Preparation and 3D Tracking of Catalytic Swimming Devices
Summary
A method to prepare catalytically active Janus colloids that can “swim” in fluids and determine their 3D trajectories is presented.
Abstract
We report a method to prepare catalytically active Janus colloids that “swim” in fluids and describe how to determine their 3D motion using fluorescence microscopy. One commonly deployed method for catalytically active colloids to produce enhanced motion is via an asymmetrical distribution of catalyst. Here this is achieved by spin coating a dispersed layer of fluorescent polymeric colloids onto a flat planar substrate, and then using directional platinum vapor deposition to half coat the exposed colloid surface, making a two faced “Janus” structure. The Janus colloids are then re-suspended from the planar substrate into an aqueous solution containing hydrogen peroxide. Hydrogen peroxide serves as a fuel for the platinum catalyst, which is decomposed into water and oxygen, but only on one side of the colloid. The asymmetry results in gradients that produce enhanced motion, or “swimming”. A fluorescence microscope, together with a video camera is used to record the motion of individual colloids. The center of the fluorescent emission is found using image analysis to provide an x and y coordinate for each frame of the video. While keeping the microscope focal position fixed, the fluorescence emission from the colloid produces a characteristic concentric ring pattern which is subject to image analysis to determine the particles relative z position. In this way 3D trajectories for the swimming colloid are obtained, allowing swimming velocity to be accurately measured, and physical phenomena such as gravitaxis, which may bias the colloids motion to be detected.
Introduction
Catalytic swimming devices are small scale, untethered colloids capable of autonomously generating motion in fluidic environments.1,2 These devices are attracting significant research interest as they have the potential to enable exciting new functions such as drug delivery,3 lab-on a chip transport4 and environmental remediation.5 One widely studied example are catalytic "Janus" swimmers.6 These particles get their name from having two distinct sides, or faces (Janus is a two faced Roman god). One side is catalytically active and able to perform a decomposition reaction, while the other is inert. In the presence of suitable dissolved fuel molecules, the resulting asymmetric chemical reaction creates gradients around the colloids which can produce motion via self-diffusiophoresis/electrophoresis.7
Characterizing the motion for these rapidly moving objects is challenging and many experimental observations to date have been limited to 2D. However, eventual applications are likely to exploit catalytic swimming devices ability to move throughout bulk solutions in 3D.8 To address this, here we describe a protocol that allows accurate 3D trajectories for swimming devices to be determined. This method is based on interpreting the ring structures produced by out of focus fluorescent colloids observed with a fixed focus objective,9 and is easy to apply using conventional unmodified microscopes. By clearly describing this method here, other researchers in this field will benefit by being able to access such 3D information. This will aid future insights into motion characteristics for swimming devices. Evidence of this potential is given by the recent report of swimming devices being directed by gravity,10,11 behavior which can most easily be visualized through the application of 3D tracking.11
This paper also clearly documents a method to manufacture catalytic Janus particle swimming devices, which will be of further benefit to standardize methods across the existing research groups investigating these devices, and additionally guide new researchers interested in making and investigating swimming devices.
Protocol
CAUTION: Please consult all relevant material safety data sheets before use. Hydrogen peroxide used in this protocol is harmful, and the evolution of oxygen gas when exposed to platinum poses an explosion risk. Use all appropriate safety controls during this protocol including engineering controls while handling peroxide solutions (fume hood) and personal protective equipment (safety glasses, gloves and lab coat).
1. Making Catalytic Janus Particles
- Prepare glass slide substrate
- Clean an unused standard microscope slide using sequential washing for a few minutes each in deionized (DI) water, glass decontamination solution and DI water. Then wash with a 70:30 v:v mixture of ethanol:DI water, and finally blow dry in a clean air/nitrogen stream.
- Examine the glass slide under a microscope to verify the surface is clean and free from evidence of particulate contamination. Repeat step 1.1.1 if required.
- Prepare colloidal dispersion for deposition
- Pipette 10 µl of aqueous stock fluorescent colloid solution (10% wt.) into 990 µl of ethanol. Adjust volumes as required depending on stock solution concentration used to arrive at approximately 0.1% wt colloidal suspension.
- Vortex mix for 10 sec.
- Spin coat colloidal dispersion onto glass slide substrate
- Equip a spin coater with the clean glass slide. Fill a pipette tip with 100 µl of the diluted colloidal solution prepared above. Program spin coater to execute a 30 sec, 2,000 rpm cycle. Start spin coater, and when up to speed continuously pipette the prepared solution onto the center of the spinning glass slide.
- Remove glass slide from the spin coater, return to the optical microscope and verify that an even dispersion of mainly non-touching separate colloids covers the central region of the glass slide.
- Vacuum evaporate platinum metal onto colloid decorated glass slide
- Insert the colloid coated glass slide into a metal evaporator. Ensure the colloid decorated side is facing the evaporation source. Install a platinum metal evaporation source and deposit 15 nm of platinum metal.
Note: Following metal deposition, samples should be stored under an inert atmosphere.
- Insert the colloid coated glass slide into a metal evaporator. Ensure the colloid decorated side is facing the evaporation source. Install a platinum metal evaporation source and deposit 15 nm of platinum metal.
2. "Swimming" Janus Particles
- Re-suspend Janus Colloids into a peroxide containing solution
- Cut a 1 x 1 cm square of lens tissue, and dampen the end with 10 µl of DI water. Hold with tweezers, and gently rub along the surface of the platinum coated colloid decorated glass slide (this step physically removes colloids from the substrate).
- Insert the lens tissue into 1.5 ml of DI water and manually shake vigorously for 30 sec in a sealed tube. Remove lens tissue.
- Pipette 1 ml of the colloid containing solution into a new container, filled with 1 ml of 30% w/v H2O2 stock solution. Gently mix the solutions, and then place in an ultrasonic bath at room temperature for 5 min, followed by another 25 min unstirred incubation period.
CAUTION: This solution may evolve oxygen; do not seal. - Dry 100 µl of the remaining aqueous colloidal solution onto a scanning electron microscope (SEM) stub to enable SEM verification of the Janus colloid structure.14
- Prepare analysis cuvette
- Add an additional 1 ml of DI water to the incubated solution containing peroxide and platinum coated colloids to arrive at a suitable fuel strength (10%) to allow fast propulsion.
Note: The prior incubation stages were carried out in higher concentration fuel concentrations to clean the platinum catalyst surface. - Fill a low volume rectangular quartz glass cuvette with the incubated solution. Loosely fit a push-in cap.
CAUTION: Explosion risk — do not use a screw cap.
- Add an additional 1 ml of DI water to the incubated solution containing peroxide and platinum coated colloids to arrive at a suitable fuel strength (10%) to allow fast propulsion.
3. Microscopic Observation
- Locate particle of interest
- Load cuvette into a fluorescent microscope equipped with a suitable objective (e.g., 20X) and excite fluorophore emission using a suitable combination of filters (excitation 450-490 nm, emission >515 nm).
- Manually search for fluorescent colloids within the cuvette.
Note: Adjusting colloid density while maintaining the peroxide concentration may be required. For example, dilution is recommended if the colloid density is high, and numerous oxygen bubbles producing flows are present. A colloid volume concentration of about 0.003% is a recommended starting point. - Optimize optical settings for 3D tracking: under suitable illumination conditions, in focus colloids will appear as sharp circular objects. However, as propulsion moves the colloids in and out of the focal plane a distinctive size changing bright ring centered around the sphere will be observed, this is used to determine the z coordinate to enable 3D tracking.
- Record video
- Before starting video capture, focus the microscope so that the particle of interest produces a concentric ring, with the particle "under" the focus position. Do not move the focus plane during video capture.
- Record videos of particles of interest. Use 30 sec video durations with frame rates in excess of 30 Hz to allow detailed trajectory reconstruction.
- 3D trajectory reconstruction
- Calibrate z-axis
- Make up a 2% wt. gellan gum solution in water containing a suspension of fluorescent Janus particles at 60 °C, place in an equivalent cuvette to that used above, and allow to set to form a stiff transparent gelled sample containing fixed static colloids.
- Focus on a single fixed colloid using the same illumination conditions selected above, now record a series of still images as the z-focus is raised by known displacements relative to this plane.
- Determine the radius of the ring at each known focus position11.
Note: This is most efficiently performed by using an image analysis algorithm that can be applied as a batch process to all the calibration still frames, and videos. A typical approach involves smoothing the image, thresholding to identify an approximate location of the objects' center, and then locating the actual x and y coordinates of the ring center by measuring the distance between intensity peaks either side of the ring. The mean radial distance to the intensity peaks from the ring center can then be found.11 This allows both the radius of the bright ring and the x-y coordinate to be determined with sub-pixel accuracy. By locating the x, y, z position of a Janus sphere fixed in gellan gum 30 µm from the focal plane, in a time sequence of images, particles can be located with an error of ± 25 nm along each axis. The error can be attributed to noise in the images. The signal to noise ratio and therefore, the accuracy of the location algorithms is dependent upon the intensity of the detected fluorescence light. When a Janus sphere is far from the focal plane its intensity becomes too weak to accurately track it, e.g., for a 4.8 µm diameter colloid a z-range of around 200 µm is possible. An alternative non-algorithmic method is to use simple manual measurement of x,y center and radius, however this will reduce accuracy. - Plot a calibration curve to relate radius to z-position, and fit to an appropriate function (e.g., cubic equation) to allow interpolation.11
- Calibrate x,y axis
- Record a still frame optical microscope image of a spatial calibration graticule using the same microscope conditions chosen in 3.2.
- Measure the "in pixels" dimension of an object with known real world size from the image of the spatial calibration graticule and use this to establish a pixels to micron conversion factor for the x,y image plane.
- Reconstruct trajectory
- Determine the x and y coordinates and radius for each frame of the video sequence as described in 3.3.1.3, use the function found in 3.3.1.4 to convert radius into z, and the calibration factor found in 3.3.2.2 to convert x and y pixel coordinates into microns. This procedure will result in an accurate x, y, z coordinate for the propulsive particles location as a function of time.11 This procedure can be implemented using an algorithm, or manually.
- Determine the derived properties such as average velocity to quantify the extent of observed catalytic swimming.
- Calibrate z-axis
Representative Results
Figure 1 shows a typical dispersion of colloids on a clean glass slide prior to depositing platinum. Figure 2 shows a typical back-scattered SEM image for a half platinum coated Janus swimmer, under this imaging mode the platinum coated region produces bright contrast. The desired hemispherical platinum layer is apparent. Figure 3 displays the appearance of a typical fluorescent Janus swimmer under optimum illumination conditions fixed in gellan gum. The swimmer appears as a symmetrical ring feature, and it is the radius of the ring that can be used to determine the z-position of the colloid relative to the focus position. Figure 4 shows representative cross-sections for the radial brightness intensity distribution that is used in combination with image analysis algorithms to accurately locate the center and apparent radius of the colloid. Figure 5 contains a calibration curve obtained using a fixed colloidal sample and a calibrated microscope z-translation stage to relate apparent colloidal size and distance from focus position. This curve is fitted to a cubic function, which is used to convert apparent radius into z-coordinate. Finally, Figure 6 displays a typical x,y,z trajectory for a fluorescent Janus particle swimmer.
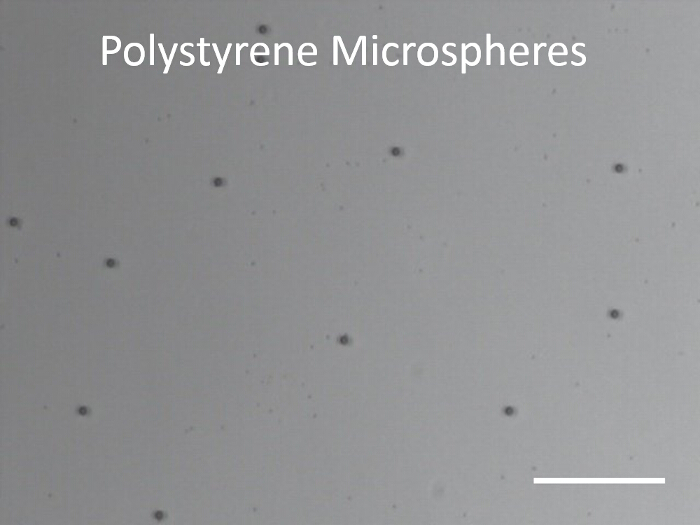
Figure 1. Optical image of 1.9 µm diameter Polystyrene microspheres. Microspheres are dispersed on a cleaned glass slide before Platinum deposition. Scale bar represents 40 µm. Please click here to view a larger version of this figure.
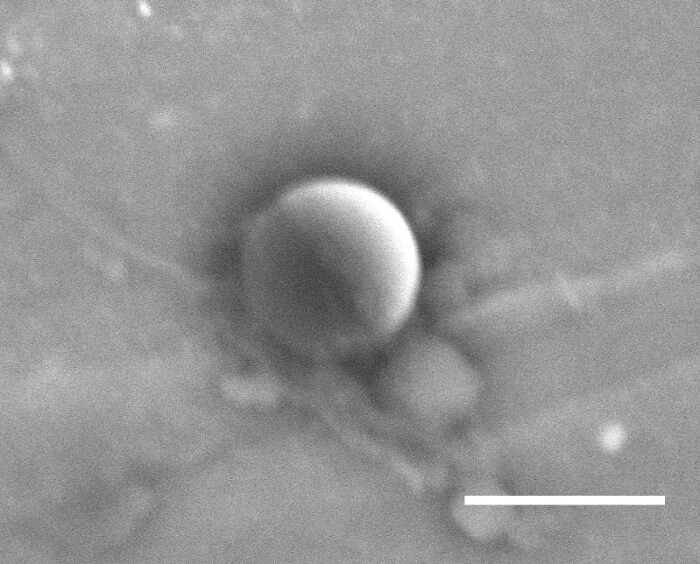
Figure 2. SEM backscatter image of a 1.9 µm diameter Polystyrene microspheres. Microspheres are shown after platinum deposition. Scale bar represents 2 µm. Please click here to view a larger version of this figure.
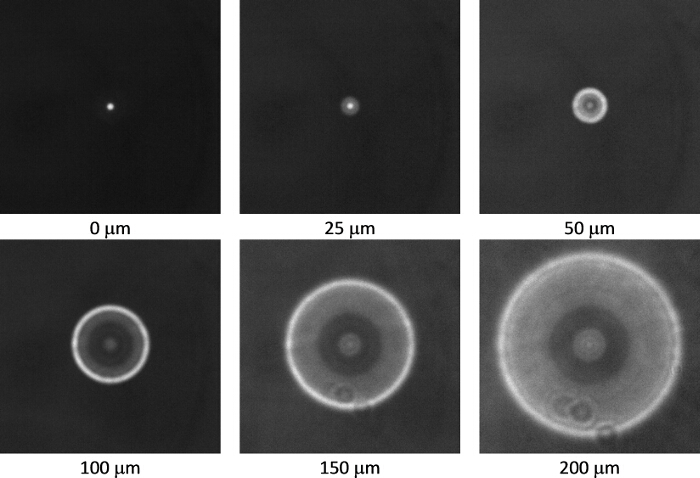
Figure 3. Calibration images of a 4.8 µm diameter fluorescent polystyrene sphere fixed in gellan gum recorded using a 20X objective (0.4 N.A.). The distances below each image indicate the distance of the focal plane of the objective above the sphere. As the image is defocused from 0 µm to 200 µm the in focus image of a bright disc changes to a bright ring, the radius of which is dependent upon magnification, the sphere size and its distance from the focal plane. Please click here to view a larger version of this figure.
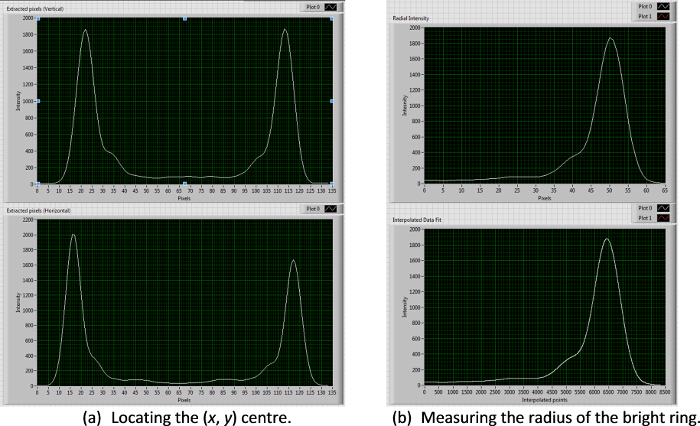
Figure 4. X, y, z particle tracking procedure. A set of self-written algorithms is used to first locate the (x, y) center of the bright ring by extracting a series of vertical and horizontal lines and finding the mean mid-point between the bright peaks (a). The ring radius is then calculated from the peak intensity of a spline fitted to the average pixel grey-values radiating out from the ring center (b). Please click here to view a larger version of this figure.
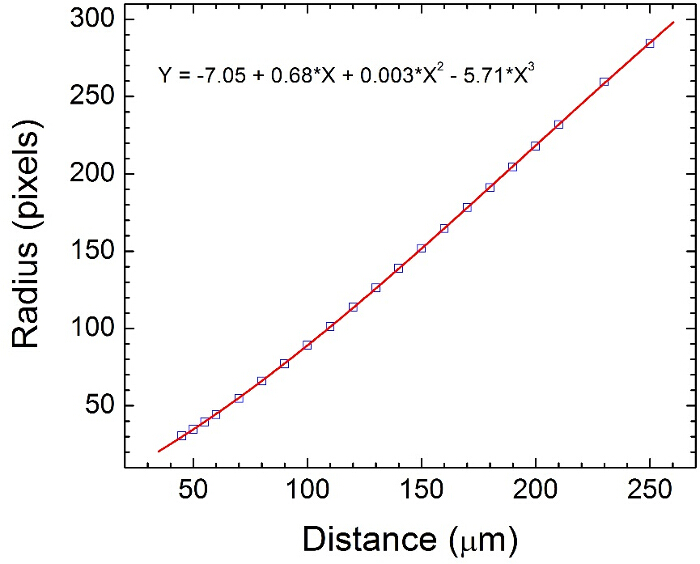
Figure 5. Z-coordinate calibration chart for Janus spheres obtained by measuring the bright ring radius of spheres fixed in gellan gum (see Figures 3 and 4). The calibration chart is used by our algorithms to convert the measured ring radius to a z-coordinate. Please click here to view a larger version of this figure.
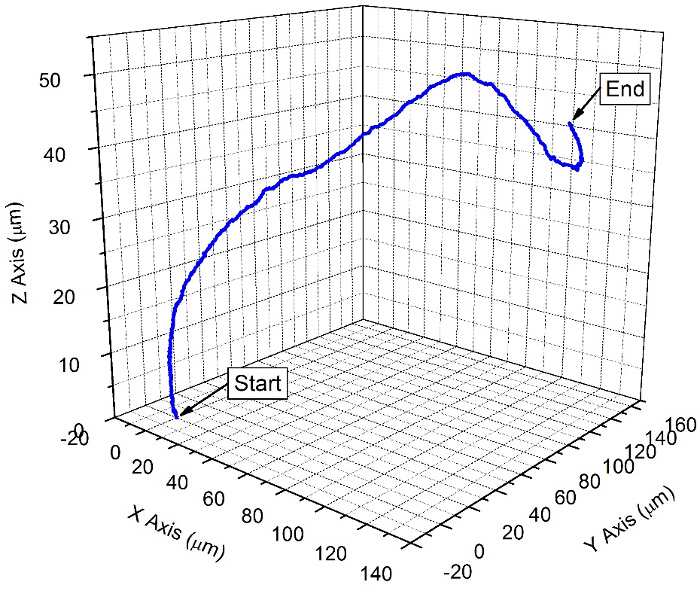
Figure 6. A trajectory of a typical fluorescent Janus sphere swimming device. A sequence of images of the moving swimming device was recorded over a period of 30 sec at a frame rate of 33 Hz. The (x, y, z) coordinates of the trajectory were obtained by locating the bright ring center (Figure 4 (a)) and comparing the measured ring radius to the calibration chart for each image in the sequence (Figures 4 (b) and 5). Please click here to view a larger version of this figure.
Discussion
Many variables in the preparation protocol for platinum Janus particles will affect the observed trajectories. The parameters as described using 2 µm diameter particles will give propulsion velocities in the order of 10 µm per second. If smaller particles are used, velocities will increase, while increasing particle size will decrease propulsion velocity.12 The details of the evaporation protocol will also alter the trajectories observed. In this current protocol, a sparse distribution of colloids is recommended, together with metal evaporation normal to the slide orientation. These conditions result in symmetrical Janus structures as shown in Figure 2, which lead to linear trajectories within the limits of Brownian rotational diffusion.13 Conversely, if tight packed colloids are subject to glancing angle deposition, then the symmetry of the Janus cap can be broken, to induce spinning behavior.14 The particles produced here display relatively isotropic motion in all three dimensions; however if thicker platinum coatings, or larger particles are used, an upwards bias or gravitaxis can be imparted.11 Details of the storage of the Janus colloids after manufacture may also effect the swimming speeds observed. The high surface energy clean platinum surface emerging from the evaporation stage is susceptible to surface contamination for example from hydrocarbons, and in particular thiols.15
In addition, the solution properties in which the Janus colloids are re-suspended are critical for observing propulsion. Low peroxide concentrations will result in slower velocities, as the rate of the decomposition reaction producing motion reduces.6 In addition, low concentrations of salts will result in a dramatic reduction in propulsion velocity.7
A key feature of the colloids produced here is their neutral buoyancy, which makes them suitable for 3D tracking. In general the field of swimming devices has paid little attention to 3D effects, partly due to some prominent examples being made from dense metals, causing them to rapidly sediment,16 but also due to the difficulties and expense associated with making the required measurements. Clear drawbacks for some established 3D tracking methods exist for these rapidly moving colloids, for example, confocal scanning laser microscopy can lack the temporal resolution to record sufficient numbers of images to resolve trajectories. In this context, the method we present here has the significant advantage of only requiring a single frame to allow estimation of z-coordinate, which consequently allows high frame rates. Also, as z-coordinate reconstruction only relies on the relative contrast of the out-of-focus colloid in single frames, rather than the absolute fluorescence intensity, it is resilient to quenching and blinking effects in the fluorophore. These advantages are possible at the expense of a reduced depth of field over which 3D trajectory reconstruction is possible, and the requirement for well separated non-overlapping colloids. We hope that describing the protocol will allow other research groups with an interest in 3D behavior for their swimming devices to access this information straightforwardly and with a high degree of precision. It is clear that expanding the understanding of these devices to 3D will open up a significant range of interesting future phenomena and applications. Readers interested in further details of trajectory analysis are directed towards Reference 17 which describes common artifacts in propulsive systems and how to ensure accurate quantification of propulsion velocities.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by EPSRC Career Acceleration grant EP/J002402/1.
Materials
| Evaporator | Moorfield (UK) | Minilab 80 e-beam evaporator | |
| Microscope | Nikon | Eclipse LV100 | |
| Fluorescence light source | Nikon | Nikon B2A filter cube | |
| Objective | Nikon | x20, 0.45 NA | |
| Cuvette | Hellma | fused quartz, 40 x 10 x 1 mm | |
| Vortex mixer | IKA | Lab Dancer S2 | |
| Spin coater | Laurell Technologies Corp. | Model WS-400BZ-6NPP/Lite | |
| Ultrasonic bath | Eumax | 2 litre | |
| Lens tissue | Whatman | 2105 841 | |
| Hydrogen Peroxide | Sigma-Aldrich | 31642-1L | 30 wt% |
| Platinum | Sigma-Aldrich | 267171 | 0.25 mm, 99.99% |
| Colloids | Thermo Scientific | Fluoro-Max PS microspheres, d= 1.9 microns | |
| Glass decontamination solution | Fisher Scientific | D/0025/15 | Decon 90 |
| Ethanol | Fisher Scientific | E/0600DF/17 | Absolute Ethanol |
| DI water | Elga | Purelab Option filtration system (15 MW) | |
| Gellan gum | Sigma-Aldrich | P8169-100G | "Phytagel" |
References
- Ebbens, S. J., Howse, J. R. In pursuit of propulsion at the nanoscale. Soft Matter. 6 (4), 726 (2010).
- Wang, W., Duan, W., Ahmed, S., Mallouk, T. E., Sen, A. Small power : Autonomous nano- and micromotors propelled by self-generated gradients. Nano Today. 8 (5), 531-554 (2013).
- Gao, W., Dong, R., et al. Artificial Micromotors in the Mouse’s Stomach : A Step toward in Vivo Use of Synthetic Motors. ACS nano. 9 (1), 117-123 (2015).
- Restrepo-Pérez, L., Soler, L., Martìnez-Cisneros, C., Sánchez, S., Schmidt, O. G. Biofunctionalized self-propelled micromotors as an alternative on-chip concentrating system. Lab Chip. 14 (16), 2914-2917 (2014).
- Soler, L., Sánchez, S. Catalytic nanomotors for environmental monitoring and water remediation. Nanoscale. 6 (13), 7175-7182 (2014).
- Howse, J., Jones, R., Ryan, A., Gough, T., Vafabakhsh, R., Golestanian, R. Self-Motile Colloidal Particles: From Directed Propulsion to Random Walk. Phys. Rev. Letts. 99 (4), 8-11 (2007).
- Ebbens, S., Gregory, D. A., et al. Electrokinetic effects in catalytic platinum-insulator Janus swimmers. EPL. 106 (5), 58003 (2014).
- Orozco, J., Jurado-Sánchez, B., et al. Bubble-propelled micromotors for enhanced transport of passive tracers. Langmuir. 30 (18), 5082-5087 (2014).
- Peterson, S. D., Chuang, H. -. S., Wereley, S. T. Three-dimensional particle tracking using micro-particle image velocimetry hardware. Meas. Sci. and Technol. 19 (11), 115406 (2008).
- Ten Hagen, B., Kümmel, F., Wittkowski, R., Takagi, D., Löwen, H., Bechinger, C. Gravitaxis of asymmetric self-propelled colloidal particles. Nat. Commun. 5, 4829 (2014).
- Campbell, A. I., Ebbens, S. J. Gravitaxis in spherical janus swimming devices. Langmuir. 29 (46), 14066-14073 (2013).
- Ebbens, S., Tu, M. -. H., Howse, J. R., Golestanian, R. Size dependence of the propulsion velocity for catalytic Janus-sphere swimmers. Physical Review. E Stat. Nonlin. Soft Matter Phys. 85, 020401 (2012).
- Ebbens, S. J., Howse, J. R. Direct observation of the direction of motion for spherical catalytic swimmers. Langmuir. 27 (20), 12293-12296 (2011).
- Archer, R. J., Campbell, a. I., Ebbens, S. J. Glancing angle metal evaporation synthesis of catalytic swimming Janus colloids with well defined angular velocity. Soft Matter. 11, 6872-6880 (2015).
- Zhao, G., Sanchez, S., Schmidt, O. G., Pumera, M. Poisoning of bubble propelled catalytic micromotors: the chemical environment matters. Nanoscale. 5 (7), 2909-2914 (2013).
- Wang, Y., Hernandez, R. M., et al. Bipolar electrochemical mechanism for the propulsion of catalytic nanomotors in hydrogen peroxide solutions. Langmuir. 22 (25), 10451-10456 (2006).
- Dunderdale, G., Ebbens, S., Fairclough, P., Howse, J. Importance of particle tracking and calculating the mean-squared displacement in distinguishing nanopropulsion from other processes. Langmuir. 28 (30), 10997-11006 (2012).

