Energy Dispersive X-ray Tomography for 3D Elemental Mapping of Individual Nanoparticles
Summary
The use of energy dispersive X-ray tomography in the scanning transmission electron microscope to characterize elemental distributions within single nanoparticles in three dimensions is described.
Abstract
Energy dispersive X-ray spectroscopy within the scanning transmission electron microscope (STEM) provides accurate elemental analysis with high spatial resolution, and is even capable of providing atomically resolved elemental maps. In this technique, a highly focused electron beam is incident upon a thin sample and the energy of emitted X-rays is measured in order to determine the atomic species of material within the beam path. This elementally sensitive spectroscopy technique can be extended to three dimensional tomographic imaging by acquiring multiple spectrum images with the sample tilted along an axis perpendicular to the electron beam direction.
Elemental distributions within single nanoparticles are often important for determining their optical, catalytic and magnetic properties. Techniques such as X-ray tomography and slice and view energy dispersive X-ray mapping in the scanning electron microscope provide elementally sensitive three dimensional imaging but are typically limited to spatial resolutions of > 20 nm. Atom probe tomography provides near atomic resolution but preparing nanoparticle samples for atom probe analysis is often challenging. Thus, elementally sensitive techniques applied within the scanning transmission electron microscope are uniquely placed to study elemental distributions within nanoparticles of dimensions 10-100 nm.
Here, energy dispersive X-ray (EDX) spectroscopy within the STEM is applied to investigate the distribution of elements in single AgAu nanoparticles. The surface segregation of both Ag and Au, at different nanoparticle compositions, has been observed.
Introduction
The aim of this method is to provide accurate determination of the three dimensional distribution of elements within single nanoparticles. This is carried out through the use of energy dispersive X-ray (EDX) spectroscopy in conjunction with a tomographic reconstruction performed in the scanning transmission electron microscope (STEM).
Energy dispersive X-ray spectroscopy has long been used as a technique to quantify and spatially map elements present in transmission electron microscopy samples. With the advent of high angle annular dark field (HAADF) STEM tomography for the three dimensional imaging of crystalline materials1, energy dispersive X-ray tomography was also proposed as a method to allow the determination of elemental distributions in three dimensions2. However, early studies were limited due to the design of X-ray detectors within the transmission electron microscope. Specifically these traditional detector designs had relatively low collection efficiencies and measured no signal at a large range of tilt angles due shadowing from the sample holder2,3. The introduction of new geometrical designs of X-ray detectors within the (scanning) transmission electron microscope has made energy dispersive X-ray tomography a viable technique and has led to a number of recent studies4-6.
HAADF STEM imaging is a widely used electron tomography imaging mode and is able to provide compositional information in specific situations based on the atomic number sensitivity of the HAADF signal intensity. For example, HAADF tomography is well suited to the study of nanoparticles with discrete elemental regions, e.g., well defined core-shell morphologies7, but cannot be used when elements have a more complicated distribution. Electron energy loss spectroscopy (EELS) provides a complementary approach for determining three dimensional elemental distributions within the STEM8. In this technique the energy losses of the incident electron beam are used to determine the composition of the sample and this has the advantage of higher signal-to-noise ratio than is often obtained by EDX spectroscopy9. The disadvantage of EELS is that multiple scattering considerations impose stringent limits on the specimen thickness, and in several situations analysis is complicated by the presence of delayed onset edges or overlapping spectral features. Thus, EDX spectroscopy is often better suited to studying heavy elements such as those often associated with catalytic or plasmonic nanoparticle systems9. Additionally, as a full spectrum image is collected in EDX spectroscopy it is straightforward to retrospectively identify unexpected elements, which is more difficult in energy filtered transmission electron microscopy (EFTEM) and EELS due to the frequency of elemental information overlapping or being outside the spectral range of the data set.
The ideal sample geometry for EDX tomography consists of a needle shaped sample suspended in a vacuum and oriented along the tomographic tilt axis4. This situation ensures that there is no shadowing of the EDX detectors at any tilt angle by either the sample or the specimen holder. However, assembly of the required needle shaped samples for nanoparticle systems is challenging10 and sample preparation usually consists of simply transferring nanoparticles onto a thin carbon film TEM support grid. These grids are used with a tomography specimen holder specially designed so that it can be tilted to large angles (≈±75°) but shadowing of the EDX detectors within this range of specimen tilt angles is unavoidable and could degrade the quality of the resulting tomographic reconstruction. This shadowing is characteristic of a particular microscope-detector-holder setup and can therefore be determined by measurement of a proper calibration sample before acquisition11. Single spherical nanoparticles are an ideal calibration specimen as the intensity of X-ray counts from these samples should remain constant over all tilt angles. The detector shadowing can then be compensated by either varying the acquisition time at each angle or by multiplication with correction factor after data acquisition. The former approach is used as this minimizes electron dose while maximizing signal to noise ratio.
Protocol
1. Nanoparticle Synthesis
- Dissolve 10 g of Polyvinyl-pyrrolidone (PVP) (MW = 10,000 g/mol) in 75 ml of ethylene glycol at RT. Add 400 mg of AgNO3 to this solution.
- Stir solution until AgNO3 is completely dissolved and subsequently heat on a hotplate to 100 °C at a constant rate of 1 °C min-1. Let reaction proceed at 100 °C for 1.5 hr.
- Add 175 ml of distilled water and cool to RT. Centrifuge at 8,000 x g, remove supernatant and redisperse nanoparticles in 50 ml of distilled water. Repeat three times.
- Dissolve 500 mg of PVP (55,000 g/mol) in 500 ml of ethylene glycol at RT. Add this solution and 27.8 ml of the Ag nanoparticle suspension from the previous step to a 1,000 ml round bottom flask. Heat the flask at 100 °C for 10 min.
- Add hydrogen tetrachloroaurate trihydrate to 1,000 ml of water to form a 0.2 mM solution of AuCl4–(aq). Add 100, 200, 300 or 400 ml aliquots of the 0.2 mM AuCl4–(aq) dropwise to the solution obtained in the previous step to obtain AgAu nanoparticles of average composition Ag93Au7 (Ag 93 at% Au 7 at%), Ag82Au18 (Ag 82 at% Au 18at%), Ag78Au22 (Ag 78 at% Au 22 at%) and Ag66Au34 (Ag 66 at% Au 18 at%) respectively.
- Cool to RT and then wash with distilled water three times by successive rounds of centrifugation at 8,000 x g, removal of the supernatant and redispersion in 50 ml of distilled water. Add 0.05 ml of the nanoparticle solution to 10 ml of DI water.
2. TEM Sample Preparation
- Pipette approximately 0.05 ml of the nanoparticle solution on to a holey/continuous carbon TEM grid. Use a metal support used for TEM grids of Beryllium to limit spurious X-rays, although a Cu or Au grid will suffice if there are no overlapping peaks of interest with Cu or Au in the sample's EDX spectrum. Use larger mesh sizes (e.g., 200 mesh) to minimize the possibility of shadowing from the grid.
- After the nanoparticle solution has dried, clean the TEM grids by washing the grids in pure methanol or ethanol. Pipette 10-20 drops over TEM grid while it is held using anti-capillary crossover tweezers. Remove any excess liquid after each drop from the grid using filter paper touched gently on to the edge of the grid.
- Anneal grid at around 80 °C (preferably in vacuum) to reduce contamination under the electron beam by removing or immobilizing any remaining contamination.
3. Characterization of Detector Shadowing
- Load nanoparticles on TEM support grid into tomography holder and then insert holder into the TEM.
- Wait for the microscope vacuum to reach a suitable stable value (below approximately 1.8 x 10-7 Torr). Open column valves, and ensure the electron beam can be seen on the microscopes fluorescent screen.
- Align the STEM.
Note: Here, alignments for a Titan G2 probe-side aberration corrected STEM instrument operated at 200 kV together with a 2020 tomography holder are described. Optimal alignment procedures for other microscopes may vary slightly from those described.- In STEM diffraction mode, use an out-of-focus shadow image to bring the sample to eucentric height. To do this, tilt the stage between ± 15° by pressing the Alpha Wobbler button in the Search tab and minimize any movement of features in the sample by adjusting the z-height.
- Move the sample so that the beam is over a hole in the sample and ensure that the appropriate condenser aperture is correctly aligned by centering the shadow of the aperture in an under focused image of the probe. Wobble the intensity and ensure a focused probe does not move to check for beam tilt misalignment.
- Move the sample so that an area of amorphous carbon is in view and focus the beam in diffraction mode to obtain the Ronchigram. Wobble focus to observe aberrations within the Ronchigram, and correct for these (e.g., astigmatism and axial coma) if necessary.
- Trace the outline of the grid square (one close to the center of the grid) by selecting 'Show Tracks' in the 'Search' tab and following the outline of the grid square whilst imaging. This makes it straightforward to ensure subsequent measurements are performed for nanoparticles that are in the middle of the grid square in order to reduce the likelihood of shadowing from the support grid. If the acquisition is performed on a particle close to a grid bar one pair of detectors will be shadowed over a much larger range of specimen tilt angles.
- Find a representative particle within the middle portion of the grid square. Tilt to maximal tilt range of the holder (usually ±70°) whilst imaging to ensure that the nanoparticle is not obscured by the grid bars at large holder tilts.
- Acquire HAADF and EDX spectrum images using a constant acquisition time (e.g., 5 min) over the full range of tilt angles (usually ± 70°) using angular increments of between 5-10°. First, acquire an overview HAADF image by clicking Acquire in the imaging pane. Select a mapping window around the nanoparticle by dragging the box over this image and press Acquire.
- Extract characteristic X-ray counts from EDX spectrum images to determine time intervals for data acquisition by opening the spectral datacubes in the image processing software (as RAW files). Then, sum the intensity of slices of the datacube that correspond to the channels of the energy of the peaks of interest by using the slice2D and sum functions, scripts included as supplemental code 1-3.
4. EDX Tomography Acquisition
- Repeat steps 3.1-3.5 for the nanoparticle specimen of interest.
- Acquire HAADF and EDX spectrum images with the time intervals determined from the detector shadowing characterization of the previous section for a range of specimen tilt angles (usually ± 70°), in the same manner as step 3.6.
5. Reconstruction and Visualization
- Compile tilt series of HAADF images into mrc file format through one of the following methods:
- Use MRC writer in image visualization software12 (File > Save As > MRC writer) after importing HAADF tifs as an 'Image Sequence'.
- Alternatively, use the tif_to_mrc function in the tomography software package13.
- Cross-correlate HAADF images in the chosen software to get a rough alignment of the tilt series. Save the alignment data as either a .sft file or as the file xcorr.txt. In the tomography software set up a filter in the Setup Filter window that provides the sharpest peak in the Cross Correlation pane.
- Subsequently, press Proceed in the Calculate Alignment Shifts window to perform cross correlation for the entire tilt series. Repeat the alignment until local shifts are below 1 pixel, ensuring to save all shift files at each step as text.
Note: Careful setup of the filter used when performing cross-correlation is sometimes necessary to ensure a good alignment of the tilt series data.
- Subsequently, press Proceed in the Calculate Alignment Shifts window to perform cross correlation for the entire tilt series. Repeat the alignment until local shifts are below 1 pixel, ensuring to save all shift files at each step as text.
- In the image processing software, load the acquired spectral datacubes and extract maps that are the summed slices of energy channels corresponding to the elements of interest using the slice3d and sum scripting functions, scripts included as supplemental code 4 & 5.
- Apply the alignments determined from the HAADF images to the extracted EDX elemental maps. Perform this in tomography reconstruction software in the Apply Alignment tab by selecting "Use Shifts from Alignment Data View".
- If necessary, perform tilt axis adjustment in the tomography reconstruction software on the HAADF image series and apply to both HAADF images and EDX maps by checking "Use Corrections from Tilt Axis Adjustment Task" in the 'Apply Alignment' tab.
- Reconstruct tomographic data set for each of the extracted EDX elemental signals using a simultaneous iterative reconstruction technique (SIRT) algorithm implemented within a tomography software package, ensuring that the dimensions of the reconstruction are the same for all elements. In the tomography reconstruction software, set up the parameters in the Reconstruct Volume window (size of volume, 20 iterations, etc.) and press Proceed. Ensure that the reconstruction parameters are the same for each element.
- Extract orthoslices of the separate elemental reconstructions by selecting Image > Stacks > Orthogonal Views in the image visualization software12.
- Construct and visualize further orthoslices, volume, and surface renderings from the reconstructions using the visualization software. Load all elemental reconstructions and ensure that the scale is set correctly as this is often not transferred from .rec files.
- Select the reconstruction object in the object pool and right-click and select the orthoslice module to extract an orthoslice. Right-click on the reconstruction and select volume rendering to extract a volume rendering. Extract an isosurface by right-clicking on the reconstruction and selecting isosurface.
Note: Automatic segmentation through thresholding is a more robust method but where the signal to noise ratio is poor manual segmentation may be necessary to remove noisy voxels from outside the nanoparticle volume for isosurface visualization.
- Select the reconstruction object in the object pool and right-click and select the orthoslice module to extract an orthoslice. Right-click on the reconstruction and select volume rendering to extract a volume rendering. Extract an isosurface by right-clicking on the reconstruction and selecting isosurface.
Representative Results
The characterization of detector shadowing for a 2020 tomography holder in a Titan G2 ChemiSTEM is displayed in Figure 1A. The detectors employed here are that of the Super-X detector, in which four detectors are employed at equal azimuthal angles of 90° around the optic axis, resulting in a detector solid angle of larger than 0.6 sR14. Characterization of the detectors allowed determination of compensated tomographic acquisition times displayed in Figure 1B. After application of these acquisition times the counts at each tilt angle should remain roughly constant for single nanoparticles, as shown in Figure 1C.
Bimetallic AgAu nanoparticles, synthesized by the galvanic replacement reaction, were investigated using EDX tomography in the STEM. In this reaction, a solution of AuCl4– is added to Ag nanoparticle seeds. Au is reduced on the surface of the nanoparticles as Ag is oxidized, resulting in a bimetallic composition and hollowing of the initial seed. Previously, it was thought that Ag and Au formed a homogeneous alloy throughout this process and that variations in catalytic and optical properties were simply due to hollowing and bulk compositional variations. However, three dimensional elemental mapping performed using EDX tomography revealed surface segregation within the AgAu nanoparticles synthesized by the galvanic reaction (Figure 2). At compositions of low Au, the nanoparticles display clear Au surface segregation. However, as the Au content increases this surface segregation switches so that for the highest Au contents there is clear Ag surface segregation (Figure 3). This switching of surface segregation correlates with changes in the yield of propargylamines in a three component coupling reaction synthesized by different compositions of these AgAu nanoparticles.
Slices through the reconstruction that are normal to the direction of the electron beam provide a comparison to standard two dimensional elemental maps. The initial maps contain information on the composition summed along the direction of the beam and this can often be tricky to interpret due to overlapping regions of differing compositions or, in the case of the AgAu nanoparticles investigated here, due to inclusion of the top and bottom surfaces in projection (Figure 3A-C). Taking slices through the reconstruction allows the removal of the intensity associated with the top and bottom surfaces of the particles and therefore results in a much clearer display of surface segregation in this case (Figure 3D-F).
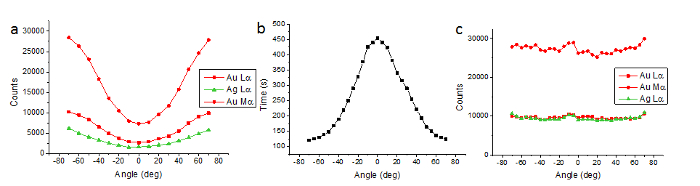
Figure 1. Characterization of detector shadowing using a single AgAu nanoparticle. (A) The counts of Ag and Au X-ray peaks as a function of tilt angle from a single AgAu nanoparticle when employing a set acquisition time (5 min). (B) The acquisition times determined from (A) and subsequently used to acquire the tilt series. (C) The counts of Ag and Au X-ray peaks as a function of tilt angles from a single AgAu nanoparticle when employing the acquisition times from (B). The counts remain roughly constant over all tilt angles. From Slater et al.15. Please click here to view a larger version of this figure.
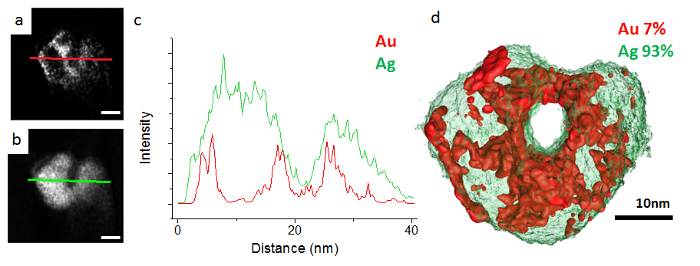
Figure 2. Reconstruction of a low Au content AgAu nanoparticle. (A) Orthoslice through the Au reconstruction. (B) Orthoslice through the Ag reconstruction. (C) Line profiles taken through orthoslices (A) and (B) displaying the clear Au surface segregation in this nanoparticle. (D) Surface visualization of the Ag and Au reconstructions. From Slater et al.15. Please click here to view a larger version of this figure.
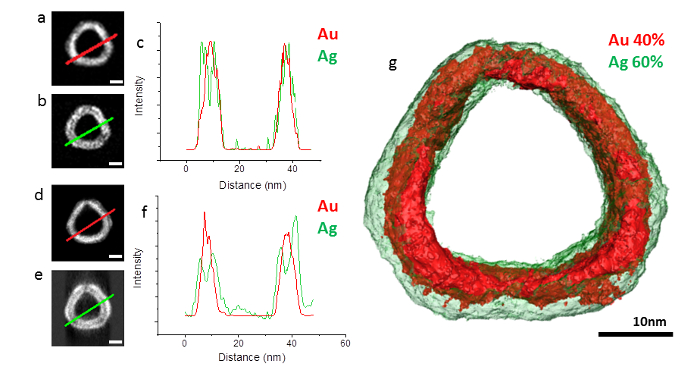
Figure 3. Reconstruction of a high Au content AgAu nanoparticle. (A) 2D EDX map of Au and (B) 2D EDX map of Ag. (C) Line profile taken through the 2D EDX maps (A) and (B) showing the difficulty in determining the surface segregation in this nanoparticle from 2D maps alone. (D) Orthoslice through the Au reconstruction. (E) Orthoslice through the Ag reconstruction. (F) Line profiles taken through orthoslices (D) and (E) displaying the clear Ag surface segregation in this nanoparticle. (G) Surface visualization of the Ag and Au reconstructions. From Slater et al.15. Please click here to view a larger version of this figure.
Discussion
The protocol presented here provides a method to determine the elemental distribution of any multi-element nanoparticle in three dimensions. In the case of the AgAu nanoparticles presented here, surface segregation of both elements is clearly identified and is shown to be correlated to catalytic yield in a three component coupling reaction. This clearly demonstrates the utility of this technique in helping to explain the physical and chemical properties of nanoparticle systems.
As is always the case in the TEM, care should be taken in sample preparation to ensure the best possible results. Thorough washing and annealing of the grids after depositing the nanoparticle solution is particularly important to avoid build-up of carbon contamination through the large electron dose needed for EDX tomography. The large dose employed can also result in severe damage to holey carbon films, particularly if on thin sections often found between holes, but silicon nitride support films can favor oxidation of the nanoparticles16.
Correction of detector shadowing effects is important to produce an accurate reconstruction, particularly if the technique is to be applied for quantitative mapping of elemental distributions in the future. This can be achieved through accurate characterization of the detector shadowing and subsequently varying the electron dose to the nanoparticle. Alternatively, shadowing can be compensated by multiplying spectrum images by a correction factor after acquisition. However, applying this technique to provide quantitative information in three dimensions is not yet feasible due to electron beam damage of nanoparticles that limits the X-ray counts achievable in each spectrum image.
Calibration is required in order to compensate for EDX detector shadowing as a function of tilt angle for a particular microscope-detector-holder combination. The shadowing should initially be determined using a sample that gives no variation in X-ray counts for different specimen tilt angles and individual spherical nanoparticles are expected to satisfy this criteria, when their composition is stable under the electron beam over the time taken to acquire the tilt series. In addition, for crystalline nanoparticles, any tilt angles at which the electron beam is oriented along a major zone-axis of the nanoparticle should be removed and the nanoparticle should be small enough to avoid significant X-ray absorption. Therefore, when EDX spectrum images of a single nanoparticle are acquired over the full range of possible specimen tilt angles using a constant acquisition time, any variations in the measured characteristic X-ray intensity will be due to detector shadowing alone. The acquisition times, and therefore the dose, is then varied in subsequent acquisitions to compensate for the shadowing meaning that the total signal counts are approximately constant for all spectrum images acquired in the tilt series.
In comparison to HAADF or EELS imaging modes, EDX tomographic data acquisition is still in its very early stages. Despite the introduction of X-ray detectors with higher solid angles the major limitation of EDX tomography, as is often the case for two-dimensional EDX imaging, is the low signal. Despite this, one advantage that EDX spectroscopy may hold over EELS for some nanoparticle systems is in the determination of small amounts of heavy elements in fairly large nanoparticles. Larger multicomponent nanoparticles (>100 nm) are often well suited to EDX studies as they provide more counts and there are fewer issues with deconvolving spectral overlaps, but care should be taken to use high-energy X-ray peaks that undergo little absorption.
Overall, EDX tomography is an excellent method of determining elemental distributions within nanoparticles in three dimensions, although limited to nanoparticles that can withstand a relatively high electron dose without significant damage. Further increases in the solid angles of X-ray detectors within the STEM and further optimization of tomographic specimen holders will allow this technique to advance even further and become an important method in the characterization of individual nanoparticles.
Disclosures
The authors have nothing to disclose.
Acknowledgements
TJAS and SJH thank the UK Engineering and Physical Sciences Research Council, (Grant numbers EP/G035954/1 and EP/L01548X/1) for funding support. The authors wish to acknowledge the support from HM Government (UK) for the provision of the funds for the Titan G2 80-200 S/TEM associated with research capability of the Nuclear Advanced Manufacturing Research Centre.
Materials
| Titan G2 80-200 STEM | FEI | With Super-X detector | |
| 2020 tomography holder | Fischione | ||
| Carbon film on 200 mesh copper grid | Agar Scientific | AGS160 | |
| EDX Acquisition software | Bruker | Esprit | |
| Tomographic alignment and reconstruction software | FEI | Inspect3D, alternatives available | |
| Tomographic alignment and reconstruction software package | University of Colorado | IMOD, alternatives available | |
| Visualisation software | FEI | Avizo, alternatives available | |
| Image processing software | Gatan | Digital Micrograph, alternatives available | |
| Image visualisation software | Open Source | Fiji, alternatives available | |
| Polyvinyl-pyrrolidone | Sigma-Aldrich | 856568 | |
| Ethylene glycol | Sigma-Aldrich | V900208 | |
| Silver nitrate | Sigma-Aldrich | 209139 | |
| Benchtop Centrifuge | Thermo Scientific | 75007200 | |
| Round bottom flask | Sigma-Aldrich | Z41,452-2 | 1000mL |
| Hydrogen tetrachloroaurate trihydrate | Sigma-Aldrich | 520918 |
References
- Midgley, P. A., Weyland, M. 3D electron microscopy in the physical sciences: the development of Z-contrast and EFTEM tomography. Ultramicroscopy. 96, 413-431 (2003).
- Mobus, G., Doole, R. C., Inkson, B. J. Spectroscopic electron tomography. Ultramicroscopy. 96, 433-451 (2003).
- Kotula, P., Brewer, L., Michael, J., Giannuzzi, L. Computed Tomographic Spectral Imaging: 3D STEM-EDS Spectral Imaging. Microsc. Microanal. 13, 1324-1325 (2007).
- Lepinay, K., Lorut, F., Pantel, R., Epicier, T. Chemical 3D tomography of 28 nm high K metal gate transistor: STEM XEDS experimental method and results. Micron. 47, 43-49 (2013).
- Genc, A., et al. XEDS STEM tomography for 3D chemical characterization of nanoscale particles. Ultramicroscopy. 131, 24-32 (2013).
- Goris, B., Polavarapu, L., Bals, S., Van Tendeloo, G., Liz-Marzan, L. M. Monitoring galvanic replacement through three-dimensional morphological and chemical mapping. Nano Lett. 14, 3220-3226 (2014).
- Goris, B., et al. Three-dimensional elemental mapping at the atomic scale in bimetallic nanocrystals. Nano Lett. 13, 4236-4241 (2013).
- Jarausch, K., Thomas, P., Leonard, D. N., Twesten, R., Booth, C. R. Four-dimensional STEM-EELS: Enabling nano-scale chemical tomography. Ultramicroscopy. 109, 326-337 (2009).
- von Harrach, H., et al. Comparison of the Detection Limits of EDS and EELS in S/TEM. Microsc. Microanal. 16, 1312-1313 (2010).
- Tedsree, K., et al. Hydrogen production from formic acid decomposition at room temperature using a Ag-Pd core-shell nanocatalyst. Nat. Nanotechnol. 6, 302-307 (2011).
- Slater, T. J. A., Camargo, P. H., Burke, M. G., Zaluzec, N. J., Haigh, S. J. Understanding the limitations of the Super-X energy dispersive x-ray spectrometer as a function of specimen tilt angle for tomographic data acquisition in the S/TEM. J. Phys. Conf. Ser. 522, 012025 (2014).
- Schindelin, J., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9, 676-682 (2012).
- Kremer, J. R., Mastronarde, D. N., McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71-76 (1996).
- von Harrach, H., et al. An integrated Silicon Drift Detector System for FEI Schottky Field Emission Transmission Electron Microscopes. Microsc. Microanal. 15, 208-209 (2009).
- Slater, T. J. A., et al. Correlating Catalytic Activity of Ag-Au Nanoparticles with 3D Compositional Variations. Nano Lett. 14, 1921-1926 (2014).
- Lewis, E., et al. Real-time imaging and elemental mapping of AgAu nanoparticle transformations. Nanoscale. 6, 13598-13605 (2014).

