Precision-cut Mouse Lung Slices to Visualize Live Pulmonary Dendritic Cells
Summary
We describe a method for generating Precision-cut Lung Slices (PCLS) and immunostaining them to visualize the localization of various immune cell types in the lung. Our protocol can be extended to visualize the location and function of many different cell types under a variety of conditions.
Abstract
Inhalation of allergens and pathogens elicits multiple changes in a variety of immune cell types in the lung. Flow cytometry is a powerful technique for quantitative analysis of cell surface proteins on immune cells, but it provides no information on the localization and migration patterns of these cells within the lung. Similarly, chemotaxis assays can be performed to study the potential of cells to respond to chemotactic factors in vitro, but these assays do not reproduce the complex environment of the intact lung. In contrast to these aforementioned techniques, the location of individual cell types within the lung can be readily visualized by generating Precision-cut Lung Slices (PCLS), staining them with commercially available, fluorescently tagged antibodies, and visualizing the sections by confocal microscopy. PCLS can be used for both live and fixed lung tissue, and the slices can encompass areas as large as a cross section of an entire lobe. We have used this protocol to successfully visualize the location of a wide variety of cell types in the lung, including distinct types of dendritic cells, macrophages, neutrophils, T cells and B cells, as well as structural cells such as lymphatic, endothelial, and epithelial cells. The ability to visualize cellular interactions, such as those between dendritic cells and T cells, in live, three-dimensional lung tissue, can reveal how cells move within the lung and interact with one another at steady state and during inflammation. Thus, when used in combination with other procedures, such as flow cytometry and quantitative PCR, PCLS can contribute to a comprehensive understanding of cellular events that underlie allergic and inflammatory diseases of the lung.
Introduction
Following inhalation of pro-inflammatory stimuli such as lipopolysaccharide (LPS), there is a coordinated movement of immune cells into, within, and from the lung. For example, neutrophils are rapidly recruited to the lung parenchyma and airway. In addition, some professional antigen presenting cells known as conventional dendritic cells (cDCs) undergo a relatively complex migration pattern1,2. cDCs can be identified using flow cytometry, based in part on their display of the surface marker, CD11c. Distinct subsets of DCs can be distinguished by the differential surface expression of CD103 and CD11b3. Upon acquiring inhaled antigen, some cDCs exit the lung and migrate through the lymphatic vessels to lung-draining Lymph Nodes (LNs) where they present peptides to antigen-specific T cells4. This is a critical early event in the initiation of adaptive immune responses. For unknown reasons, however, not all cDCs that acquire inhaled antigens leave the lung, and many of these cells remain in that organ for several months5,6. This observation can be partly explained by the developmental ancestry of these cells because monocyte-derived CD11c+ cells lacking the chemokine receptor, CCR7, are unable to migrate to regional LNs7,8. It seems likely that the migration potential of cDCs is also determined, at least in part, by their anatomical position within the lung. However, the precise localization of these different populations of cDCs in the lung is not fully characterized. An improved knowledge of immune cell localization within the lung, and of the molecules that direct it, is needed for a better understanding of how the immune system of the lung becomes activated.
PCLS are being increasingly used as an ex vivo approach to visualize cellular positioning and cell-cell interactions, while maintaining the structural integrity of the lung architecture9,10. PCLS have been used to study lungs of many species, including mice, cattle, monkeys, sheep, horses, and humans11. A major advantage of this technique is that approximately 20 slices can be prepared from a single lobe of a mouse lung, thereby reducing the number of animals needed for individual experiments. Virtually all immune cell types, including DCs, macrophages, neutrophils, and T cells, are present in PCLS and maintain their normal structures.
PCLS can also be used to study calcium signaling and contractility of airway and smooth muscle cells after treatment with acetylcholine12 or methacholine13. In this approach, only a small portion of the lung is analyzed microscopically, but one study reported that measurements of airway contraction in PCLS vary only about 10% from slice to slice, and this variance is comparable to that seen using lung function tests in intact animals14. Other investigators have used PCLS as an ex vivo approach to study changes in cytokine expression and cell surface markers after incubation with LPS15. PCLS have also been used in an ex vivo model of hypoxic pulmonary vasoconstriction in small intra-acinar arteries. These vessels are located in the part of the lung that cannot be reached using other procedures, including recordings from dissected arterial segments or analysis of subpleural vessels16. Our lab has primarily used PCLS to visualize immune cell localization in live lung tissue at steady state and following an in vivo inflammatory stimulus. The procedures we have developed for this are as follows.
Protocol
Animal experimental procedures described in this paper were approved by the NIEHS Animal Care and Use Committee (IACUC).
1. Lung Preparation
- House mice between 6 and 12 weeks of age in specific pathogen-free conditions in accordance with the guidelines provided by the Institutional Animal Care and Use Committees.
NOTE: Imaged mice can be either naïve, or treated, depending on each researcher's specific interests. Here, we describe immune cell localization in naïve mice and in mice treated with 100 µg ovalbumin (OVA) and 0.1 µg lipopolysaccharide (LPS), using phosphate buffered saline (PBS) as a vehicle in a total volume of 50 µL. To oropharyngeally instill OVA/LPS into the airways, animals were anesthetized with isoflurane inhalation and vertically suspended by their teeth with a rubber band. The tongue was gently grasped with forceps and held to one side to prevent swallowing, and 50 µL of the OVA/LPS solution deposited at the back of the oral cavity as previously described17. - Euthanize the mouse with intraperitoneal injection of sodium pentobarbital (100 mg/kg), and pin the animal to a polystyrene base.
- Open the abdominal cavity with scissors by cutting the skin and peritoneum from the middle of the abdomen up to the jaw. Pull the intestines aside with forceps or the dull edge of the scissors, and snip the inferior vena cava to drain blood away from lungs. Puncture the diaphragm with the sharp tip of the scissors to allow expansion of the rib cage, being careful not to cut the lungs.
- Clear salivary glands and other tissue away from the trachea by grabbing the tissue and manually pulling it away from the underlying trachea with forceps. Make a small incision in the trachea on the anterior side of the thickest band of cartilage using fine forceps or scissors, being careful not to cut all the way though the trachea. The incision will be just large enough to allow a 20 G needle to pass through.
- Slide a 1.5 inch, 20 G needle onto a section of polyethylene tubing, leaving approximately 1 cm of tubing beyond the end of the needle. Cut the tubing at an angle of approximately 45° to bevel the end, attach the needle to a 1 mL syringe, and load the syringe by drawing 0.8 mL of warm (40 °C) 2% low melting point agarose up through the needle.
- Place the end of the tubing into the incision of the trachea and slowly inject the agarose into the lungs. Without moving the needle or syringe, tape the syringe to the polystyrene base to ensure the needle remains in the trachea.
NOTE: After injection, the lungs will be larger and inflated within the chest. The right lobes expand first, then the left lobe. If only one lobe expands, the needle has been inserted too deeply (past the bronchus), and it will be necessary to pull it out slightly before proceeding. This part of the procedure needs to be done relatively quickly, before the agarose starts to solidify. - Place the mouse, with the base and the syringe still inserted in the trachea, in a cold room or refrigerator for at least 10 min. If necessary, the animal can be kept there for up to several hours, even if proceeding to image cell migration by video microscopy.
- Once the mouse is cold, carefully excise the agarose-inflated lungs using scissors, and place them in a 3 cm dish with ice cold PBS. Keep on ice.
- Choose the desired lobe for tissue slicing (e.g., the right superior lobe). Make sure the interior part of the lung (where it connects to trachea) is face down in the dish.
NOTE: The lobe used and its orientation on the plunger will determine the size of vessels and airways that will be visible. The orientation described here is ideal for visualization of large airways, and parenchyma. Mice have one large left lobe and four smaller lobes on the right. For PCLS imaging following introduction of allergens into the lungs via oropharyngeal or intranasal aspiration, the right superior lobe is typically sectioned, in part because it is a convenient size, but also because inhaled agents disperse uniformly within it. However, other lobes, especially the left lobe, can also be studied. When comparing mouse strains or treatments, it is important to compare the same lobe.
2. Lung Slicing
NOTE: Make slices using an automated slicer, metal cooling block, plunger and metal syringe, according to the manufacturer's instructions.
- Put a small drop of all-purpose, no-run gel superglue onto the plunger and spread the glue in a circular motion, taking care to not let the glue touch the sides of the metal syringe. If the glue touches the sides of the syringe, it will glue the syringe and plunger together.
- Immediately after covering the plunger with glue, gently grasp the excised lobe with forceps with the trachea side down, dab it on a tissue to remove excess liquid, and carefully place it on top of the plunger. Trim off any extra tissue extending beyond the edge of the plunger.
- Move the plunger down so that the sides of the metal syringe move up and over the tissue, creating a well with the lung glued at the bottom, no more than several centimeters deep. Tape around the bottom of the metal syringe to hold this position in place.
- Carefully pour 2% low melt agarose at 40 °C into the well so it just covers the top of the lobe.
- Surround the metal syringe with the ice-cold chilling block and cool the lobe and agarose for 1 – 2 min.
NOTE: A shorter time cooling of the agarose surrounding the tissue could lead to disintegration of the agarose during slicing, which might result in an unevenly cut PCLS. - Load the specimen syringe into the automated slicer and fill the buffer tank with ice cold PBS. Align the step motor drive with the specimen syringe and remove the piece of tape from the bottom. Turn the switch to the fast forward (FF) position until the step motor drive just touches the back of the plunger.
- Align the fresh blade with the specimen syringe. Set tissue thickness, continuous/single slice, oscillation and speed in the slicer. For example, tissue thickness: 150 µm, continuous cutting (cont.), oscillation: 9, and speed: 3 – 4. Press start. The above settings need to be adjusted to the specific automated slicer specifications.
- Using a thin spatula or paintbrush, collect the PCLS one at a time as they fall into the buffer tank and place them in a 24-well plate containing ice cold PBS.
NOTE: It is important to keep the slices in order to maintain consistency between experiments. Although every lung is different depending on age, gender and weight, the 10th slice, for example, generally yields similarly sized airways.
3. Antibody Staining
- Design a panel of fluorescently tagged antibodies based on the cell type of interest, taking into account potential fluorescent spectral overlap and the detection capabilities of the microscope.
NOTE: As an example, a panel for DC subset detection is as follows: CD11c/alpha X integrin – BrilliantViolet (BV) 605 (Excitation: 405 nm, Peak emission: 605 nm), CD324/E-cadherin – Alexa 488 (AF488) (Excitation: 488 nm, Peak emission: 519 nm), CD88/C5Ra1 – Phycoerythrin (PE) (Excitation: 561 nm, Peak emission: 578 nm), CD103/alpha E integrin – Allophycocyanin (APC) (Excitation: 633 nm, Peak emission: 660 nm). Fluorescence spectral viewer tools (see Materials List) are useful to design panels. - Make an antibody cocktail containing the desired antibodies.
- For static PCLS imaging, add 800 µL staining buffer, 100 µL Fc blocker, 50 µL normal mouse serum, and 50 µL normal rat serum to a 1.5 mL microcentrifugation tube for a total volume of 1 mL. For live cell imaging, use 800 µL Leibovitz's medium (1x) containing 10% FBS (Leibovitz-10) instead of staining buffer.
- Add antibodies to adjust a desired final concentration of each antibody. Transfer the antibody solution to a 3 cm dish, and keep in the dark until ready to use.
NOTE: The final concentrations need to be optimized for each individual antibody (usually 1 – 5 µg/mL).
- Choose a PCLS with an anatomical area of interest, such as large airways or periphery of the lung, and place in the 3-cm dish containing 1 mL antibody solution.
- Stain PCLS in the dark, on ice, rocking slowly for 30 or 60 min.
- For static PCLS imaging, proceed to the section 4. For live cell imaging, proceed to section 5.
4. Static Imaging of PCLS
- After 60 min incubation of PCLS with antibodies, remove the antibody solution from the dish and rinse the slices twice with 1 mL ice cold PBS.
- Pipette 50 µL PBS onto a 3 cm round glass-bottom dish. Using a spatula or paintbrush, transfer the stained PCLS to the drop, gently manipulating the slice until it is flat and spread out. Remove the PBS with a pipette. The slice should lie as flat as possible. The above procedures need to be performed quickly to avoid photobleaching.
- Place 1 drop of room temperature mounting medium onto the slice and gently drop a glass coverslip into the well of the plate. Keep PCLS in the dark at 4 °C for up to an hour before imaging.
- When viewing under a confocal microscope, use a 20X objective (see Materials List). Use the "tile" tool to locate and mark the edges of the tissue in the "convex hull" setting. This will help reduce the time of the tile scan.
- While setting the z-stack range, verify at multiple areas of the slice, especially along the edges, that the set ranges are appropriate.
NOTE: The z-range will likely need to be greater than the thickness of the tissue itself because it is very difficult to get the tissue to lay completely flat. For example, with a 150 µm thick PCLS, the z-stack range will likely need to be set to 200 – 250 µm to accommodate bumps and curves in the slice. Depending on the z-stack range and the size of the tissue, each full lung scan will take between 7 – 12 h with a 20X objective. Each lung is unique and the settings must be adjusted for every experiment.
5. Live Cell Imaging of PCLS
- After 30 min incubation of PCLS with antibodies, remove the antibody solution from the dish and rinse the slices twice with 1 mL cold Leibovitz-10.
- Pipette fresh 50 µL Leibovitz-10 into one slot of a chambered coverglass (8 slots). Use a spatula to transfer the PCLS to the medium, gently manipulating it until the slice is flat and spread out. The above procedures need to be performed quickly to avoid photobleaching.
- Remove the 50 µL medium using a pipette. The slice is to be lying as flat as possible.
NOTE: It is acceptable if the edges of the slice are flipped up vertically against the wall of the chamber. Furthermore, if available, a metal platinum weight can be used to anchor the slice before proceeding to the next step. See the attached Materials List for one example of platinum wire that can be cut and bent into small weights. - In a 4 °C cold room, mix 100 µL of growth factor-reduced, phenol red-free extracellular matrix with 100 µL Leibovitz-10. Use precooled pipette tips for this, or the extracellular matrix will solidify in the tip.
NOTE: This ratio of matrix and medium (1:1) creates a gel dense enough to hold the PCLS down in tissue-culture conditions. - Gently pipette the matrix to mix, and carefully pipette on top of the PCLS, making sure that the gel goes on top of the lung slice, and that the slice does not float up on top of the gel. The gel should work as a weight, anchoring the PCLS down onto the bottom of the plate.
- Carefully transfer the chambered coverglass to a 37 °C incubator and let the matrix solidify for ~5 min.
- Transfer the entire chambered coverglass onto the pre-warmed stage of a confocal microscope. Maintain the chamber at 37 °C throughout imaging. CO2 supply is not required during the cell incubation when using Leibovitz's medium.
- Set the z-stack range and tile range based on the desired interval between frames. When viewing under a confocal microscope, use a 20X objective (see Materials List). Use the "tile" tool to denote the area of interest in the tissue, using the "centered grid" setting (e.g., a 2×2 centered grid).
- While setting the z-stack range, verify at multiple areas of the slice that the set ranges are appropriate.
NOTE: The z-range will likely equal the thickness of the tissue. Each lung is unique and the settings must be adjusted for every experiment. 2×2 tiles and 10 z-stacks will generally result in ~1 frame/2 min. Ensure the auto-focus function is activated before starting the experiment to minimize x-y and z-drift during the imaging period.
Representative Results
To identify the location of two DC subsets, CD11bhi cDCs and CD103+ cDCs, PCLS from C57BL/6 mice were cut and stained with monoclonal antibodies (mAbs) specific to CD11c, CD88, CD103, and CD324 (E-cadherin). Antibodies to CD324 stain airway epithelial cells, and CD88 is displayed on macrophages and neutrophils, but not cDCs8. This allowed us to distinguish cDCs from CD11c+ macrophages, and to observe the spatial relationship of each cell type to the airways (Figure 1). We found that CD11bhi cDCs localize in the parenchyma, whereas CD103+ cDCs reside primarily around the airways and the subpleural area. Although CD103+ cDCs were readily detected by direct staining of cell surface CD103 in this experiment, the detection of CD11bhi cDCs relied on the absence of CD88 and CD103 staining. To directly identify CD11bhi cDCs, we first attempted to stain PCLS using an anti-CD11b mAb (clone M1/70), which is widely used in immunohistochemistry and flow cytometry. However, this mAb had a high background and low specificity in this application, regardless of which fluorochromes were conjugated to the mAb (Figure 2A, and data not shown). By contrast, antibodies to CD172a (SIRP1α) stained CD11bhi cDCs18, but not structural cells or CD103+ cDCs (Figure 2B, and data not shown). On their own, antibodies to CD172a could not discriminate between cDCs and monocytes or macrophages. However, by co-staining with antibodies against CD88 and CD172a, we were able to distinguish alveolar macrophages (CD172a–CD88+) from interstitial macrophages (CD172a+CD88+), and CD11bhi cDCs (CD172a+CD88–), and show that the latter cells preferentially localize in the parenchyma, not the sub-epithelial area (Figure 2C – F), in agreement with our results obtained by indirect staining of these cells.
Study of intra-tissue migration of leukocytes requires visualization of structural cells, including those of the lymphatics, through which DCs traffic from peripheral tissues to regional LNs. In the skin and LNs, lymphatic vessels are often identified using mAbs to LYVE-1 or Podoplanin. However, we found that in PCLS, mAbs to LYVE-1 and Podoplanin stain a variety of other cell types, including vascular endothelium and alveolar epithelium (data not shown). These mAbs are therefore of limited practical use for specifically identifying lymphatic vessels in PCLS. However, mAbs to CD90.2 (Thy1.2), a well-known T cell marker, labeled lymphatic structures, as shown previously in other tissues19,20 (Figures 3A, B), as did TdTomato fluorescent protein encoded by a transgene under transcriptional control of the Prox1 promoter21 (Figure 3A, C). PROX1 is a master transcription factor that is necessary for lymphangiogenesis22. Prox1TdTomato transgenic mice exhibit brightly fluorescent lymphatic vessels in the lungs and other tissues such as the liver, lens, dentate gyrus and neuroendocrine cells of the adrenal medulla21. Indeed, these two labeling methods gave overlapping staining patterns (Figure 3D). Using these labeling approaches, we were able to determine that CD103+ cDCs were present in several anatomical areas, including CD324-expressing airway epithelium, the lymphatics, and the subpleural area (Figure 3A, E).
In addition to static imaging, cell movement and cell to cell interactions can be recorded using live cell imaging of PCLS (Movie 1). PCLS were freshly prepared from mice that had received injections of GFP-expressing OT-II T cells and instillation of OVA and LPS, stained with mAbs against CD103 (red) and the airway epithelium (CD324, blue), and were imaged to track the movement of CD103+ cDCs and T cells 16 h after OVA/LPS instillation. Over the course of the video (4 h), the dynamic movement of CD103+ DCs (red) and the adoptively transferred OVA-specific T cells (green) and their interaction are clearly visible (Movie 1).
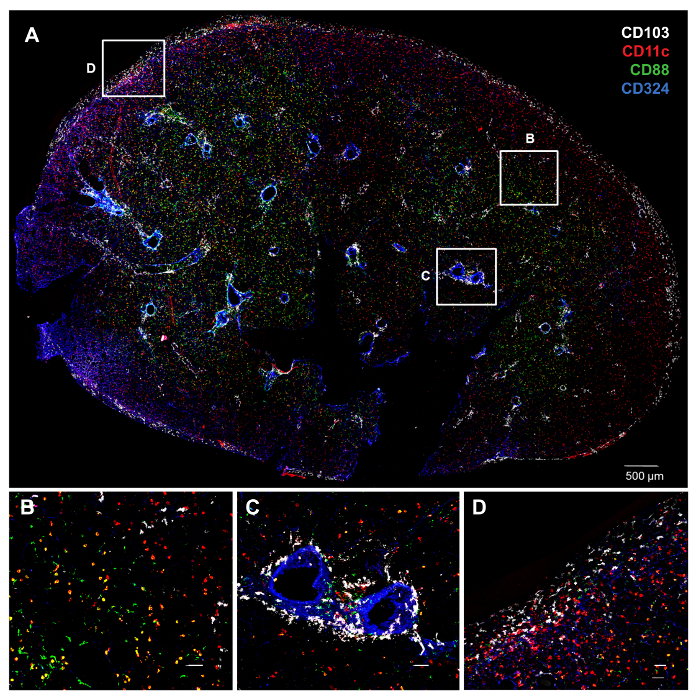
Figure 1. Distinct Anatomic Locations of cDC Subsets as Revealed by PCLS Staining and Confocal Microscopy. A) Whole PCLS from an untreated C57BL/6 mouse lung stained with various antibodies. The color of each molecule is indicated by its font color (top right). Individual and combinatorial staining gives the following colors for cell types of interest: CD103+ cDCs (CD103+CD11c+CD88–; white), CD11bhi cDCs (CD11c+CD103–CD88–; red), macrophages (CD11c+CD88+; yellow), neutrophils (CD11c–CD88+; green) and airway epithelial cells (CD324+; blue). B–D) Higher magnification of the insets in A. B) CD11bhi cDCs localize in the parenchyma. C) CD103+ cDCs around the airways. D) CD103+ cDCs underneath the visceral pleura. White bars denote 50 µm. Please click here to view a larger version of this figure.
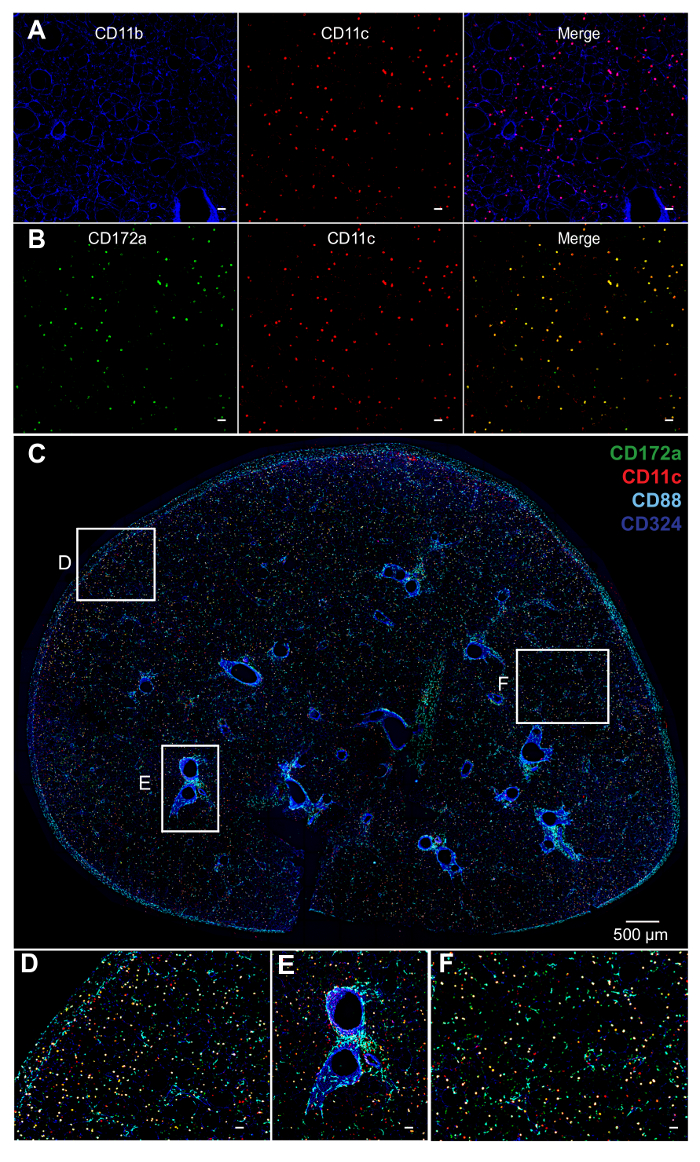
Figure 2. Staining of DCs and Macrophages. A) Staining of PCLS with antibodies to CD11b (blue) gives a high background in PCLS. B) Antibodies to CD172a (green) have a relatively low background in PCLS and greater specificity for CD11b+ cells. C) Whole PCLS of C57BL/6 mice stained with mAbs whose color is indicated by font color (top right). CD11c (red), CD88 (cyan), CD172a (green) and CD324 (blue). CD11bhi cDCs (CD11c+CD88–CD172a+; yellow), alveolar macrophages (CD11c+CD88+CD172a–; pink), interstitial macrophages (CD11c+CD88+CD172a+; white), neutrophils (CD11c–CD88+; cyan) and airways (CD324+; blue). D – F) Higher magnification of the insets depicted in C. D) Sub-pleural area. E) Sub-epithelial area. F) Interstitium. Interstitial macrophages localize in the parenchyma, and alveolar macrophages are in alveoli. Unlabeled white bars denote 50 µm. Please click here to view a larger version of this figure.
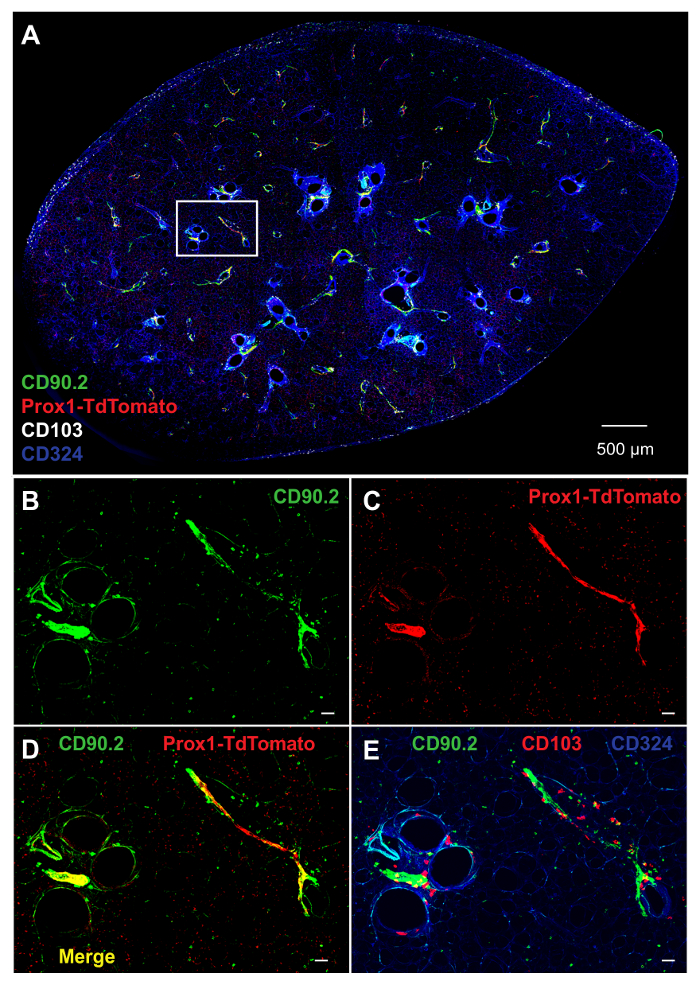
Figure 3. Labeling of Structural Cells in PCLS. A) Whole PCLS prepared from Prox1tdTomato mouse, and stained with mAbs to CD90.2 (green), CD103 (white) and CD324 (blue). Prox1-expressing cells (red) are genetically labeled with TdTomato under the regulation of Prox1 promoter (Prox1-TdTomato). B–E) Higher magnification of the inset in A. B) CD90.2 (green), C) Prox1-TdTomato (red), and D) combination of CD90.2 and Prox1-TdTomato. E) Combination of CD90.2 (green), CD103 (red) and CD324 (blue). Unlabeled white bars denote 50 µm. Please click here to view a larger version of this figure.
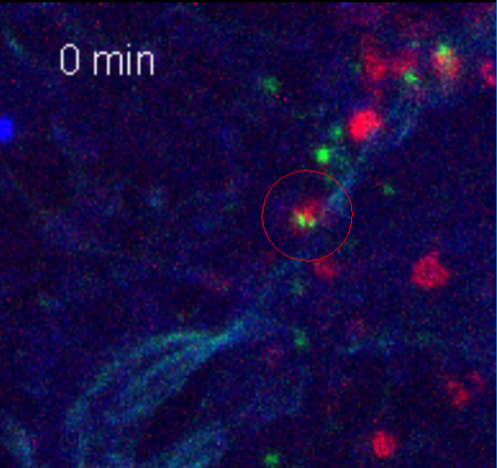
Movie 1. T Cells Interact with CD103+ cDCs in the Lung. Live cell imaging of CD103+ cDCs (red) interacting with T cells (green). CD4+ T cells isolated from OVA-specific OT-II Nur77gfp mice were stimulated with splenic DCs and OVA in vitro, and adoptively transferred into Rag -/- mouse. 2 h later, the recipient mouse was treated with OVA/LPS instillation to the airway. 16 h after OVA/LPS treatment, PCLS were made, stained and imaged for 4 h at 37 °C. Time recording image (min) is shown. Please click here to view this video. (Right-click to download.)
Discussion
The protocol described here was originally developed to visualize the locations of two subsets of cDCs within the lung. However, this protocol can be readily adapted to study many different cell types, while maintaining cell viability and the three-dimensional architecture of the lung. The latter feature is an important advantage over cell culture systems and facilitates identification of rare cell types. The method relies on the generation of PCLS from the lung, and an appropriate combination of antibodies to identify specific cell types while minimizing background staining. To a large extent, this is an empirical exercise because antibodies that work well in other applications, including flow cytometry, do not necessarily work well for staining PCLS. We have already made considerable progress in this regard, but investigators that wish to study cell types not addressed here may have to test different antibodies and optimize fluorochromes and concentrations.
Using the protocol described here, we found that CD103+ cDCs and CD11bhi cDCs reside at distinct localizations in the lung. At steady state, CD103+ cDCs were detected around the airways and in the subpleural region, whereas CD11bhi cDCs were primarily found in the parenchyma. Although the protocol we describe here is not suitable for staining intracellular molecules, it is useful for imaging cell surface markers, as well as secretory molecules. Thus, we were able to detect the chemokines, CCL21 and CCL19, in PCLS (data not shown). If only weak fluorescent signals are seen using primary antibodies, it is possible to use secondary antibodies to amplify those signals.
A major challenge when conducting live cell video-imaging of PCLS was to sufficiently anchor the tissue in culture medium to prevent significant x-y- and z-drift during the 4-h microscopy period. To address this, we experimented with different ratios of Leibovitz's medium to extracellular matrix, which is solid at room temperature, and determined a 1:1 ratio maintained a stable, 3D structure with cell viability. During post-image processing, we also utilized a series of ImageJ plug-ins, called TurboReg and StackReg that eliminate any additional incidental x-y or z-shift (http://www.einstein.yu.edu/segallLab/page.aspx?id=18226). Using the auto-focus tool of the confocal microscope, the region of interest was kept in focus, especially in the z-range, during the entire scanning period. Another challenge during the live cell imaging of PCLS was utilizing the tile scan, z-stack, time series, and autofocus functions of the microscope. To minimize the time between each frame, a balance must be struck between the time spent scanning each tile. To optimize the scan time in each research project, z-stack number, tiles, and scanning speeds need to be adjusted.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Jeff Tucker, Erica Scappini, and Agnes Janoshazi for their help with microscopy, Ligon Perrow for her management of the mouse colony, and Jun Chen and Michael Sanderson for help with the tissue slicer, and Michael Fessler and Derek Cain for critical reading of the manuscript. This work was funded by the intramural branch of the NIEHS, NIH (ZIA ES102025-09), which is in turn sponsored by the Department of Health and Human Services.
Materials
| C57BL/6J mice | Jackson Laboratory | 000664 | |
| Prox1-TdTomato transgenic mice | Jackson Laboratory | 018128 | B6;129S-Tg(Prox1-tdTomato)12Nrud/J |
| OT-II OVA-specific TCR x Nur77-GFP transgenic mice | Jackson Laboratory | 004194, 006617 | B6.Cg-Tg(TcraTcrb)425Cbn/J x C57BL/6-Tg(Nr4a1-EGFP/cre)820Khog/J |
| Rag1 knock-out mice | Jackson Laboratory | 002216 | B6.129S7-Rag1tm1Mom/J |
| Ovalbumin, Low Endo, Purified | Worthington Biochemical Corporation | LS003059 | |
| Lipopolysaccharides from Escherichia coli | Sigma-Aldrich Co. | L2630-25MG | |
| Polyethylene tubing (Non-Sterile) 100 ft | BD Diagnostic Systems | 427421 | 0.86 mm inside diameter, 1.27 mm outside diameter |
| GeneMate Sieve GQA Low Melt Agarose | BioExpress | E-3112-125 | 2% solution dissolved in PBS at 70 °C and held at 40 °C. |
| Compresstome VF-300 | Precisionary Instruments, Inc. | VF-300 | |
| Double Edge Stainless Razor Blade | Electron Microscopy Sciences | 72000 | Disposable; 250/box. Blade should be changed for every lung. |
| Krazy Glue All Purpose Instant Gel | VWR | 500033-484 | Commonly available for $3/tube in local drugstores |
| Leibovitz's L-15 Medium, no phenol red | ThermoFisher Scientific | 21083027 | |
| Normal Rat Serum | Jackson ImmunoResearch Inc. | 012-000-120 | |
| Normal Mouse Serum | Jackson ImmunoResearch Inc. | 015-000-120 | |
| Fetal bovine serum | Hyclone | SH30071.03HI | |
| Staining Buffer | Made in House | N/A | PBS w/ 0.5% bovine serum albumin, 0.1% NaN3, pH 7.4 |
| Fc Blocker (anti-CD16/32 antibodies) | Made in House | N/A | Supernatant of cultured hybridoma cell line 2.4G2 |
| Anti-mouse CD11b eFluor 450 | eBioscience | 48-0112-80 | Anti-mouse CD11b eFluor 450 (clone: M1/70) |
| Anti-mouse CD11c Brilliant Violet 605 | BioLegend | 101237 | Brilliant Violet 605 anti-mouse CD11c (clone: M1/70) |
| Anti-mouse CD11c Phycoerythrin | eBioscience | 12-0114-82 | PE conjugated anti-mouse CD11c (clone: N418) |
| Anti-mouse CD11c Allophycocyanin | BD Phamingen | 550261 | APC-labeled anti-mouse CD11c 9clone: HL3) |
| Anti-mouse CD88 Phycoerythrin | BioLegend | 135806 | PE anti-mouse CD88 (clone: 20/70) |
| Anti-mouse CD103 Allophycocyanin | eBioscience | 17-1031-82 | Anti-mouse CD103 APC (clone: 2E7) |
| Anti-mouse CD90.2/Thy1.2 eF450 | eBioscience | 48-0902-82 | Anti-mouse CD90.2 eFluor 450 (clone: 53-2.1) |
| Anti-mouse CD172a/Sirp1a Allophycocyanin | eBioscience | 17-1721-82 | Anti-mouse CD172a APC (clone: P84) |
| Anti-mouse CD324 Brilliant Violet 421 | BD Horizon | 564188 | BV421 mouse anti-E Cadherin (clone: 5E8 also known as 5E8-G9-B4) |
| Anti-mouse CD324 Alexa Fluor 488 | eBioscience | 53-3249-82 | Anti-CD324(E-Cadherin) Alexa Flour 488 (clone: DECMA-1) |
| Anti-mouse CD324 Alexa Fluor 647 | eBioscience | 51-3249-82 | Anti-CD324(E-Cadherin) Alexa Flour 647 (clone: DECMA-1) |
| Glass Bottom Microwell Dishes 35mm petri dish, 14mm Microwell, No. 1.5 coverglass | MatTek Corperation | P35G-1.5-14-C | |
| Nunc Lab-Tek Chambered Coverglass | ThermoFisher Scientific | 155411PK | Pack of 16 |
| 15 mm Coverslip, No. 1.5 Glass Thickness | MatTek Corperation | PCS-1.5-15 | |
| Bare Platinum Wire | World Precision Instruments | PTP201 | 0.020" (0.5mm) diameter cut into ~1 cm long pieces and bent into an "L" shape |
| ProLong Gold Antifade Mountant | ThermoFisher Scientific | P36934 | Keep at 4 °C, warm to room tempterature before use. |
| Matrigel Growth Factor Reduced (GFR) Basement Membrane Matrix, Phenol Red-Free, *LDEV-Free | Corning | 356231 | |
| Zeiss 880 multi-photon laser-scanning microscope | Carl Zeiss | Zen Black software version 8.1, 2012 (Zeiss) | |
| Plan-Apochromat 20x/0.8 M27 objective lends | Carl Zeiss | 420650-9901-000 |
References
- Schneider, T., van Velzen, D., Moqbel, R., Issekutz, A. C. Kinetics and quantitation of eosinophil and neutrophil recruitment to allergic lung inflammation in a brown Norway rat model. Am J Respir Cell Mol Biol. 17 (6), 702-712 (1997).
- Vermaelen, K. Y., Carro-Muino, I., Lambrecht, B. N., Pauwels, R. A. Specific migratory dendritic cells rapidly transport antigen from the airways to the thoracic lymph nodes. J Exp Med. 193 (1), 51-60 (2001).
- Sung, S. S., et al. A major lung CD103 (alphaE)-beta7 integrin-positive epithelial dendritic cell population expressing Langerin and tight junction proteins. J Immunol. 176 (4), 2161-2172 (2006).
- Steinman, R. M. Lasker Basic Medical Research Award. Dendritic cells: versatile controllers of the immune system. Nat Med. 13 (10), 1155-1159 (2007).
- Jakubzick, C., et al. Lymph-migrating, tissue-derived dendritic cells are minor constituents within steady-state lymph nodes. J Exp Med. 205 (12), 2839-2850 (2008).
- Julia, V., et al. A restricted subset of dendritic cells captures airborne antigens and remains able to activate specific T cells long after antigen exposure. Immunity. 16 (2), 271-283 (2002).
- Nakano, H., et al. Migratory properties of pulmonary dendritic cells are determined by their developmental lineage. Mucosal Immunol. 6 (4), 678-691 (2013).
- Nakano, H., et al. Complement receptor C5aR1/CD88 and dipeptidyl peptidase-4/CD26 define distinct hematopoietic lineages of dendritic cells. J Immunol. 194 (8), 3808-3819 (2015).
- Sanderson, M. J. Exploring lung physiology in health and disease with lung slices. Pulm Pharmacol Ther. 24 (5), 452-465 (2011).
- Liberati, T. A., Randle, M. R., Toth, L. A. In vitro lung slices: a powerful approach for assessment of lung pathophysiology. Expert Rev Mol Diagn. 10 (4), 501-508 (2010).
- Parrish, A. R., Gandolfi, A. J., Brendel, K. Precision-cut tissue slices: applications in pharmacology and toxicology. Life Sci. 57 (21), 1887-1901 (1995).
- Bergner, A., Sanderson, M. J. ATP stimulates Ca2+ oscillations and contraction in airway smooth muscle cells of mouse lung slices. Am J Physiol Lung Cell Mol Physiol. 283 (6), L1271-L1279 (2002).
- Martin, C., Uhlig, S., Ullrich, V. Videomicroscopy of methacholine-induced contraction of individual airways in precision-cut lung slices. Eur Respir J. 9 (12), 2479-2487 (1996).
- Henjakovic, M., et al. Ex vivo testing of immune responses in precision-cut lung slices. Toxicol Appl Pharmacol. 231 (1), 68-76 (2008).
- Henjakovic, M., et al. Ex vivo lung function measurements in precision-cut lung slices (PCLS) from chemical allergen-sensitized mice represent a suitable alternative to in vivo studies. Toxicol Sci. 106 (2), 444-453 (2008).
- Paddenberg, R., et al. Hypoxic vasoconstriction of partial muscular intra-acinar pulmonary arteries in murine precision cut lung slices. Respir Res. 7, 93 (2006).
- Wilson, R. H., et al. Allergic sensitization through the airway primes Th17-dependent neutrophilia and airway hyperresponsiveness. Am J Respir Crit Care Med. 180 (8), 720-730 (2009).
- Schlitzer, A., et al. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat Immunol. 16 (7), 718-728 (2015).
- Baluk, P., et al. Preferential lymphatic growth in bronchus-associated lymphoid tissue in sustained lung inflammation. Am J Pathol. 184 (5), 1577-1592 (2014).
- Jurisic, G., Iolyeva, M., Proulx, S. T., Halin, C., Detmar, M. Thymus cell antigen 1 (Thy1, CD90) is expressed by lymphatic vessels and mediates cell adhesion to lymphatic endothelium. Exp Cell Res. 316 (17), 2982-2992 (2010).
- Truman, L. A., et al. ProxTom lymphatic vessel reporter mice reveal Prox1 expression in the adrenal medulla, megakaryocytes, and platelets. Am J Pathol. 180 (4), 1715-1725 (2012).
- Wigle, J. T., Oliver, G. Prox1 function is required for the development of the murine lymphatic system. Cell. 98 (6), 769-778 (1999).

