Self-assembly of Complex Two-dimensional Shapes from Single-stranded DNA Tiles
Summary
DNA tiling is an effective approach to make programmable nanostructures. We describe the protocols to construct complex two-dimensional shapes by the self-assembly of single-stranded DNA tiles.
Abstract
Current methods in DNA nano-architecture have successfully engineered a variety of 2D and 3D structures using principles of self-assembly. In this article, we describe detailed protocols on how to fabricate sophisticated 2D shapes through the self-assembly of uniquely addressable single-stranded DNA tiles which act as molecular pixels on a molecular canvas. Each single-stranded tile (SST) is a 42-nucleotide DNA strand composed of four concatenated modular domains which bind to four neighbors during self-assembly. The molecular canvas is a rectangle structure self-assembled from SSTs. A prescribed complex 2D shape is formed by selecting the constituent molecular pixels (SSTs) from a 310-pixel molecular canvas and then subjecting the corresponding strands to one-pot annealing. Due to the modular nature of the SST approach we demonstrate the scalability, versatility and robustness of this method. Compared with alternative methods, the SST method enables a wider selection of information polymers and sequences through the use of de novo designed and synthesized short DNA strands.
Introduction
Previous nucleic acid self-assembly work1-25 has led to the successful construction of a variety of complex structures, including DNA2–5,8,10–13,17,23 or RNA7,22 periodic3,4,7,22 and algorithmic5 two-dimensional lattices, ribbons10,12 and tubes4,12,13, 3D crystals17, polyhedra11 and finite, 2D shapes7,8. A particularly effective method is scaffolded DNA origami, whereby a single scaffold strand is folded by many short auxiliary staple strands to form a complex shape9,14–16,18–21,25.
We recently reported a method for constructing discrete nanostructures with prescribed 2D shapes using single-stranded tiles (SST), and demonstrated structures with complexity comparable to DNA origami26. This article is an adaptation of our earlier work26 and describes detailed protocols for arranging individually addressable SSTs into sophisticated finite 2D shapes with precisely prescribed dimensions (widths and lengths) and morphologies. One key advantage of the SST method is its modularity. Every component SST of a structure serves as a modular construction unit in the assembly, and different subsets of these SSTs produce distinct shapes. Thus, we established a general platform to construct nanostructures with prescribed sizes and shapes from short, synthetic DNA strands.
SSTs contain four domains, each 10 or 11 nucleotides long (Figure 1A). The SSTs bind such that their parallel helices create a DNA lattice held together by crossover linkages. Each crossover is the phosphate between domains 2 and 3. The phosphate is stretched artificially in the diagrams for visual clarity. The crossovers are spaced two helical turns (21 bases) apart (Figure 1B). The composite rectangles are referred to by their dimensions in the number of helices and helical turns. For example, a rectangle that is six helices wide and eight helical turns long is referenced as a 6H × 8T rectangle. SSTs can be left out, added, or otherwise rearranged to create structures of arbitrary shapes and sizes (Figure 1C). For instance, a rectangular design can be rolled into a tube with a desired length and radius (Figure 1D).
Alternatively, the rectangular SST lattice can be viewed as a molecular canvas made up of SST pixels, each 3 nm by 7 nm. In this study, we use a molecular canvas of 310 full-length internal SSTs, 24 full-length SSTs making up the left and right boundaries, and 28 half-length SSTs forming the top and bottom boundaries. The canvas has 24 double helices linked by crossovers and each helix contains 28 helical turns (294 bases) and is therefore referred to as a 24H × 28T rectangular canvas. The 24H × 28T canvas has a molecular weight similar to that of a DNA origami structure created from an M13 phage scaffold.
Protocol
1. DNA Sequence Design
- Use UNIQUIMER software27 to design a SST-finite structure by specifying the number of double helices, lengths of top and bottom helix for each double helix, and the crossover pattern to create a 24H × 28T canvas. After defining these parameters, the overall architecture (strand composition and the complementarity arrangement) is illustrated graphically in the program.
- Generate sequences for the strands of the specified structure to meet the complementarity arrangement and additional requirements (if any). Design DNA sequences by minimizing the sequence symmetry28 (for most of the structures).
- Generate nucleotides (A, C, G and T) randomly one by one.
- Match nucleotides complementary to those generated following the base-pairing rule: A to T and vice versa; C to G and vice versa.
- Do not use repeating segments beyond eight or nine nucleotides. When such repeating segments emerge during design, mutate the most recently generated nucleotides until the repeating-segment requirement is satisfied.
- Do not use four consecutive A, C, G or T bases.
- Use pre-specified nucleotides at the single-stranded linkage points (for example T and G as the twenty-first and twenty-second nucleotides, respectively, for most of the strands) to avoid sliding bases around the linkage points (e.g., if both twenty-first and twenty-second nucleotides were T, it was possible for the twenty-first nucleotide to bind to the nucleotide that the twenty-second nucleotide is supposed to bind to).
- Modify DNA sequences manually for streptavidin labeling, the transformation of rectangles into tubes, the formation of different rectangles and tubes across different scales, and to prevent aggregation caused by exposed domains on SST.
- Replace the exposed domains on the boundary of the molecular canvas with poly-T tracts.
- For streptavidin labeling, design the handle segments (the extra segment appended to the component strand that can bind to a complementary counterpart such as an anti-handle strand) such that they can accommodate an anti-handle strand with 3' biotin modification.
- For the conversion of a rectangle into a tube, remove the top and bottom rows from the rectangle and insert a new row whose SSTs have specific complementarity to the corresponding tiles on the second to top row and the second to bottom row.
- For an exposed single stranded domain of different shapes, replace with poly-T segments of 10-11 nucleotides length, or cover by edge protectors that are complementary to the exposed domain and terminate with a 10-11 nucleotide long poly-T segments.
2. Preparation of the Molecular Canvas
- Obtain DNA component strands of specified sequences dissolved in RNase-free water from an oligonucleotide manufacturer in V bottom 96 well plates with oligonucleotides synthesized at a 10 nmol scale with standard desalting. Centrifuge the 96 well plates with the DNA strands for roughly 10 s at 600 x g.
- Pipette 1 μl from each of the 362 wells with DNA strands for the 24H × 28T rectangle (Figure 3A) at 100 μM per strand (manufacturer specification) into a 2 ml test tube. Add 138 μl of distilled deionized H2O to the 2 ml test tube to make a stock solution of the strand mixture at a concentration of 200 nM per strand.
- Add 50 μl of the 200 nM stock solution of strand mixture, 10 μl of 10X annealing buffer A (50 mM Tris, pH 7.9, 10 mM EDTA, 250 mM MgCl2), and 40 μl of distilled deionized H2O to a 0.2 ml PCR tube. This makes a 100 μl sample of the molecular canvas in 1X annealing buffer A (5 mM Tris, pH 7.9, 1 mM EDTA, 25 mM MgCl2).
- Anneal the sample for 17 hr in a thermal cycler cooling from 90 °C to 25 °C. Program the thermal cycler as follows: ramp down from 90 °C to 61 °C at a constant speed of 5 min per °C; ramp down from 60 °C to 25 °C at a constant speed of 20 min per °C; and incubate at 4 °C until the sample can be taken out of the thermal cycler.
- Prepare a native 2% agarose gel.
- Measure 2.4 g of agarose into a 600 ml beaker. Add 120 ml of 0.5X TBE buffer (44.5 mM Tris-Borate, pH 8.3, 1 mM EDTA) and 30 ml of distilled deionized water to the beaker.
- Microwave for 3 min (or 40 – 60 sec more after boiling). Replenish the water lost during evaporation by addition of 30 ml of water, which helps to maintain the concentration of agarose in the gel at 2%.
- Swirl the contents of the beaker for 1 min in a cold water bath. Add 1 ml of 1.2 M MgCl2 (to make a 10 mM MgCl2 concentration in the gel) and pre-stain the gel with 6 μl of SYBR Safe dye (1:20,000).
- Pour the gel solution into a dry gel box and insert a 1.5 mm thick 12 well gel electrophoresis comb. Let the solution solidify for 15 – 30 min on an even platform.
- Pour gel running buffer (44.5 mM Tris-Borate, pH 8.3, 1 mM EDTA, and 10 mM MgCl2) into the gel box. Load 2 – 3 μl of 1 kb DNA ladder into a well of the agarose gel. Mix 4 μl of 6X bromophenol blue dye (0.25% bromophenol blue, 5 mM Tris, 1 mM EDTA, 10 mM MgCl2 and 30% glycerol) with 20 μl of the sample of the molecular canvas and load the solution into another well of the agarose gel.
- Run the native 2% agarose for 2 hr at 100 V in an ice water bath (bromophenol blue, shown in blue, runs faster than component strands so it can serve as an indicator that if 2 hr running time is appropriate).
- Image the gel using a gel scanner with an appropriate filter for the SYBR safe stain (Figure 2B).
- On a blue light transilluminator, excise the dominant band with similar mobility to the band of 1,500 base pairs of the 1 kb DNA ladder using a sharp clean razor blade. Place the desired gel piece(s) in a spin column and then crush the gel into fine pieces using a microtube pestle. Centrifuge at 438 x g for 3 min at 4 °C.
- Measure the concentration of the DNA using an ultra violet spectrophotometer at 260 nm.
- Use 2 μl of ultrapure DNase/RNase-Free distilled water as the blank to calibrate the machine.
- Use an equivalent volume of the collected sample from the spin column to get an estimate of the DNA concentration. The measured concentration value (“a” = ng/μl) obtained at a UV absorption of 260 nm will help determine the dilution factor for AFM and TEM imaging.
- Convert the measured concentration from “a” (ng/μl) to “b” (nM) following b = a × 106 / (330 × number of nucleotides).
NOTE: The molecular weight of a nucleotide is 330 g/mol. The calculated molar concentration value will determine the dilution factor for AFM and TEM imaging. As for the case of a 24H × 28T rectangle, the common measurement for the purified sample is around 10 – 60 ng/μl. As the number of nucleotides for such a structure is 14616 Da, the molar concentration is approximately 2 – 12 nM. The final concentration is determined by the collection yield (~20 – 50%) from centrifugation and the volume of the gel band excised out (the higher the volume, the more dilute the purified sample).
3. Atomic Force Microscopy Imaging
- Peel off mica attached to a specimen metallic disc using scotch tape to get a flat surface and place the specimen disc onto the imaging stage.
- Add 40 μl of 1X annealing buffer A followed by 5 μl (2 – 5 nM) of the purified sample of the 24H × 28T rectangle to the freshly cleaved mica surface. Allow the mixture to settle for approximately 2 min.
- Install AFM silicon nitride cantilever chip onto a cantilever holder. Use a C triangular tip (resonant frequency, f0 = 40 – 75 kHz; spring constant, k = 0.24 Nm-1) for imaging.
- Image a sample in the fluid-tapping mode. Use the following imaging parameters for scanning: scan size of 2 μm, 1024 lines for the resolution, and a scan rate of 0.5 – 1 Hz (Figure 2C).
- If necessary, add 10 μl or more of 10 mM NiCl2 solution to increase the strength of DNA-mica binding29.
4. Sample Preparation for Streptavidin Labeling
- Manually design the 24H × 28T rectangle such that handle strands at specific locations are incorporated to be complementary to the anti-handle strands with biotin modification, which in turn are able to bind to streptavidin specifically.
- Modify the molecular canvas by attaching a 3' 17-nucleotide segment that consists of a two-nucleotide sequence (TT) as a spacer and a 15-nucleotide handle (GGAAGGGATGGAGGA) to the 14 tiles on the top row and 14 tiles on the bottom row of the rectangle (Figure 3A) to label the boundary. This sequence is complementary to a 3' biotin modified anti-handle strand (TCCTCCATCCCTTCC-biotin).
- For internal labeling, attach the 3' 17 nucleotide segment to 8 internal tiles and 6 boundary tiles of the rectangle (Figure 4A).
- Mix the 28 strands with handles (boundary labeling) with the rest of the component strands of the 24H × 28T rectangle (334 strands) to make a 200 nM stock solution. Mix the 14 strands with handles (internal labeling) with the rest of the component strands of the 24H × 28T rectangle (348 strands) to obtain a 200 nM stock solution.
- For boundary labeling, add 50 μl of the 200 nM stock solution of the strands, 6 μl of the 100 μM biotin modified anti-handle strands, 10 μl of the 10X annealing buffer A, and 34 μl of distilled deionized water to a 0.1 – 0.2 ml PCR test tube. The final concentration of the component strand is 100 nM and the final concentration of the anti-handle biotin modified strand is 6,000 nM. Note that 28 molecules of the anti-handle biotin modified strand bind to a 24H × 28T rectangle so the anti-handle biotin modified strand is in excess of 100% to make the binding favorable.
- For internal labeling, add 50 μl of the 200 nM stock solution of the strands, 3 μl of the internal anti-handle biotin modified strand at 100 μM, 10 μl of the 10X annealing buffer A, 37 μl of distilled deionized water to a 0.1 – 0.2 ml PCR test tube. The final concentration of the component strand is 100 nM and the final concentration of the anti-handle biotin modified strand is 3,000 nM. Note that 14 molecules of the anti-handle biotin modified strand bind to a 24H × 28T rectangle so anti-handle biotin modified strand is in excess of 100% to make the binding favorable.
- Anneal both samples in a thermal cycler for 17 hr using the steps described in 2.3.
- Prepare a native 2% agarose gel as described in steps 2.4.
- Purify both samples as described in steps 2.5.
- Measure the concentrations of the desired DNA structures as described in steps 2.6.
5. Atomic Force Microscopy For Streptavidin Labeling
- Image each sample using an AFM as described in step 3 (Figures 3B and 4B).
- After the first round of imaging, add 40 μl of 1X annealing buffer A followed by 1 μl of streptavidin at 10 mg/ml to the sample. Allow the mixture to settle for approximately 2 min before reimaging (Figures 3C and 4C).
6. Converting A Rectangle Into A Tube
- In order to design a tube based on the rectangle, keep all of the component SSTs in place except the top and bottom rows. Introduce a new row whose SSTs are designed by concatenating the top and bottom half-tiles of their respective columns to cyclize the original rectangle into a tube conformation (Figure 5A).
- Mix the new row of strands (14 strands) with the component strands of the 24H × 28T rectangle excluding the top and bottom rows (336 strands) to obtain a 200 nM stock solution.
- Follow the gel purification steps 2.2 – 2.7 (Figure 5B).
7. Transmission Electron Microscope Imaging
- Weigh 0.06 g of uranyl formate in 3 ml of distilled water to prepare a 2% aqueous uranyl formate stain solution. Filter the 2% aqueous uranyl formate stain solution using a 0.2 µm filter attached to a syringe.
- Add 5 µl of 5 N NaOH to the 1 ml of filtered stain solution. Briefly vortex the solution and centrifuge at 20,000 x g.
- Glow discharge the carbon-coated grids (coated side facing up) with the following settings: 25 mA, 45 s, 0.1 mBar, and negative High Tension polarity.
- Use self-closing forceps to grab a glow discharger treated grid from the edge.
- Pipette 3.5 µl of sample onto the grid for 4 min. Use a piece of filter paper to wick off the sample by bringing the filter paper in contact with the grid from the side.
- Immediately add 3.5 µl of the stain solution onto the grid for 1 min.
- Wick off the stain as in 7.4 and hold the filter paper against the grid for 1 – 2 min.
- Transfer the grid to a TEM specimen holder and image using a JEOL JEM-1400 operated at 80 kV with magnification ranging from 10 K to 80 K (Figure 5C).
8. Constructing Arbitrary Shapes Using the Molecular Canvas
- Identify a shape and select the strands that correspond to the shape on the molecular canvas. For a triangle shape for example, select 206 out 362 strands for the molecular canvas.
- Replace the exposed domain (that is, along the hypotenuse of the triangle) with a poly-T segment 10 – 11 nucleotides long (design 1 in Figure 6A), or add an edge protector that is complementary to the exposed domain and terminate it with a 10 – 11 nucleotide long poly-T segment (design 2 in Figure 6A) to prevent aggregation.
NOTE: Simply annealing the SSTs that correspond to the pixels of the triangle (without using protector strands) will lead to aggregation and no distinct product band formation on a gel. Use the edge protector design for various shapes as it is more resource efficient; it requires only four additional sets of strands (Figure 6C) as compared to 14 additional sets in replacement design (Figure 6B). - Create a strand library for the 310-pixel molecular canvas which includes the core set (the 362 SST canvas), Set 1* (edge protectors that bind to domain 1 of a SST), Set 2* (edge protectors that bind to domain 2 of a SST), Set 3* (edge protectors that bind to domain 3 of a SST), and Set 4* (edge protectors that bind to domain 4 of a SST) to give a total of 1344 edge protectors.
- Centrifuge the 96 well plates for the different sets for roughly 10 s. Pipet out the desired strands from the plates in order to make a stock solution for a certain shape.
- For the shape of a triangle with edge protectors, pipette 1 μl of 206 strands from the core set, 0 from set 1*, 0 from set 2*, 0 from set 3* and 24 strands from set 4* into a 2 ml centrifuge tube. Add 20 μl of distilled deionized water to the tube to make a 400 nM stock solution of the DNA strands.
- Add 50 μl of the 400 nM stock solution of DNA strands, 10 μl of the 10X annealing buffer B (50 mM Tris, pH 7.9, 10 mM EDTA, 125 mM MgCl2) stock solution and 40 μl distilled deionized H2O to a 0.2 ml PCR tube. This makes a 100 μl sample of the triangle in 1X annealing buffer B (5 mM Tris, pH 7.9, 1 mM EDTA, 12.5 mM MgCl2) with strands at a concentration of approximately 200 nM per strand.
- Anneal the mixture in a thermal cycler for 17 hr as described in step 2.3.
- Prepare a native 2% agarose gel as described in step 2.4.
- Purify the sample as described in step 2.5.
- Measure the concentration of the DNA as described in step 2.6.
- Image the sample under AFM as described in step 3 (Figure 7C and 7D).
- Pick and mix strands for different shapes using the same strand library. Anneal the different solutions and combine purified samples of different shapes to save time during AFM imaging.
9. Optional: Robot Automation Of Shape Design and Liquid Mixing
- Use a custom MATLAB program to aid the design of complex shapes.
- Use the software to deliver instructions in the form of a pipetting sequence to a robot liquid handler. Use the robot liquid handler to select and mix the strands that constitute the target shape.
NOTE: Shape design and strand mixing was automated to reduce human error and make the construction less labor intensive.
10. Rectangles and Tubes Across Different Scales
- Construct different sized rectangles (Figure 10) by simply altering the number of parallel helices (H) and the number of helical turns (T).
- Follow Step 3 for a specific size rectangle.
- Follow Step 6 for a specific size tube.
Representative Results
The self-assembly of SSTs (Figure 1) will yield a 24H × 28T rectangle, as illustrated in Figure 2. DNA sequences for the different SSTs can be modified/optimized to enable streptavidin labeling (Figure 3 and 4), the transformation of a rectangle into a tube (Figure 5), the programmable self-assembly of SSTs to form tubes and rectangles of varying sizes (Figure 10), and the construction of 2D arbitrary shapes using the molecular canvas (Figure 8). Two designs (domain substitution design and edge protector design) were tested as solutions to aggregation along exposed domains of arbitrary shapes (Figure 7). Both designs have comparable gel yield and structural integrity, but the edge protector design is more cost effective since it requires fewer auxiliary species (Figure 6). Strand picking and mixing can be automated, as shown in Figures 9 to reduce human error and save time.
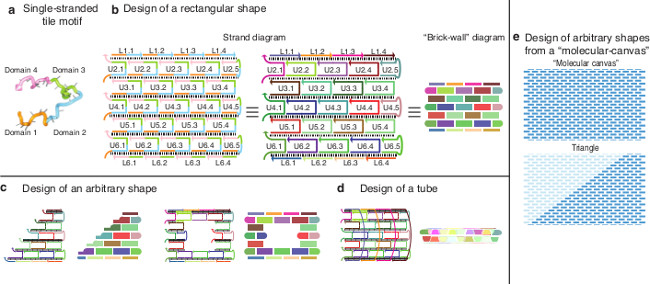
Figure 1. Self-assembly of Molecular Shapes Using Single-stranded Tiles. (A) The canonical SST motif, adapted from12. (B) Left and middle: Two depictions of assembled SSTs. Inner SSTs have a full length of 42 bases and are labeled “U” while boundary SSTs have 21 bases and are labeled “L”. In the left diagram, colors distinguish different domains. In the middle diagram, colors distinguish different strands. Right: A schematic of the brick wall view of SST structures. Thick bricks are full SSTs and thin bricks are boundary SSTs. Rounded edges indicate no complementary pairing. As in the middle figure, colors distinguish individual tiles. In all diagrams, each tile has a unique sequence. (C) Withholding an appropriate selection of strands from the SST collection making up the rectangle design in figure b results in the formation of a different desired shape such as a triangle (left) or a rectangular ring (right). (D) The design of a tube with a predetermined width and length. (E) Given a pre-synthesized pool of SST sequences (top), arbitrary shapes (bottom) can be designed by choosing a subset of strands to use in the strand mixture (dark blue pixels) and eliminating the rest (light blue pixels). This figure has been modified from a previously published figure26. Please click here to view a larger version of this figure.
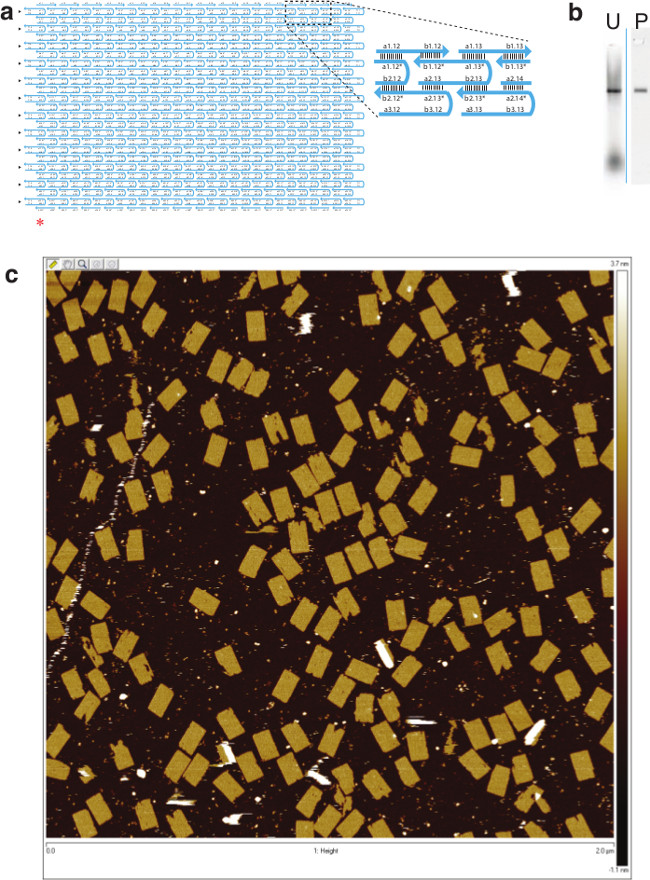
Figure 2. Design and AFM Image of the 24H × 28T Rectangle. (A) Schematic drawing of the 24H × 28T rectangle. A zoomed-in view is also shown for the detailed local structure. The individual SST segment arrangement is either 10 nt – 11 nt – 11 nt – 10 nt (e.g., a2.13-b2.13-a1.13*-b1.12*) or 11 nt – 10 nt- 10 nt – 11 nt (e.g., a3.12-b3.12-a2.13*-b2.12*). Triangles on the left hand side of the rectangle indicate rows with a 10nt-11nt-11nt-10nt SST arrangement; the other internal SSTs have 11 nt – 10 nt -10 nt- 11 nt instead (nt is an acronym for nucleotide). Since the 24H × 28T SST rectangle has similar dimensions to a DNA origami structure, the criterion introduced in the DNA origami work and consider an SST rectangle “well-formed” if it has no defect in the expected outline greater than 15 nm in diameter was adopted. Additionally, a “well-formed” rectangle structure has no holes in its interior larger than 10 nm in diameter was required. Following the above criteria, a “well-formation” ratio of 55% (N = 163) was obtained. A red asterisk (*) indicates the bottom left corner of the rectangle here. (B) 2% native agarose gel electrophoresis. U, unpurified; P, purified (by gel extraction from lane U). (C) AFM image of the lattice structure (scanning size: 2 μm×2 μm). This figure has been modified from a previously published figure26. Please click here to view a larger version of this figure.
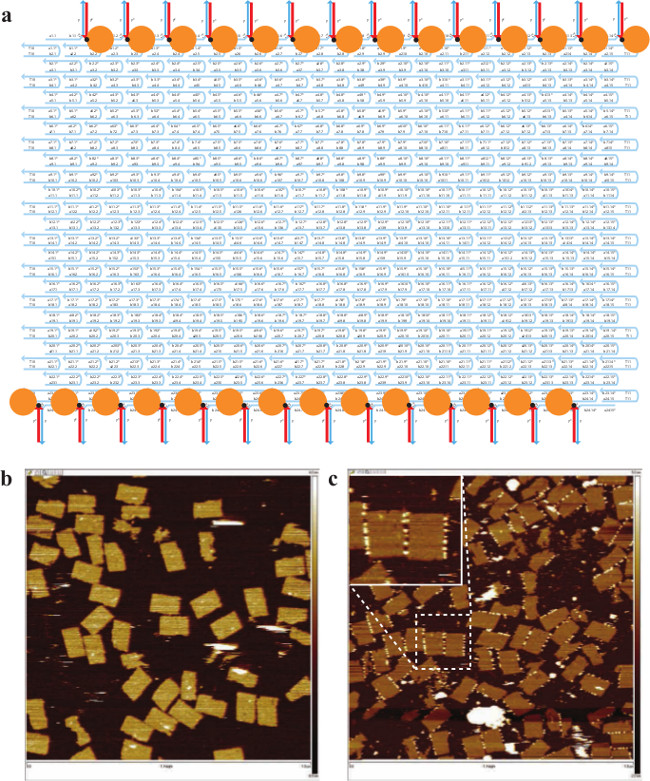
Figure 3. Boundary Labeling of 24H × 28T Rectangle. (A) Schematic drawing of the specific biotin-labeled 24H × 28T rectangle. The strands highlighted in blue are the handle strands. The strands highlighted in red are the anti-handle strands labeled with 3' biotin (black dots). Streptavidin is depicted as an orange ball. (B) AFM image before adding streptavidin (scanning size: 1 μm×1 μm). (C) AFM image after adding streptavidin (scanning size: 1 μm×1 μm). Inset, a zoomed-in view showing successful labeling. Note that streptavidin appeared as either white dots or stripes due to their raised heights. This figure has been modified from a previously published figure26. Please click here to view a larger version of this figure.
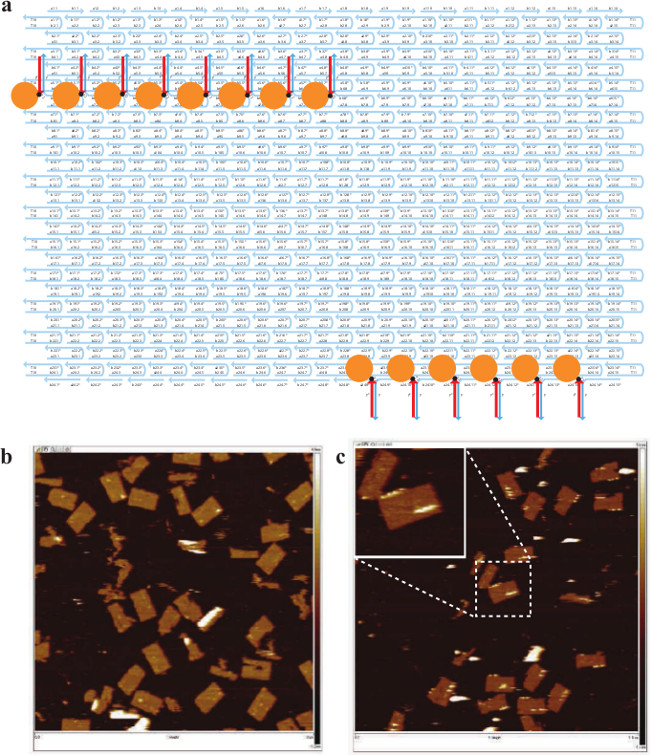
Figure 4. Internal Labeling of 24H × 28T Rectangle. (A) Schematic drawing of the specific biotin labeled 24H × 28T rectangle. The strands highlighted in blue are the handle strands. The strands highlighted in red are the anti-handle strands labeled with 3' biotin (black dots). Streptavidin is depicted as an orange ball. (B) AFM image before adding streptavidin (scanning size: 1 μm×1 μm). (C) AFM image after adding streptavidin (scanning size: 1 μm×1 μm). Inset, a zoomed-in view showing successful labeling. Note that streptavidin appeared either as white dots or stripes due to the raised heights. This figure has been modified from a previously published figure26. Please click here to view a larger version of this figure.
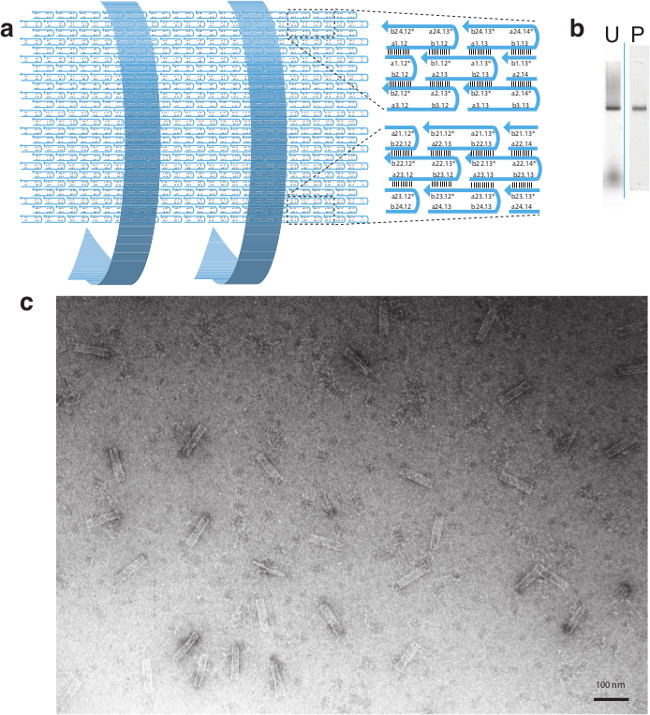
Figure 5. Design and TEM Image of the 24H × 28T Tube. (A) Schematic drawing of the 24H × 28T barrel. Two zoomed-in views at the top and the bottom show detailed segment identities. Note that segments a24.x* (e.g., a24.13*) and b24.x* (e.g., b24.12*) of the top row are complementary to segments a24.x (e.g., a24.13) and b24.x (e.g., b24.12) of the bottom row; such complementarity is expected to result in the formation of the tubular structure. (B) 2% native agarose gel electrophoresis. U, unpurified; P, purified (by gel extraction from lane U). (C) TEM image of the barrel structure (scale bar: 100 nm). This figure has been modified from a previously published figure26. Please click here to view a larger version of this figure.
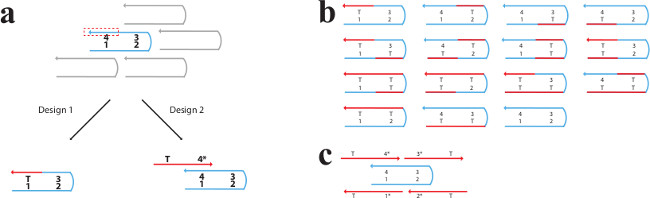
Figure 6. Two Designs to Prevent Aggregation Caused by Exposed Sticky Domains. (A) A side-by-side demonstration of two methods to cover exposed domains. The unpaired sticky domain (domain 4) is indicated with a red, dashed box. Design 1 is the domain substitution design, in which the unpaired domain is substituted by a poly-T domain (shown in red and labeled “T”). Design 2 is the edge protector design, which involves covering the unpaired domain with an edge protector strand (shown in red and labeled “T-4*”). The edge protector strand is composed of a poly-T section and a complement to the exposed domain. (B) The different possible boundary configurations associated with domain substitution (design 1). There are 14 different ways an internal SST (shown in blue) can leave an exposed domain or domains. The appropriate strand substitutions are shown in red for each situation. (C) The edge protector design (design 2). One internal SST (blue) requires only four edge protector strands (red) to cover exposed domains using this design. This figure has been modified from a previously published figure26. Please click here to view a larger version of this figure.
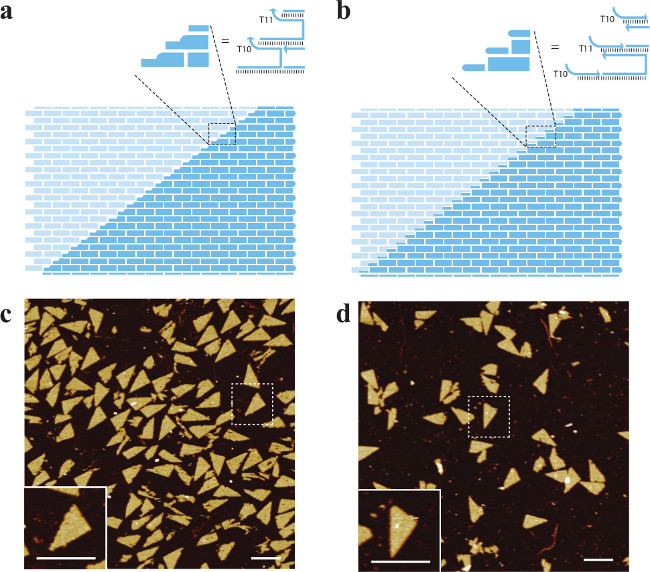
Figure 7. Two Designs for the SST Triangle. (A) and (B) depict schematics based on design 1 (domain substitution) and design 2 (edge protector) in Figure 8, respectively. A poly-T region (T10 or T11 in the figure) is depicted as a rounded corner in the block diagram. The inset shows a magnified view of the structure indicated with the dashed box. Scale bars, 100 nm. (C) and (D) are the AFM images. This figure has been modified from a previously published figure26.

Figure 8. Diagrams and AFM Images of 100 Different Shapes. Structure designs are displayed in the top panels. Light blue coloring indicates the canvas strands left out of the mixture while dark blue coloring indicates the strands included. For clarity, edge protector strands are omitted. A corresponding AFM image of each structure is shown below its design. Each image is 150 nm×150 nm in area. There are 100 unique shapes. Included are 10 Arabic numerals, the 26 capital letters of the Latin alphabet, 23 punctuation marks and keyboard symbols, 10 emoticons, 9 astrological symbols, 6 Chinese characters, and several miscellaneous symbols. This figure has been modified from a previously published figure26. Please click here to view a larger version of this figure.
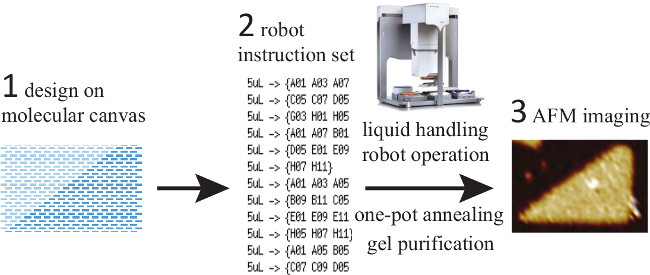
Figure 9. Robot Automation Flow Diagram. This diagram depicts the workflow used to automate the mixing process. First, a custom MATLAB program is used to design the desired shape. Once again, dark blue rectangles indicate the strands to be included in the mixture. After the design is completed, the program outputs a set of pipetting instructions for the robotic liquid handler. The robot pipettes the correct concentrations of the appropriate strands for one particular shape into a plate well. The mixture then undergoes one pot annealing, followed by AFM imaging of the resulting shapes. (This figure has been modified from a previously published figure26). Please click here to view a larger version of this figure.
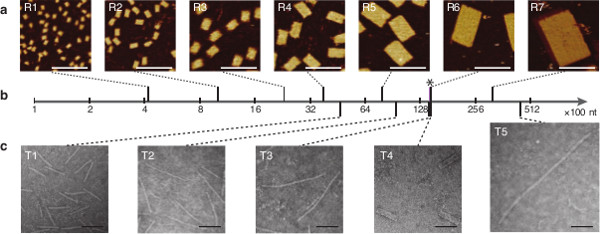
Figure 10. Self-Assembly of Rectangles and Tubes Across Different Scales. (A) AFM images of SST rectangles with the following designed dimensions (from left to right): 4H × 4T, 6H × 7T, 10H × 10T, 12H × 14T, 18H × 20T, 24H × 28T, and 36H × 34T. (B) The molecular weights of the structures are on a logarithmic scale. The asterisk indicates the weight of a typical M13 DNA origami structure as a reference. (C) TEM images of SST tubes with the following designed dimensions (from left to right): 8H × 28T, 8H × 55T, 8H × 84T, 24H × 28T, and 12H ×177T. All scale bars show 100 nm. This figure has been modified from a previously published figure26. Please click here to view a larger version of this figure.
Discussion
In the structure formation step, it is important to keep an appropriate concentration of magnesium cations (e.g., 15 mM) in the DNA strand mixture to self-assemble DNA nanostructures. Similarly, in the agarose gel characterization/purification step, it is important to keep an appropriate magnesium cation concentration (e.g., 10 mM) in the gel and the gel running buffer to retain the DNA nanostructures during electrophoresis. For the 24H×28T rectangle structure, we tested annealing in different Mg++ concentrations and found that the structure typically formed in the range of 10 – 30 mM Mg++ (with the best formation yield with around 20 mM Mg++).
Unlike the scaffolded origami structure, the SST structure has a modular architecture. Different shapes can be designed from the same canvas by selecting the appropriate subset of SST tiles. Additionally, the SST structure is assembled from only short synthetic DNA strands and does not involve a long scaffold that is used in the origami approach. Therefore, the size of the SST structure is not restricted by the length of the available scaffold. However, while DNA origami approach can often be optimized to produce either folded structures or unfolded monomers, SST may result in partially formed structures and thus may require gel purification for subsequent use. Additionally, due to the lack of a scaffold strand, an SST structure can be more “brittle” than its DNA origami counterpart. Both the SST approach and the DNA origami approach are fast evolving, and a user is advised to choose an approach that best suits his or her particular functional needs.
It is striking that hundreds of small monomers mediated only by local binding interactions can self-assemble into a prescribed global shape, as demonstrated by the SST approach. The fundamental principles shown in the SST approach should be generalizable to materials beside the DNA strands, and the basic method described in this article, after appropriate modification, should enable the constructions of complex structures self-assembled from diverse materials.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was funded by the Office of Naval Research Young Investigator Program Award N000141110914, Office of Naval Research Grant N000141010827, NSF CAREER Award CCF1054898, NIH Director’s New Innovator Award 1DP2OD007292 and a Wyss Institute for Biologically Inspired Engineering Faculty Startup Fund (to P.Y.) and Tsinghua-Peking Center for Life Sciences Startup Fund (to B. W.).
Materials
| Name of Material/ Equipment | Company | Catalog Number | Comments/Description |
| DNA Strands | Integrated DNA Technology | Section 3.1 | |
| SYBR Safe DNA gel stain | Invitrogen | S33102 | Section 3.4.2 |
| Freeze'N Squeeze DNA Gel Extraction Spin Columns | BIO-RAD | 731-6166 | Section 3.6 |
| Bruker's Sharp Nitride Lever Probes | Bruker AFM Probes | SNL10 | Section 4.3 |
| Safe Imager 2.0 Blue Light Transilluminator | Invitrogen | G6600 | Section 3.6 |
| Centrifuge 5430R | Eppendorf | 5428 000.414 | Section 3.6 |
| Transmission Electron Microscope | Jeol | Jem 1400 | Section 7.4 |
| Multimode 8 | Veeco | Section 4 | |
| Typhoon FLA 9000 Laser Scanner | GE Heathcare Life Sciences | 28-9558-08 | Section 3.5 |
| Ultrapure Distilled water | Invitrogen | 10977-023 | Section 3.7.1 |
| Mica disk | SPI Supplies | 12001-26-2 | Section 4.1 |
| Steel mounting disk | Ted Pella, Inc. | 16218 | Section 4.1 |
| carbon coated copper grid for TEM | Electron Microscopy Sciences | FCF400-Cu | Section 7.2 |
| tweezers | Dumont | 0203-N5AC-PO | Section 7.31 |
| glow discharge system | Quorum Technologies | K100X | Section 7.2 |
| DNA Engine Tetrad 2 Peltier Thermal Cycler | BIO-RAD | PTC–0240G | Section 3.3 |
| Owl Easycast B2 Mini Gel Electrophoresis Systems | ThermoScientific | B2 | Section 3.4.3 |
| Seekam LE Agarose 500G | Lonza | 50004 | Section 3.4.1 |
| GeneRuler 1kb Plus DNA Ladder, Ready-To-Use 75-20000bp | ThermoScientific | SM1333 | Section 3.4.4 |
| Nanodrop 2000c UV-vis Spectrophotometer | ThermoScientific | Section 3.7 | |
| 0.2 um filter | Corning Inc. | 431219 | Section 7.1.2 |
References
- Seeman, N. C. Nucleic acid junctions and lattices. J. Theor. Biol. 99 (2), 237-247 (1982).
- Fu, T. J., Seeman, N. C. DNA double-crossover molecules. Biochemistry. 32 (13), 3211-3220 (1993).
- Winfree, E., Liu, F., Wenzler, L. A., Seeman, N. C. Design and self-assembly of two-dimensional DNA crystals. Nature. 394 (6693), 539-544 (1998).
- Yan, H., Park, S. H., Finkelstein, G., Reif, J. H., LaBean, T. H. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science. 301 (5641), 1882-1884 (2003).
- Rothemund, P. W. K., Papadakis, N., Winfree, E. Algorithmic self-assembly of DNA Sierpinski triangles. PLoS Biol. 2 (12), e424 (2004).
- Shih, W., Quispe, J., Joyce, G. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature. 427 (6975), 618-621 (2004).
- Chworos, A., et al. Building programmable jigsaw puzzles with RNA. Science. 306 (5704), 2068-2072 (2004).
- Park, S. H., et al. Finite-size, fully-addressable DNA tile lattices formed by hierarchical assembly procedures. Angew. Chem. Int. Ed. 45 (5), 735-739 (2006).
- Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature. 440 (7082), 297-302 (2006).
- Schulman, R., Winfree, E. Synthesis of crystals with a programmable kinetic barrier to nucleation. Proc. Natl Acad. Sci. USA. 104 (39), 15236-15241 (2007).
- He, Y., et al. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature. 452 (7184), 198-201 (2008).
- Yin, P., et al. Programming DNA tube circumferences. Science. 321 (5890), 824-826 (2008).
- Sharma, J., et al. Control of self-assembly of DNA tubules through integration of gold nanoparticles. Science. 323 (5910), 112-116 (2009).
- Douglas, S. M., Dietz, H., Liedl, T., Högberg, B., Graf, F., Shih, W. M. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 459 (7245), 414-418 (2009).
- Andersen, E. S., et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature. 459 (7243), 73-76 (2009).
- Dietz, H., Douglas, S. M., Shih, W. M. Folding DNA into twisted and curved nanoscale shapes. Science. 325 (5941), 725-730 (2009).
- Zheng, J. P., et al. From molecular to macroscopic via the rational design of self-assembled 3D. DNA crystal. Nature. 461 (7260), 74-77 (2009).
- Han, D., et al. DNA origami with complex curvatures in three-dimensional space. Science. 332 (6024), 342-346 (2011).
- Liu, W., Zhong, H., Wang, R., Seeman, N. C. Crystalline two-dimensional DNA origami arrays. Angew. Chem. Int. Ed. 50 (1), 264-267 (2011).
- Zhao, Z., Liu, Y., Yan, H. Organizing DNA origami tiles into larger structures using preformed scaffold frames. Nano Lett. 11 (7), 2997-3002 (2011).
- Woo, S., Rothemund, P. Programmable molecular recognition based on the geometry of DNA nanostructures. Nat. Chem. 3 (8), 620-627 (2011).
- Delebecque, C. J., Lindner, A. B., Silver, P. A., Aldaye, F. A. Organization of intracellular reactions with rationally designed RNA assemblies. Science. 333 (6041), 470-474 (2011).
- Lin, C., Liu, Y., Rinker, S., Yan, H. DNA tile based self-assembly: building complex nanoarchitectures. ChemPhysChem. 7 (8), 1641-1647 (2006).
- Seeman, N. C. Nanomaterials based on DNA. Annu. Rev. Biochem. 79 (1), 65-87 (2010).
- Voigt, N. V., Nangreave, J., Yan, H., Gothelf, K. V. DNA origami: a quantum leap for self-assembly of complex structures. Chem. Soc. Rev. 40 (12), 5636-5646 (2011).
- Wei, B., Dai, M., Yin, P. Complex Shapes self-assembled from single stranded DNA tiles. Nature. 485 (7400), 623-626 (2012).
- Wei, B., Wang, Z., Mi, Y. Uniquimer: software of de novo DNA sequence generation for DNA self-assembly: an introduction and the related applications in DNA self-assembly. J. Comput. Theor. Nanosci. 4 (1), 133-141 (2007).
- Seeman, N. C. De novo design of sequences for nucleic acid structural engineering. J. Biomol. Struct. Dyn. 8 (3), 573-581 (1990).
- Hansma, H. G., Laney, D. E. DNA binding to mica correlates with cationic radius: assay by atomic force microscopy. Biophys. J. 70 (4), 1933-1939 (1996).
- Seelig, G., Soloveichik, D., Zhang, D. Y., Winfree, E. Enzyme-free nucleic acid logic circuits. Science. 314 (5805), 1585-1588 (2006).
- Yin, P., Choi, H. M. T., Calvert, C. R., Pierce, N. A. Programming biomolecular self-assembly pathways. Nature. 451 (7176), 318-322 (2008).

