cell Biol. 176 (4), 435-444 (2007).
- Thornberry, N. A., et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem. 272 (29), 17907-17911 (1997).
- Bird, C. H., et al. Selective regulation of apoptosis: the cytotoxic lymphocyte serpin proteinase inhibitor 9 protects against granzyme B-mediated apoptosis without perturbing the Fas cell death pathway. Mol Cell Biol. 18 (11), 6387-6398 (1998).
- Bots, M., Medema, J. P. Serpins in T cell immunity. J Leukoc Biol. 84 (5), 1238-1247 (2008).
- Sun, J., et al. A new family of 10 murine ovalbumin serpins includes two homologs of proteinase inhibitor 8 and two homologs of the granzyme B inhibitor (proteinase inhibitor 9). J Biol Chem. 272 (24), 15434-15441 (1997).
- Bird, C. H., Hitchen, C., Prescott, M., Harper, I., Bird, P. I. Immunodetection of granzyme B tissue distribution and cellular localisation. Methods Mol Biol. 844, 237-250 (2012).
- Jenkins, M. R., et al. Visualizing CTL activity for different CD8+ effector T cells supports the idea that lower TCR/epitope avidity may be advantageous for target cell killing. Cell Death Differ. 16 (4), 537-542 (2009).
- Edwards, K. M., Kam, C. M., Powers, J. C., Trapani, J. A. The human cytotoxic T cell granule serine protease granzyme H has chymotrypsin-like (chymase) activity and is taken up into cytoplasmic vesicles reminiscent of granzyme B-containing endosomes. J Biol Chem. 274 (43), 30468-30473 (1999).
- Sutton, V. R., et al. Initiation of apoptosis by granzyme B requires direct cleavage of bid, but not direct granzyme B-mediated caspase activation. J Exp Med. 192 (10), 1403-1414 (2000).

