Using Fluorescence Activated Cell Sorting to Examine Cell-Type-Specific Gene Expression in Rat Brain Tissue
Summary
The goal of this protocol is to use the fluorescence activated cell sorting (FACS) technique to sort specific types of neural cells for subsequent analysis of cell-type-specific gene expression, epigenetic markers, and or protein expression.
Abstract
The brain is comprised of four primary cell types including neurons, astrocytes, microglia and oligodendrocytes. Though they are not the most abundant cell type in the brain, neurons are the most widely studied of these cell types given their direct role in impacting behaviors. Other cell types in the brain also impact neuronal function and behavior via the signaling molecules they produce. Neuroscientists must understand the interactions between the cell types in the brain to better understand how these interactions impact neural function and disease. To date, the most common method of analyzing protein or gene expression utilizes the homogenization of whole tissue samples, usually with blood, and without regard for cell type. This approach is an informative approach for examining general changes in gene or protein expression that may influence neural function and behavior; however, this method of analysis does not lend itself to a greater understanding of cell-type-specific gene expression and the effect of cell-to-cell communication on neural function. Analysis of behavioral epigenetics has been an area of growing focus which examines how modifications of the deoxyribonucleic acid (DNA) structure impact long-term gene expression and behavior; however, this information may only be relevant if analyzed in a cell-type-specific manner given the differential lineage and thus epigenetic markers that may be present on certain genes of individual neural cell types. The Fluorescence Activated Cell Sorting (FACS) technique described below provides a simple and effective way to isolate individual neural cells for the subsequent analysis of gene expression, protein expression, or epigenetic modifications of DNA. This technique can also be modified to isolate more specific neural cell types in the brain for subsequent cell-type-specific analysis.
Introduction
The purpose of the protocol described below is to isolate individual cell types from a heterogeneous population of neural tissue for the subsequent analysis of cell-type-specific gene expression, protein expression, or even epigenetic markers. The brain is comprised of many cell types that are derived from distinct progenitor cells and that have cell-type-specific properties and functions. Despite these differences, these distinct neural cell types can express similar receptors and intracellular signaling molecules which make the analysis of these more ubiquitous proteins difficult to measure or interpret in a cell-type-specific manner using conventional methods. Neuroscientists must identify the function and activation of these distinct cell types in the brain and how they can individually impact behavior as this will ultimately be the first step to identifying more specific drugs and therapeutic targets for neurological and neuropsychiatric diseases. Despite this overarching goal of neuroscience research, it can been difficult to isolate individual neural cell types from the brain for the analysis of gene or protein expression, the activation of signaling molecules, or the modification of epigenetic markers on DNA. Of the techniques that are currently used, immunohistochemistry can identify the expression of proteins in a cell-type-specific manner when combined with additional staining of a cell-type-specific marker, though this relies on specific antibodies that can properly stain for the proteins of interest and it can be difficult to quantify. In situ hybridization can identify the specific localization of messenger ribonucleic acid (mRNA) in individual cells in the brain, but this is a laborious process that also limits the co-analysis of specific cell types and only allows for the analysis of one or maybe a few genes of interest. Laser capture micro-dissection uses a laser to isolate subpopulations of cells that are visualized via microscopy; however the time-consuming nature of this process and the relatively low yield can significantly limit the subsequent analysis of proteins or mRNA levels, particularly if the expression of these molecules is low to begin with. Fluorescence activated cell sorting (FACS) is a relatively novel technique in the field of neuroscience to isolate individual cell types from the brain for subsequent analysis of gene expression1 and/or epigenetic targets2. This process can also be used to sort specific types of neural cells for subsequent analysis of cell-type-specific gene expression, protein expression, or epigenetic markers. FACS has been used in a number of medical research fields such as cancer and immunology for decades to count and sort different cells based on either physical or biochemical characteristics3. In addition, flow cytometry has classically been used to analyze protein expression on a per cell basis, using specific antibodies. The procedure described below, takes advantage of classical flow cytometry techniques to isolate individual cell types for subsequent analysis of molecular biology endpoints. The flow cytometer can analyze several thousand cells in a second, which makes it a quick and efficient alternative to the techniques described above. In addition, cells can be isolated based on the cellular expression of a specific protein (for example a neurotransmitter receptor) or a combination of two or more proteins (colocalization of multiple proteins in a specific cell type). This allows the user to isolate very selective neural cell types based on their molecular properties to identify their function in the brain.
To perform FACS, neural cells are prepared into a single-cell suspension which is passed through a flow cell that carries and aligns the cells so that they pass single-file through a light beam and lasers for analysis. A computer acquires the data from each cell and plots it on a histogram for analysis of specified parameters (size, granularity, and fluorescence). Based on these parameters, the cells can immediately be sorted into separate tubes for their recollection and subsequent analysis of any endpoint desired. The protocol described below utilizes three antibodies to sort neurons (using a Thymocyte antigen 1, Thy1 antibody), astrocytes (using a glial glutamate transporter, GLT1 antibody), and microglia (using a cluster of differentiation molecule 11B, CD11b antibody). This protocol can be used as described below or modified with different antibodies depending on the cell type that one would like to isolate for his own experiments.
There are a few caveats to consider when determining whether this protocol is appropriate for specific experiments. One major caveat may concern the specific cell type that one would like to isolate. In this protocol, the three antibodies that are used are extracellular antibodies, which allow the experimenter to keep the cell types intact during the staining procedure, thus preserving the integrity of the RNA, DNA and proteins inside. It is possible that one may wish to isolate a specific neural cell type using an antibody that identifies a protein that is only expressed inside that particular cell. For example, one might want to isolate dopaminergic neurons using an antibody to tyrosine hydroxylase or isolate acetylcholine neurons using an antibody for choline acetyltransferase. These proteins are intracellular and thus would require fixation and permeabilization of the cell membrane for subsequent staining with the appropriate antibodies. While this has been done before 4,5, this process may significantly decrease the yield of RNA or DNA from these permeabilized cells. Another caveat may be that not all antibodies are appropriate for FACS. For example, one may currently use an antibody that works very well for western blot, a technique that requires the denaturation of proteins. This antibody may not necessarily be suitable for identification of these cell types using FACS given that the proteins are not denatured at any point in this protocol and thus the antibody may have no way to bind to its inherent antigen. Companies provide specification sheets which identify the applications for which an antibody has been approved. If an antibody has not been approved for flow cytometry, it should not discourage one from trying this protocol with a particular antibody; however, one should be aware that it isn’t guaranteed to work for FACS. A third caveat of this protocol has to do with the number of cells that one is trying to isolate. FACS is an excellent technique to yield the most cells possible from even a small piece of tissue, but it is also possible that if one would like to isolate a relatively sparse population of cells from a relatively small brain region, the yield from one animal will be inherently low. In this case, it may be necessary to pool the brain tissue from a few animals in the same treatment group in order to yield the number of cells necessary for subsequent analysis; however, a recent publication has used gene-targeted pre-amplification of cDNA for subsequent analysis of gene expression from a small number of activated neurons (5-6%) from a larger set sorted neurons, indicating that it is possible to analyze gene expression from even small subsets of neural cells without pooling large numbers of animals6A final caveat of this technique is that one should have access to a cell sorter that is not too far away. The cell sorter is a complicated machine that requires significant training in order to use it properly. Thus these machines are often run by a qualified technician in a core facility. In addition, the goal of this procedure is to dissociate the neural tissue, stain it for specific antibodies, and immediately take those samples to a sorter within a short period of time (perhaps a half day). This timeframe will help to increase the survival and yield of isolated cells and maintain the integrity of the cells for subsequent processing and analysis. If all of these parameters described above are met, FACS is an excellent method to analyze cell-type specific expression of genes and proteins from neural tissue.
All experiments were performed in accordance with the Institutional Animal Care and Use Committee (IACUC) of the University of Delaware and the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Protocol
1. Preparation for Tissue Collection (15 – 30 min)
- Fire polish three sets of glass Pasteur pipettes in decreasing diameters and number them “1” = approximately 1 mm diameter, “2” = approximately 0.75 mm diameter, and “3” = approximately 0.50 mm diameter.
- Resuspend lyophilized Enzyme A with all of Buffer A according to the protocol provided with the Neural Dissociation Kit (see Table of Materials). Do not vortex. Aliquot the solution into 50 µl aliquots and store at -20 ºC for later use.
- Make 1 L of myelin removal buffer and vacuum filter for sterilization [myelin removal buffer = 1,000 ml of 0.1 M phosphate-buffered saline (PBS), 2 ml of 500 mM Ethylenediaminetetraacetic acid (EDTA), and 10 g of Bovine Serum Albumin (BSA)].
- Cut nylon mesh sheet into small 1 x 1 inch (2.5 x 2.5 cm) squares that will be used for filtering cellular debris and autoclave in a small beaker covered with aluminum foil.
- Prepare 1 L of wash buffer [wash buffer = 1,000 ml of 0.1 M PBS, 2 ml of 500 mM EDTA, and 10 g BSA].
2. Tissue Collection
- Turn on water bath to 37 ºC.
- Prepare Buffer X from the Neural Dissociation Kit by adding beta-mercaptoethanol to Buffer X to a final concentration of 0.067 mM (for 4 samples, add 13.5 µl of 50 mM beta-mercaptoethanol to 10 ml of Buffer X).
- Prepare Enzyme Mix 1 by mixing 50 µl of Enzyme P with 1,900 µl of Buffer X for each sample as described in the protocol for the Neural Dissociation Kit For Example, for 4 samples, mix 200 µl of Enzyme P with 7,600 µl of Buffer X.
- Place Buffer X in the water bath at 37 ºC.
- Place 2 ml of cold Hank’s Buffered Salt Solution (HBSS) (without Ca++ and Mg++) into one well of a sterile 6-well culture plate for each sample that will be collected and keep on ice for tissue dissection.
3. Tissue Dissection (45 min – 1 hr)
- Inject the animal with appropriate volume of euthanasia solution (commercial barbiturate solution, e.g., Euthasol), according to Institutional Animal Care and Use Committee (IACUC) guidelines, and perfuse out all blood with cold 0.9% saline solution.
- Administer Rats with an overdose of euthanasia solution.
- Monitor anesthesia via breathing and via toe-pinch. When the animal is sufficiently anesthetized (no response to the toe-pinch and deep, slow breathing), the rat can be perfused.
- To perfuse the rat, cut the chest cavity open with a pair of heavy-duty scissors. Cut the diaphragm to allow access up to the heart. Cut the ribs along both sides up to towards the top of the chest, to allow full access to the heart. At that point, the rib cage can be held off to the side using a pair of hemostats.
- Perform cardiac perfusion by inserting an 18 G needle (for adult rats) attached to an electric pump into the left ventricle of the heart. The pump is drawing up cold 0.9% saline solution.
- Then cut open the right atrium with a small pair of scissors to allow for the blood and liquid perfusate to drain from the body.
- Perfuse rats with approximately 10 ml (for adult rats) of 0.9% saline to remove all peripheral immune cells and factors.
- Dissect the desired part of the brain region using sterile razor blades and appropriate dissection, then place the tissue pieces into the cold HBSS without Ca2+ and Mg2+.
NOTE: The protocol requires HBSS without Ca2+ and Mg2+ in the early steps of the protocol as these ions may interfere with the enzymes required for neural dissociation. - After all tissue samples have been collected, return to the clean bench or sterile culture hood for tissue processing.
4. Neural Dissociation (1 hr)
- Dice each tissue sample into small pieces using sterile razor blades on the lid of the 6-well culture plate. This protocol has been used for tissue samples of approximately 30 – 100 mg, but is approved for tissue samples of up to 400 mg. For tissue samples larger than 400 mg, the reagent volumes must be scaled up appropriately.
- Use 1 ml of fresh HBSS without Ca2+ and Mg2+ to transfer the diced pieces of tissue with a pipette into a sterile 2 ml centrifuge tube. Repeat steps 4.1 and 4.2 for all sample until all samples have been transferred into a 2ml centrifuge tube.
- Centrifuge the samples at 300 x g for 2 min at RT and aspirate supernatant.
- Add 1,900 µl of warm Enzyme Mix 1, as prepared in Steps 2.2 and 2.3, to each sample and close the tubes.
- Incubate the samples for 15 min in the 37 ºC water bath, inverting the tubes several times every 5 min to re-suspend the settled pieces of tissue.
- Meanwhile, prepare Enzyme Mix 2 by mixing 20 µl of Buffer Y with 10 µl of thawed Enzyme A (for 4 samples, mix 80 µl of Buffer Y with 40 µl of thawed Enzyme A).
- Add 30 µl of Enzyme Mix 2 to each sample and invert gently. Do not vortex.
- Dissociate each tissue sample with Pasteur pipette “1”, triturating up and down 30 times.
- Incubate the samples for 15 min in the 37 ºC water bath; invert the tubes several times every 5 min to re-suspend the settled pieces of tissue.
- Dissociate each tissue sample with Pasteur pipette “2”, triturating up and down 30 times. Avoid generating bubbles.
- Dissociate each tissue sample with Pasteur pipette “3”, triturating up and down 30 times. Avoid generating bubbles.
- Incubate the samples for 10 min in the 37 ºC water bath; invert the tubes several times every 5 min to re-suspend the settled cells.
- Apply single-cell suspension to an 80 micron cell strain placed on top of a 15 ml falcon tube to remove any large pieces of debris and wash the filter and cells with 10 ml of HBSS (with Ca2+ and Mg2+) to stop the enzyme reactions. Repeat until each sample is transferred into a clean 15 ml falcon tube.
- Centrifuge the samples at 300 x g for 10 min at RT and aspirate off the supernatant.
5. Myelin Depletion (45 min)
- Thoroughly re-suspend the pellet of cells with 400 µl of myelin removal buffer.
- Add specified amount of the myelin removal beads and pipette up and down to thoroughly mix.
NOTE: The volume of myelin removal beads that is used will depend upon the size of the tissue, the age of the animal, and the amount of myelin expected in that particular brain region. e.g., for an adult rat hippocampus sample (approximately 300 mg), incubate the sample with 100 µl of myelin removal beads. - Incubate for 15 min in the refrigerator at 4 ºC.
- Wash each sample with 5 ml of myelin removal buffer.
- Centrifuge the samples at 300 x g for 10 min at RT.
- While centrifuging the samples, place 1 column for each sample in the magnetic field of the magnetic sorter and place a clean 80 micron filter on top of each column.
- Prepare the columns and filter by rinsing each column with 1 ml of myelin removal buffer, 3 times (3 ml total). Collect all flow-through in a waste container such as the bottom of an empty tip box.
- Position 5 ml polystyrene round bottom tube directly underneath each column in preparation of sample collection.
- When cells have finished centrifugation, aspirate supernatant and thoroughly re-suspend in 500 µl of myelin removal buffer. Immediately apply cell suspension to the column and collect the cells in the tube below.
- Wash the filter and column with 1 ml myelin removal buffer, 4 times (4 ml total).
- Centrifuge the cell collection at 300 x g for 10 min at RT.
- Aspirate the supernatant and the cells are now ready for staining.
6. Staining Live Cells for FACS (1 hr for staining)
- Briefly vortex (2 sec) the cells to separate the pellet and immediately add 5 µl of Fc block (anti- clusters of differentiation 32, CD32), vortex again, and incubate in the refrigerator at 4 ºC for 5 min.
- Meanwhile, prepare the primary antibody mixture by adding the three primary antibodies in wash buffer as follows: 0.125 µl of allophycocyanin (APC) -conjugated CD11b antibody (1:800), 1 µl of anti-GLT1 antibody (1:100), and 0.4 µl of Fluorescein isothiocyanate (FITC) – conjugated Thy1 antibody (1:250) for every 100 µl of wash buffer. Each sample will receive 100 µl of primary antibody mixture for incubation; however, if your sample is much larger than 400 mg of tissue, you may want to consider increasing the volume of your primary antibody mixture accordingly
- Prepare tubes for single staining controls by removing just 2 µl of cells from each sample tube and adding it to each single stain control tube. The “single stain” controls include: APC-CD11b only, GLT1 only, FITC-Thy1 only, PE secondary antibody only, and the “blank” or unstained control.
NOTE: All samples should be represented in all staining controls, thus 2 µl of cells from each sample should be added to each staining control tube. - Briefly vortex the tubes (2 sec) and add 100 µl of primary antibody mixture to each sample tube and 100 µl of appropriate primary antibody to each staining control tube, vortex the tubes again, cover the tubes with aluminum foil to protect the fluorescent antibodies from any light, and incubate the tubes in the refrigerator at 4 ºC for 20 min. The samples should be kept cool (at 4 ºC) for the remainder of the procedure to maintain the integrity of the samples and the antibodies.
- Briefly vortex the tubes and add 2 ml of wash buffer to each tube.
- Centrifuge tubes at 350 x g for 5 min at 4 ºC.
- Meanwhile, prepare the secondary antibody mixture by adding 0.2 µl of PE-conjugated anti-rabbit secondary antibody for every 100 µl of wash buffer. Each sample will get 100 µl of secondary antibody mixture; however, if your sample is much larger than 400 mg of tissue, you may want to consider increasing the volume of your primary antibody mixture accordingly.
- When the centrifugation is complete, dump the supernatant off of each sample into the sink and blot the tubes upside-down on a paper towel.
- Briefly vortex the tubes and add 100 µl of secondary antibody mixture to each sample tube and the appropriate secondary antibody to each staining control, vortex the tubes again, cover the tubes with aluminum foil, and incubate the tubes in the refrigerator at 4 ºC for 15 min.
NOTE: The GLT1-only staining control tube should get the secondary antibody mixture. - After the incubation, briefly vortex the tubes and add 2 ml of wash buffer to each tube.
- Centrifuge the tubes at 350 x g for 5 min at 4 ºC.
- When the centrifugation is complete, dump the supernatant off of each sample into the sink and blot the tubes upside-down on a paper towel.
- Vortex the tubes and add 0.25 ml of sterile PBS to each tube. The samples are ready for sorting.
NOTE: More cells may require 0.5 ml of sterile PBS; however it is best to begin with a lower volume as the sample can always be diluted if it is too concentrated with cells.
NOTE: Optionally add DNase to the tube to prevent clumping of the cells and thus clogging of the sorter. Samples can also be filtered using a sterile 80 micron filter just prior to sorting in order to avoid clogging the sorter. - Take cells to the sorter on ice. Collect the cell populations in 2 ml nuclease-free centrifuge tubes with 0.5 ml of sterile PBS in each tube.
- After the sort, spin the cells down at 300 x g for 2 min at 4 ºC. Aspirate off most of the PBS and flash freeze the cells in the -80 ºC freezer until further processing for either RNA extraction (see 1 for further protocol details), DNA extraction (see 2 for further protocol details) or cell lysis for protein analysis.
Representative Results
The Importance of Myelin Depletion and Tissue Perfusion
Figure 1 depicts the importance of myelin depletion. Myelin depletion (Steps 5.1 through 5.12 of the protocol above) occurs when the single-cell suspension is incubated in the Myelin Removal Beads, washed, and subsequently passed through the column on the magnetic sorter. The purpose of these steps is to reduce the amount of cellular debris present in each sample. As neurons are dissociated and triturated, it can shear off processes that are typically myelinated, and thus may stain positive with the Thy1 antibody. This results in myelinated debris which can ultimately interfere with the antibody staining and cell sorting. In addition it can reduce the relative number of cells that are retrieved from each sample. In Figure 1, the polygons drawn in each panel of the figure depict the living cells that will be gated and analyzed for subsequent sorting. This gate for living cells is drawn based on the forward scatter (FSC), which is a measure of size, and side scatter (SSC), which is a measure of granularity, of light passing by that particle, which ultimately excludes either dead cells and/or cellular debris. Thus, living cells should have a low level of granularity (low SSC) and a reasonable (not too small or not too large) size (FSC). Sample 1 (Figure 1A) and Sample 2 (Figure 1B) represent two different hippocampal samples that were individually prepared using all steps of the protocol above. The percent of cells that are gated is 53.9% of the total events analyzed in Sample 1 (9,656 cells out of 17,916 events) and 56.1% of the total events analyzed in Sample 2 (9,492 cells out of 16,920 events). Sample 3 (Figure 1C) is also a single hippocampus that was prepared using the protocol above but without the myelin depletion performed in Steps 5.1 – 5.12. The percent of cells that are gated is 3.93% of the total events analyzed in Sample 3 (4,312 cells out of 109,742 events), which is significantly less than in Sample 1 or Sample 2.
Myelin depletion is a relatively new technique, which can be compared in its purpose to a more popular technique, using the density gradient method such as Percoll. The purpose of performing a density gradient method is to remove cellular debris and dying cells from each sample. While the density gradient method is effective to some extent in this purpose (see 4,7 for examples of cells sorted following a density gradient method); it does not work nearly as well as the method of myelin depletion. The data in Figure 1 demonstrate the importance of myelin depletion in the increased total yield of cells. In addition, antibody staining can be greatly enhanced with the removal of myelinated debris.
In this protocol, it is important to perfuse the blood from the neural tissue using cardiac perfusion prior to tissue dissection and collection. This significantly helps to reduce the amount of neural cells that die during the procedure. Typically blood is separated from neural tissue by the blood brain barrier; however, dissection and dissociation of the tissue without perfusion can allow direct contact between neural cells and blood, which can result in increased neural cell death.
Staining Controls
Figure 2 represents the examples of the staining controls that must be performed each time cells are sorted. In this protocol, representative samples were stained with three antibodies including an APC-conjugated CD11b antibody to identify microglia, a FITC-conjugated Thy1 antibody to identify neurons, and an anti-GLT1 primary antibody with a phycoerythrin (PE) -conjugated secondary antibody to identify astrocytes. First, cells are sorted based on their forward and side scatter from all possible events. This gate is identified as P1 (population 1, Figure 2A). Next, the single cells, also called singlets, are sorted based on their size from any doublets or larger clumps of cells (see Figure 2A). These cells are not stained (and thus not gated) with any of the antibodies listed above. Instead, the gates for each cell type are determined based on the staining of the “single stain” controls that are prepared for each antibody. Figure 2B and Figure 2C depict the gate for APC-CD11b positive cells that are not present in the blank (unstained) control (Figure 2B) but are present in the APC alone (single stain) control sample. Figure 2D and Figure 2E depict the gate for the PE-GLT1 positive cells that are present in the GLT1-PE single stain control but not present in the FITC-Thy1 single stain control. Figure 2F depicts the gate for PE-GLT1 positive cells and FITC-Thy1 positive cells that are not present in the blank (unstained) control (similar to Figure 2B, which contains no stained cells in the CD11b positive gate). Figure 2G depicts the lack of both GLT1-PE staining and the lack of FITC-Thy1 staining in the APC alone (single stain) control. Figure 2H represents the PE-GLT1 positive cells that are present in the GLT1-PE single stain control (similar to Figure 2D but with a different axis for comparison), while Figure 2I represents the FITC-Thy1 positive cells found in the FITC-Thy1 single stain control. These single stain and unstained controls are used to identify the positive and negative cell populations for each antibody, to perform compensation in case one fluorochrome interferes with the signal from another fluorochrome, and to confirm that these antibodies work individually before they are combined in the samples themselves.
Sorting Neurons, Astrocytes, and Microglia from Individual Samples
Figure 3 depicts the sorted populations of neural cells from a single male hippocampus obtained from a FACS machine, and Figure 4 depicts the sorted populations of neural cells from a single male cerebellum. As described above, cells are sorted based on their forward and side scatter from all possible events (see Figure 3A and Figure 4A). This gate is identified as P1 (population 1). Next, the single cells, also called singlets, are sorted based on their size from any doublets or larger clumps of cells (see Figure 3B and Figure 4B). This gate is identified as P2. It is necessary to note the importance of obtaining singlets from the cell population. In order to accurately sort individual cell types, it is critical that the cells are not bound to each other in clumps of two or more. This would ultimately prevent the proper sorting of pure populations of cells. In order to prevent the appearance of doublets or larger clumps of cells in the samples, one must be sure to triturate each sample thoroughly and consistently across all samples with the glass Pasteur pipettes (numbered 1, 2, and 3) as described in Steps 4.8 through 4.11. Third, the single cells are then gated as either APC-CD11b positive (CD11b+ gate, P4, see Figure 3C and Figure 4C) or APC-CD11b negative (CD11b- gate, P3, see Figure 3C and Figure 4C). APC-CD11b negative cells are subsequently sorted into PE-GLT1 positive cells (GLT1+ gate, P6) and FITC-Thy1 positive cells (Thy1+ gate, P5), see Figure 3D and Figure 4D. Following each sort, the analysis program will determine the total number of events in each gate or population as well as the percent of the parent population(s). NOTE: We have used the FACS software in the current examples.
Confirming the Purity of each Cell Population using Real-time Polymerase Chain Reaction (PCR) of Cell-type-specific Genes
Figure 5 depicts the real-time PCR results obtained from the male hippocampus sample depicted in Figure 3 plus three additional male hippocampal samples (n = 4 hippocampi) and from the male cerebellum sample depicted in Figure 4 plus three additional male cerebellum samples (n = 4 cerebellum). To confirm the purity of the sorted cells in this particular example, we analyzed the relative gene expression of alternative cell-type-specific genes, using ribosomal 18s as the housekeeping gene for standardization. We analyzed the relative expression of the ionized calcium-binding adapter molecule (Iba1) which is a cytoplasmic protein expressed exclusively in microglia within the brain. We also analyzed the relative expression of the glial fibrillary acidic protein (GFAP) which is an intermediate filament protein expressed in astrocytes, and we also analyzed the relative expression of the N-methyl-D-aspartate (NMDA) receptor subunit 1 (NR1), which is expressed predominantly on neurons. These real-time PCR results confirm the purity of the sorted cells and identify some interesting differences across brain regions. For example, as expected, Iba1 was expressed approximately 400-fold higher in microglia than it was in either neurons or astrocytes (see Figure 5A and Figure 5D). As expected, GFAP was expressed more than 2,000-fold higher in hippocampal astrocytes than it was in hippocampal neurons or microglia (Figure 5B). Interestingly, however, GFAP was not highly expressed in the neurons or even the GLT1+ astrocytes that were sorted from the cerebellum as expected (Figure 5E). Given that others have identified GFAP expression in the cerebellum7; there are two possible explanations for these results. First, it is possible that GLT1 is expressed on a different subtype of astrocytes, one that does not co-express GFAP in the cerebellum or it is possible that GLT1 is not expressed at all on astrocytes in the cerebellum and that a different type of cell, that is neither Thy1+ or CD11b+, has been isolated from the cerebellum using this technique. Finally, as expected, NR1 was expressed approximately 300-fold higher in neurons than it was in either astrocytes or microglia (see Figure 5C and Figure 5F). These representative results confirm and expand upon previous findings using this technique to sort neurons, astrocytes, and microglia with these antibodies from the hippocampus and the nucleus accumbens using a different machine at a different institution1.
Following confirmation of purity of the sort, one can analyze their gene of interest to determine the specific cell type in which it is expressed or whether it is altered following treatment in a cell-type-specific manner. For example, we analyzed the expression of calbindin, a calcium-binding protein that is often used to identify neurons in the hippocampus and the cerebellum. Calbindin was expressed significantly more in hippocampal neurons than in any other hippocampal cell type (Figure 6A) but that it was expressed at significantly higher levels in the neurons isolated from the cerebellum (Figure 6B) than it was from the neurons of the hippocampus.
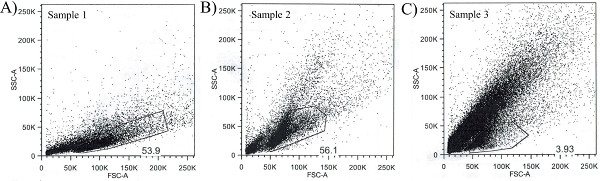
Figure 1. The effect of myelin removal on cell sorting. (A) Sample 1 and (B) Sample 2 represent two different hippocampal samples that were individually prepared using all steps of the protocol above. The percent of cells that are gated is listed inside each example. Specifically, 53.9% of the total events analyzed in Sample 1 were cells (9,656 out of 17,916 events) and 56.1% of the total events analyzed in Sample 2 were cells (9,492 out of 16,920 events). (C) Sample 3 is also a single hippocampus that was prepared using the described protocol, but without the myelin depletion performed in Steps 5.1 – 5.12. The percent of cells that are gated is 3.93% of the total events analyzed in Sample 3 (4,312 out of 109,742 events), which is significantly less than in Sample 1 or Sample 2. Please click here to view a larger version of this figure.
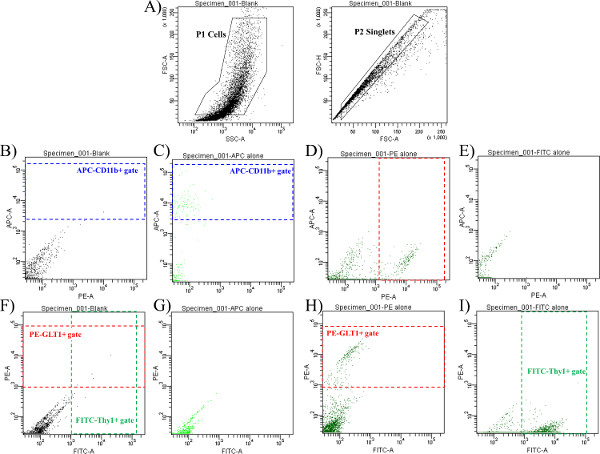
Figure 2. Staining controls must be performed for each sort. Cells were stained with three antibodies including an APC-conjugated CD11b antibody to identify microglia, a FITC-conjugated Thy1 antibody to identify neurons, and an anti-GLT1 primary antibody with a PE-conjugated secondary antibody to identify astrocytes. (A) These plots depict the forward and side scatter of cells that are not stained (and thus not gated) with any of the antibodies listed above. The gates for each cell type are determined based on the staining of the “single stain” controls that are prepared for each antibody below. (B) and (C) depict the gate for APC-CD11b positive cells that are not present in the blank (unstained) control (B) but are present in the APC alone (single stain) control sample (C). (D) and (E) depict the gate for the PE-GLT1 positive cells that are present in the GLT1-PE single stain control (D) but not present in the FITC-Thy1 single stain control (E). (F) depicts the gate for PE-GLT1 positive cells and FITC-Thy1 positive cells that are not present in the blank (unstained) control (similar to (B), which contains no stained cells in the CD11b positive gate). (G) depicts the lack of both GLT1-PE staining and the lack of FITC-Thy1 staining in the APC alone (single stain) control. (H) represents the PE-GLT1 positive cells that are present in the GLT1-PE single stain control (similar to (D) but with a different axis for comparison), while (I) represents the FITC-Thy1 positive cells found in the FITC-Thy1 single stain control. Please click here to view a larger version of this figure.
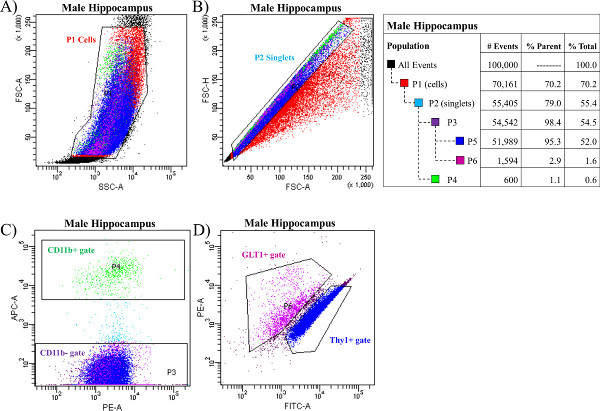
Figure 3. Neurons, astrocytes, and microglia sorted from a male hippocampus. The hippocampus from one male rat was dissociated and stained with the antibodies for CD11b, GLT1 and Thy1 and sorted using a FACS machine. (A) Cells were first sorted based on their forward and side scatter from all possible events. This gate is called P1 (population 1). (B) Next, single cells, also called singlets, were sorted based on their size from the doublets or larger clumps of cells. This gate is called P2. (C) Third, the single cells were gated as either APC-CD11b positive (CD11b+ gate, P4) or APC-CD11b negative (CD11b- gate, P3). (D) APC-CD11b negative cells were subsequently sorted into PE-GLT1 positive cells (GLT1+ gate, P6) and FITC-Thy1 positive cells (Thy1+ gate, P5). The breakdown of all events and all gates was generated from the FACS software depicted in a table which is presented on the right. Please click here to view a larger version of this figure.
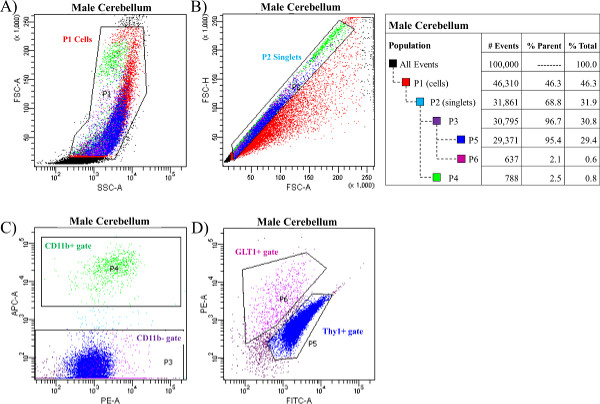
Figure 4. Neurons, astrocytes, and microglia sorted from a male cerebellum. The cerebellum from one male rat was dissociated and stained with the antibodies for CD11b, GLT1 and Thy1 and sorted using a FACS machine. (A) Cells were first sorted based on their forward and side scatter from all possible events. This gate is called P1 (population 1). (B) Next, single cells, also called singlets, were sorted based on their size from the doublets or larger clumps of cells. This gate is called P2. (C) Third, the single cells were gated as either APC-CD11b positive (CD11b+ gate, P4) or APC-CD11b negative (CD11b- gate, P3). (D) APC-CD11b negative cells were subsequently sorted into PE-GLT1 positive cells (GLT1+ gate, P6) and FITC-Thy1 positive cells (Thy1+ gate, P5). The breakdown of all events and all gates was generated from the FACS software depicted in a table which is presented on the right. Please click here to view a larger version of this figure.
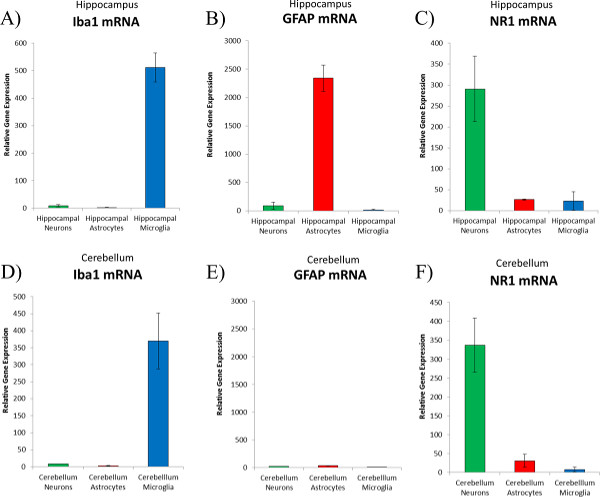
Figure 5. Real-time PCR analysis of cell-type-specific genes from sorted cells. Neurons (green bars), astrocytes (red bars) and microglia (blue bars) were sorted based on the protocol described above and mRNA was extracted for confirmation of cell-type-specific gene expression. (A) Iba1 is a calcium binding protein expressed exclusively in microglia sorted from the male hippocampus. (B) GFAP is a filament protein expressed predominantly in astrocytes sorted from the male hippocampus (C) NR1 is a ubiquitous subunit of the NMDA glutamatergic receptor that was expressed predominantly on neurons sorted from the male hippocampus. (D) Iba1 was also expressed exclusively on microglia sorted from the male cerebellum. (E) Interestingly, GFAP was not expressed in any of the cell types sorted from the male cerebellum. (F) The NR1 subunit of the NMDA receptor was also expressed predominantly on neurons sorted from the male cerebellum. Please click here to view a larger version of this figure.
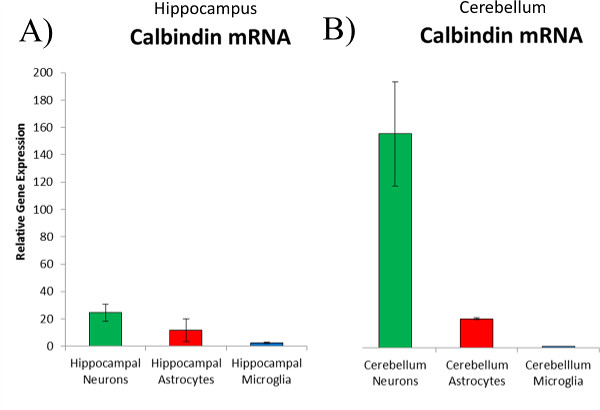
Figure 6. Real-time PCR analysis of calbindin expressed in sorted neural cells. Cells sorted using FACS can be used to analyze cell-type-specific gene expression. (A) Neurons (green bars) expressed significantly more Calbindin than either astrocytes (red bars) or microglia (blue bars) sorted from the male hippocampus. (B) Neurons sorted from the male cerebellum expressed significantly higher levels of Calbindin than either astrocytes or microglia sorted from the cerebellum, but also significantly higher levels than the neurons sorted from the hippocampus. Please click here to view a larger version of this figure.
Discussion
Similar to many other tissues and systems, the brain is comprised of a heterogeneous cell population that functions together to impact behavior. The analysis of gene expression, protein expression, or epigenetic modifications from individual cells within that heterogeneous population has the potential to reveal information about the function of the system as a whole, to identify cellular processes regulating both normal behavior and disease processes, and to provide cell-type-specific targets for potential therapies. It is possible that in the future, modifying the function of a single cell type within the brain may unlock the key to successfully treating neurodegenerative or neuropsychiatric disorders in a more specific manner, reducing non-specific side effects caused by impacting multiple cell types. Thus a key focus of future neuroscience research must seek to understand the interactions of different cell types in the brain and how they influence behavior and disease states. The goal of the FACS technique described here is to rapidly isolate individual cell populations for subsequent analysis. It can also be modified to analyze more specific cell types depending upon the research of interest.
There are a few critical steps within the protocol that will help increase the successful yield of isolated neural cells using this technique. First, it should be mentioned that the protocol described above has been optimized for the isolation of individual cell types from fresh rat brain tissue. It is possible that many of these procedures could be used for the processing and isolation of individual cell types from previously frozen or fixed tissue, but the specifics of those procedures must be optimized. We have found that perfusing the blood from the neural tissue with cold physiological saline will help to decrease neural cell death, increase the specificity of the cells isolated, and thus the yield of sorted neural cells. While this portion of the protocol may not be a common technique for most neuroscientists, it will significantly help with the successful implementation of this particular protocol. In addition, if one would like to sort neural cells based on a protein that is also expressed by circulating blood cells, it is important to remove these circulating cells from the neural tissue prior to dissociation and staining so as to eliminate the potential confound of isolating both neural and peripheral blood cells. A second critical step is the thorough trituration of the neural tissue with the Pasteur pipettes numbered as 1, 2 and 3. While this may seem simple, it is important that this is done thoroughly in order to effectively separate clumps of neural cells for sorting. The trituration should be done vigorously enough to dissociate the cells, but should avoid generating excessive air bubbles, which can also kill delicate neural cells. If this step is not done well, it will be reflected at the cell sorter by an increase in the number of “doublets” or debris seen in the sort. This will also result in a decrease in cell yield, as the sorter will only sort individual, fully dissociated cells that are stained with the appropriate antibodies to maintain the purity of the sorting process. Choosing the proper antibodies for staining is also a critical step in the successful application of this protocol. In this particular protocol, neural cells were isolated from the adult rat brain using antibodies specific to rat CD11b, rat GLT1 and rat Thy1. Thus different antibodies would need to be used to isolate these same cells using these cell-type-specific markers for mouse, non-human primate, or human neural tissue. With the appropriate species-specific antibodies, this protocol should work well in other species. In addition to the species used in this protocol, age of the animal should also be considered in this protocol. The expression of many molecules, including CD11b, GLT1 and Thy1 may change dramatically across development. If a protein is expressed at low or non-existent levels in the developing brain, it should not be used as a target for isolating cells via FACS.. Finally, as mentioned previously, it would be ideal to have access to a cell sorter that is close in proximity to the lab in order to reduce the length of time needed to complete the procedure with these delicate living cells.
The sorting of the cells is dependent, to some extent, upon the machine that is used to sort the cells. In this example, we have used a FACS using FACS software to gate and sort the cells dependent upon the strength of their fluorescent staining. This machine is located at the Helen F. Graham Cancer Center affiliated with the University of Delaware. In a previous experiment, we used a High-Speed Sorter such as MoFlo, to perform these same exact experiments. This machine was located at the Duke Cancer Institute Flow Cytometry Core found on the campus of Duke University Medical campus. For comparison of the sorts produced by these two machines, please reference the following paper1. Based on the findings from the two machines, we have determined that the machine used for analysis can impact the number of cells detected, particularly if the laser is stronger on one machine than it is on another. This was, in fact, the case for the current experiment. In the current experiment, we were able to more easily detect FITC-Thy1+ neurons because the laser was significantly stronger on the FACS than the standard laser of the High-Speed Sorter used in previous experiments1. This increased the number of FITC-Thy1+ neurons obtained from one hippocampus from approximately 3,000-5,000 neurons isolated with the High-Speed Sorter to approximately 50,000 neurons isolated with the FACS.
In this particular example, RNA was extracted from the sorted cells for subsequent analysis of gene expression. The method for extracting RNA from sorted cells has been described in a previous paper1, but can be done using any established RNA extraction protocols or RNA extraction kits. Good quality RNA can be extracted from relatively few, very specific, populations of isolated cells6. In addition to extracting RNA, it is possible to extract DNA from these isolated cells. We have also extracted DNA for analysis of DNA methylation from isolated microglia in a separate paper 2; however, these cells were isolated using a density gradient method as opposed to FACS. Despite that, the isolation of individual cells was critical in finding significant effects of neonatal handling, increased maternal care, on DNA methylation. Specifically, we found that the anti-inflammatory gene Interleukin (IL)-10 was expressed at significantly elevated levels in the brain of neonatally handled rats than control rats. To determine whether this effect was the result of epigenetic modifications to the IL-10 gene, we first extracted DNA from whole tissue for a subsequent Methylated DNA Immunoprecipitation and analysis of the IL-10 gene; however, we found no significant effects of neonatal handling on IL-10 methylation from whole tissue. Next, we isolated microglia from the brains of neonatally handled and control rats before extracting the DNA and analyzing the IL-10 gene following a Methylated DNA Immunoprecipitation (MeDIP). The results of this cell-type-specific analysis revealed a significant effect of neonatal handling as expected 2, contrary to the results obtained using whole tissue analysis. In retrospect, this is perhaps not surprising given that these cells have different lineages and different functions. DNA methylation is an important process of cellular differentiation as certain genes are silenced and others are “turned on” 9, 10. Each cell type in the brain may have different epigenetic markers indicative of their lineage and their function 11-13, and thus it is critically important to analyze the epigenetic control of gene expression, neural function and behavior from individual cell types. FACS is a simple and effective technique to analyze these differences in a cell-type-specific manner. Finally, the isolated cells can also be used to analyze protein expression in a cell-type-specific manner. Many different cell types use similar signaling molecules for very different processes. For example, the transcription factor, Nuclear factor kappa B (NF-κB), can alter neuronal morphology and behavior via its activation in the nucleus accumbens 14. NF-κB is also expressed in immune cells and is a critical transcription factor for the induction of pro- and anti-inflammatory molecules (see 15 for review). Thus it is critical to examine factors such as these in a cell-type-specific manner in order to increase our understanding of the function of these important molecules in behavior and disease processes.
As mentioned, this procedure can also be used to isolate more specific cell types than just neurons, astrocytes, and microglia in general. When considering this, one should determine approximately how many cells in the brain are of a particular phenotype. For example, if one would like to isolate only serotonergic neurons, this is a more select subset of neurons and thus the experimenter may obtain fewer cells than the numbers obtained here. In this case, it may help to pool the neural tissue from two or three rats to obtain one sample and thus enough cells for subsequent analysis. If one would like to isolate cells that have serotonergic receptors, consider whether serotonergic receptors are found only on neurons or on astrocytes and microglia too. If they are localized to multiple cell types, it might be best to sort cells that stain positive for the serotonergic receptor of interest and one of the neural cell markers used in the example here to isolate, for example, only neurons (Thy1+) that contain serotonergic receptor if that is the endpoint of interest. This technique has also been used to sort neuronal nuclei, using an antibody for neuronal nuclei (NeuN), and glial nuclei, using an antibody for the Oligodendrocyte transcription factor (Oligo2), from whole brain tissue15. This, of course, requires the fixation and permeabilization of tissue so that the antibodies can access the nuclear proteins to which they can bind. It is possible to effectively analyze RNA from cells sorted using intracellular markers such as NeuN or Oligo2 (see References 4-6 for examples); however, the caveat of this process being that it only measures RNA that remains in the nucleus and given the harsh treatment of the cells with fixation and permeabilization, this process may result in only approximately 25% of the total amount of RNA that can typically be extracted from whole tissue16. Despite this potential caveat, sorting and analyzing gene expression from individual cells based on nuclear proteins is also an important and necessary technique for understanding the processes that regulate neural function on a per cell basis.
Overall the FACS procedure presented here is a very simple and effective way to isolate individual cell types for subsequent analysis. It requires very little specialized equipment in the lab, though it does require a good connection with a core flow cytometry facility; however, this technique can easily be implemented into any lab’s ongoing experiments.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors would like to acknowledge Lynn Opdenaker at the University of Delaware Center for Translational Research at the Helen F. Graham Cancer Center for technical assistance, as well as Nancy Martin from the Duke University Cancer Institute Flow Cytometry Shared Resource, and Dr. Susan H. Smith for guidance in methods and data collection.
Materials
| Neural Dissociation Kit (P) | Miltenyi Biotec | 130-092-628 | |
| Myelin Removal Beads II | Miltenyi Biotec | 130-096-733 | |
| LS Columns | Miltenyi Biotec | 130-042-401 | |
| QuadroMACS Separator | Miltenyi Biotec | 130-090-976 | |
| MACS MultiStand | Miltenyi Biotec | 130-042-303 | |
| Nylon Mesh Sheet | Amazon | CMN-0074-10YD | 40 inch width, 80 micron size mesh |
| Fc Block / anti-CD32 | BD Biosciences | BDB550270 | reactivity for rat |
| APC-conjugated CD11b antibody | Biolegend | 201809 | reactivity for rat |
| Rabbit anti-GLT1 | Novus Biologicals | NBP1-20136 | reactivity for rat or human |
| PE-conjugated anti-rabbit secondary antibody | eBioscience | 1037259 | secondary antibody for anti-GLT1 |
| FITC-conjugated anti-rat CD90 (Thy1) mouse antibody | Biolegend | 202504 | reactivity for rat |
References
- Schwarz, J. M., Smith, S. H., Bilbo, S. D. FACS analysis of neuronal-glial interactions in the nucleus accumbens following morphine administration. Psychopharmacology. 230 (4), 525-535 (2013).
- Schwarz, J. M., Hutchinson, M. R., Bilbo, S. D. Early-life experience decreases drug-induced reinstatement of morphine CPP in adulthood via microglial-specific epigenetic programming of anti-inflammatory IL-10 expression. J Neurosci. 31 (49), 17835-1523 (2011).
- Herzenberg, L. A., Tung, J., Moore, W. A., Herzenberg, L. A., Parks, D. R. Interpreting flow cytometry data: a guide for the perplexed. Nature Immunology. 7 (7), 681-685 (2006).
- Guez-Barber, D., et al. FACS identifies unique cocaine-induced gene regulation in selectively activated adult striatal neurons. J Neurosci. 31 (11), 4251-4259 (2011).
- Fanous, S., et al. Unique gene alterations are induced in FACS-purified Fos-positive neurons activated during cue-induced relapse to heroin seeking. J Neurochem. 124 (1), 100-108 (2013).
- Liu, Q. R., et al. Detection of molecular alterations in methamphetamine-activated Fos-expressing neurons from single rat dorsal striatum using fluorescence-activated cell sorting (FACS). J Neurochem. 128 (1), 173-185 (2013).
- Guez-Barber, D., et al. FACS purification of immunolabeled cell types from adult rat brain. J Neurosci Methods. 203 (1), 10-18 (2012).
- Nolte, C., et al. GFAP promoter-controlled EGFP-expressing transgenic mice: a tool to visualize astrocytes and astrogliosis in living brain tissue. Glia. 33 (1), 72-86 (2000).
- Okana, M., Bell, D. W., Haber, D. A., Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 99 (3), 247-257 (1999).
- Bogdanović, O., Veenstra, G. J. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma. 118 (5), 549-565 (2009).
- Iwamoto, K., et al. Neurons show distinctive DNA methylation profile and higher inter-individual variations compared with non-neurons. Genome Res. 21 (5), 688-696 (2011).
- Nishioka, M., et al. Neuronal cell-type specific DNA methylation patterns of the Cacna1c gene. Int J Dev Neurosci. 31 (2), 89-95 (2013).
- Kozlenkov, A., et al. Differences in DNA methylation between human neuronal and glial cells are concentrated in enhancers and non-CpG sites. Nucleic Acid Res. 42 (1), 109-127 (2014).
- Russo, S. J., et al. Nuclear factor kappa B signaling regulates neuronal morphology and cocaine reward. J Neurosci. 29 (11), 3529-3537 (2009).
- Bhatt, D., Ghosh, S. Regulation of the NF-κB-Mediated Transcription of Inflammatory Genes. Front Immunol. 5, 71 (2014).
- Okada, S., et al. Flow cytometric sorting of neuronal and glial nuclei from central nervous system tissue. J Cell Physiol. 226 (2), 552-558 (2011).

