Small RNA Transfection in Primary Human Th17 Cells by Next Generation Electroporation
Summary
Next generation electroporation is an efficient method for transfecting human Th17 cells with small RNAs to alter gene expression and cell behavior.
Abstract
CD4+ T cells can differentiate into several subsets of effector T helper cells depending on the surrounding cytokine milieu. Th17 cells can be generated from naïve CD4+ T cells in vitro by activating them in the presence of the polarizing cytokines IL-1β, IL-6, IL-23, and TGFβ. Th17 cells orchestrate immunity against extracellular bacteria and fungi, but their aberrant activity has also been associated with several autoimmune and inflammatory diseases. Th17 cells are identified by the chemokine receptor CCR6 and defined by their master transcription factor, RORγt, and characteristic effector cytokine, IL-17A. Optimized culture conditions for Th17 cell differentiation facilitate mechanistic studies of human T cell biology in a controlled environment. They also provide a setting for studying the importance of specific genes and gene expression programs through RNA interference or the introduction of microRNA (miRNA) mimics or inhibitors. This protocol provides an easy to use, reproducible, and highly efficient method for transient transfection of differentiating primary human Th17 cells with small RNAs using a next generation electroporation device.
Introduction
CD4+ T cells are crucial orchestrators of the adaptive immune response. Naïve CD4+ T cells are capable of developing into several different effector T cells (e.g. Th1, Th2, Th17, etc.), each with their own set of characteristic cytokines and transcription factors, depending upon the local microenvironment1. The lineage decisions that T cells make are critical for both protective immunity and tolerance to self. Th17 cells are one subset of T cells known to combat extracellular bacteria and fungi, but their improper responses are also implicated in the pathogenesis of multiple autoimmune and inflammatory diseases such as multiple sclerosis and psoriasis2,3. Human Th17 cells can be generated from naïve CD4+ T cells in vitro by providing them with an appropriate polarizing environment4. Various combinations of the cytokines IL-1β, IL-23, TGFβ, and IL-6 have been used for the development of human Th17 cells. Human Th17 cells express CCR6, a chemokine receptor that is commonly used to identify this cell population and are defined by the expression of their principal transcription factor, RORγt (encoded by RORC)5,6. Th17 cells have the ability to express multiple cytokines, but IL-17A is the lineage-defining effector cytokine produced by these cells. We examined the expression of all three Th17-associated markers (CCR6, RORγt, IL-17A) to assess the robustness of our human Th17 in vitro differentiation assay. Additionally, we cultured human CD4+ T cells under non-polarizing conditions, where no cytokines or blocking antibodies were added to the culture media to use as a negative control since expression of these Th17 markers should be very low or absent.
One way to study normal human T cell development and biology is to manipulate gene expression during their development. Short-interfering RNA (siRNA) are synthetic small RNA molecules that target protein-coding mRNAs and can be utilized to reduce specific gene expression. MicroRNAs (miRNAs) are endogenous non-coding small RNAs known to modulate gene expression post-transcriptionally. miRNAs have been shown to play an important role in both murine and human T cell biology, including in Th17 cells7,8,9. It is crucial to have reliable methods of manipulating small RNA activity in human T cells to study their effects on gene expression and ultimately on human T cell biology. Here, we describe an easy-to-use, consistent and reliable protocol that we developed for introducing small synthetic RNAs and locked nucleic acids (LNAs, chemically modified nucleic acids with increased stability) into immune cells, and specifically into human Th17 cells.
There are several alternative methods of introducing small RNAs into mammalian cells, which generally fall into chemical, biological, or physical categories10. Commonly used chemical methods, including lipid-based transfections and calcium-phosphate transfections, rely on creating chemical-DNA complexes that are more efficiently taken up by cells. In general, chemical methods are not as efficient for the transfection of primary T cells. The most common biological method is to use a viral vector (e.g. retrovirus or lentivirus), which directly inserts foreign RNA into a host as a part of its natural replication cycle. Viral transduction typically takes longer to complete, especially when one factors in time for molecular cloning of proviral plasmids. Additionally, viral transduction vectors can be potentially harmful to human researchers. Electroporation is a physical method of inducing membrane permeabilization by subjecting cells to high voltage pulses, allowing nucleic acids to transiently enter into the cell where they can act on their target. Traditional electroporation instruments were not effective for transfecting primary lymphocytes. However, optimized next generation electroporation has proven to be capable of transfecting T cells at very high efficiency, especially when the material to be transfected is small RNA. The term next generation is loosely used to differentiate the two newer platforms (e.g., Neon, Amaxa) from traditional electroporation machines. Additionally, this method is easily scalable for moderate throughput screens with up to approximately 120 small RNAs in a single experiment, often using validated synthetic reagents. Importantly, successful transfections can be achieved in as little as 16 h after T cell activation. The disadvantage of this method, however, is that it does not result in stable genomic incorporation, and is therefore transient. Hence, it is worth the extra effort to create a stable expression construct that can be packaged into a viral vector and successfully expressed in T cells in cases where long-term expression of a small RNA is required.
We have used a next generation transfection (e.g., Neon) to deliver diverse synthetic single or double-stranded RNA or LNA oligonucleotide tools for different purposes11,12,13. Efficient RNA interference can be induced in primary mouse and human T cells using double-stranded short-interfering RNA (siRNA). This protocol describes optimized conditions for using this technique in human Th17 cells. In addition to siRNAs, commercially available synthetic miRNA mimics and inhibitors can be used to study miRNA gain and loss of function. miRNA mimics are double-stranded RNA molecules very similar to siRNAs, but designed with the sequence of endogenous mature miRNAs. miRNA inhibitors are chemically-modified RNA and/or LNA based single stranded oligonucleotides that bind to native miRNAs and antagonize their function. We have found that all of these tools can be used effectively in cultured primary T lymphocytes, including but not limited to human Th17 cells.
Protocol
This protocol adheres to UCSF's guidelines for human research ethics.
1. Preparation of T Cell Culture, Isolation of CD4+ T Cells, and Th17 Polarization
- On Day 0, coat 6-well tissue culture plates with 1.5 mL per well of anti-human CD3 (2 µg/mL) and anti-human CD28 (4 µg/mL) in PBS with calcium and magnesium for at least 2 h at 37 °C.
- Alternatively, coat the plates overnight at 4 °C. Wrap plates in parafilm.
- Prepare the cord blood mononuclear cells (CBMCs) by density gradient centrifugation per manufacturer's instructions.
CAUTION: Work carefully and ensure proper personal protective equipment (PPE) is worn when handling human blood to avoid any risk of exposure to blood-borne pathogens. - Once the mononuclear cells have been isolated and washed, perform human CD4+ T cell isolation by negative selection using a commercial Human CD4+ T cell kit.
NOTE: If there is red blood cell contamination after mononuclear cell isolation, an optional red blood cell lysis may be performed prior to the CD4+ T cell isolation steps. Resuspend the mononuclear cells in 1 mL of isolation buffer (2% FBS in PBS). Add 5 mL of 1x lysing solution. Incubate for 15 min at room temperature. Then add 5 mL of isolation buffer and centrifuge at 300 x g for 5 min at 4 °C to pellet the cells.- Resuspend the mononuclear cells at a density of 50 x 106 per 500 µL in the isolation buffer in a new 5 mL tube.
- Add 100 µL of FBS and 100 µL of the Antibody Mix per tube then incubate each tube at 4 °C for 20 min on an orbital shaker to mix well.
- After the incubation, add 3-4 mL of isolation buffer to wash the cells. Centrifuge the cells at 300 x g for 8 min at 4 °C to pellet the cells. Carefully aspirate the supernatant.
- During this spin, pre-wash the magnetic beads. Transfer the desired amount of magnetic beads into a new tube (used at 1:1 with the cells) and add an equal volume of isolation buffer to wash the beads. Mix well using a 1 mL micropipette and then place the tube in the magnet for at least 1 min. Carefully aspirate the supernatant. Resuspend the magnetic beads in the same volume initially transferred prior to the wash.
- Resuspend the cells in each tube with 500 µL of the isolation buffer and add 500 µL of pre-washed magnetic beads.
- Incubate the cells with the magnetic beads for 15 min on an orbital shaker at room temperature (18 °C to 25 °C) to mix well.
- After the incubation, thoroughly pipet the cells using a 1 mL micropipette at least 10 times. Then add 3-4 mL of isolation buffer and place each tube in the magnet for 2 min.
- Carefully transfer the negatively selected CD4+ T cells that are in the supernatant to a new tube.
- Once CD4+ T cell isolation is completed, count the cells with a hemocytometer and keep the cells on ice.
- Wash the antibody-coated plates two times with PBS. Then add 1.5 mL of 2x mix of Th17-polarizing media to each well: anti-human IFNγ (20 µg/mL), anti-human IL-4 (20 µg/mL), human TGFβ (10 ng/mL), human IL-1β (40 ng/mL), human IL-23 (40 ng/mL), human IL-6 (50 ng/mL) all diluted in a serum-free base media (supplemented with 2 mM L-glutamine, 100 U/mL penicillin, 100 µg/mL streptomycin, 10 mM HEPES, 1 mM sodium pyruvate and 100 µM 2-mercaptoethanol).
NOTE: Transfection and RNA interference does work in serum-containing media. The purpose of using serum-free media with this protocol is to achieve better IL-17A production. - Centrifuge the human CD4+ T cells at 500 x g for 5 min at 4 °C to pellet cells. Carefully aspirate the supernatant. Then resuspend the cells in serum-free base media and plate the cells at a density of 3 x 106 in 1.5 mL per well so that the final volume is 3 mL per well and polarizing cytokines are now at a 1x final concentration.
- Place the plates in 5% CO2, 37 °C incubator for two days.
2. Electroporation of In Vitro Polarized Human Th17 Cells
- On Day 2, coat 48-well tissue culture plates with 250 µL per well of anti-human CD3 (2 µg/mL) and anti-human CD28 (4 µg/mL) in PBS with calcium and magnesium for at least 2 h at 37 °C.
- Alternatively, coat the plates overnight at 4 °C. Wrap plates in parafilm.
- Prepare small RNAs for transfection. For each transfection, aliquot 1 µL of a 5 µM stock solution of siRNA into a 1.5 mL microcentrifuge tube. Include appropriate chemistry-matched small RNA control. Keep all tubes on ice.
- Wash the antibody-coated plates two times with PBS. Then add 500 µL of 1X Th17-polarizing media to each well: anti-human IFNγ (10 µg/mL), anti-human IL-4 (10 µg/mL), human TGFβ (5 ng/mL), human IL-1β (20 ng/mL), human IL-23 (20 ng/mL), human IL-6 (25 ng/mL) all diluted in a serum-free base media (supplemented with 2 mM L-glutamine, 100 U/mL penicillin, 100 µg/mL streptomycin, 10 mM HEPES, 1 mM sodium pyruvate and 100 µM 2-mercaptoethanol).
- After the culture plates and transfection reagents are prepared, resuspend the cells with a 1 mL micropipette, pipetting gently but ensuring that all the cells are detached from the bottom of the wells. Pool the cells into a conical tube and centrifuge at 500 x g for 5 min at 4 °C.
NOTE: For culture periods longer than the four-day protocol presented herein, the cells should be transfected approximately every 3 days using this same protocol. Step 2.4 should be modified however since the cells treated with different small RNAs should not be pooled. All of the conditions being tested must be collected, counted, and transfected separately. - Carefully aspirate the supernatant, and then resuspend the cells in at least 1 mL of PBS to wash. Count the live Th17 cells (e.g. with a hemocytometer using Trypan Blue exclusion to assess viability) and then transfer the cells to a microcentrifuge tube.
- Centrifuge at 500 x g for 5 min at room temperature to pellet the cells.
- Carefully aspirate the supernatant, and then resuspend the cells using the provided resuspension buffer from the transfection system 10 µL kit at a density of 2.5-4 x 107 cells per mL. Keep cells at room temperature.
NOTE: As a guideline, try to prepare only as many cells as can be transfected within 30 min. Multiple batches can be compared. - Add 9 µL of cells to 1 µL of small RNA in each microcentrifuge tube. Pipet once to mix the cells and small RNAs for transfection then load into the provided pipette electrode tip.
NOTE: Typically, add 9.5 µL of cell suspension to ensure there is enough of the mixture to prevent creating bubbles in the pipet electrode tip prior to transfection. - Fill the provided cuvette with 3 mL of room temperature Electrolytic Buffer E. Place the cuvette inside the pipette station and then place the pipette into position inside the cuvette.
- Immediately electroporate each 10 µL mix of cells and small RNA using the following parameters: pulse voltage 1,500-1,550 V, pulse width 10 ms, and 3 pulses total on the transfection device.
- After the electroporation is complete, directly add the cell mixture to 500 µL of 1X Th17-polarizing media in prepared wells of a culture plate. Place plates in a 5% CO2, 37 °C incubator for two more days.
NOTE: Wash the pipette electrode tip by pipetting up and down in PBS in between each transfection.
3. Harvesting Human Th17 Cells
- Resuspend the cells in culture media with a micropipette, pipetting gently but ensuring that all the cells are detached from the bottom of the wells. Prepare the cells for functional and/or gene expression assays.
NOTE: Routinely, enough cells are yielded from a single well of transfected Th17 cells to be used for flow cytometric analysis of surface marker and intracellular cytokine and transcription factor staining or for preparation of RNA for gene expression analysis.
Representative Results
The first step to developing a reliable system of successfully electroporating human Th17 cells was to generate robust in vitro differentiated human Th17 cell cultures. T cells cultured under Th17-polarizing conditions expressed the chemokine receptor CCR6 and the transcription factor RORγt (Figure 1A, left). These markers were not expressed when T cells were cultured under non-polarizing (ThN) conditions (Figure 1A, right). T cells cultured under Th17-polarizing conditions also produced IL-17A upon restimulation (Figure 1B, left) but not under ThN conditions (Figure 1B, right). Th17 differentiation still occurs after transfection with small RNAs but in some experiments, there is an observed reduction in the frequency of IL-17A production (Figure 1C). The effect of small RNA transfection on cytokine production demonstrates the importance of having a chemistry-matched small RNA control for comparison within each experiment. Importantly, viability is maintained after transfection with small RNAs and is typically greater than 70% (Figure 1D).
To determine the efficiency of this protocol, we used RNA interference. The CD45 antigen is encoded by the PTPRC (protein tyrosine phosphatase, receptor type C) gene. CD45 is expressed on all hematopoietic cells and therefore human Th17 cells should robustly express this surface marker. Transfection of human Th17 cells on day 2 with an siRNA pool targeting PTPRC strongly reduced the expression of CD45 compared to a chemistry matched control siRNA (Figure 2A). This unimodal population shift in CD45 expression indicates near 100% transfection efficiency, typical of this protocol for small RNAs. Thus, this is an important tool that can be used to assess transfection efficiency during protocol optimizations. RORC encodes RORγt, the predominant transcription factor of human Th17 cells, which directly regulates IL-17A production. Transfecting developing human Th17 cells with an siRNA pool against RORC strongly reduced RORγt expression as expected (Figure 2B). Reducing RORγt expression by RNAi had a functional effect, as these cells exhibited reduced IL-17A production (Figure 2C) compared to cells transfected with a control siRNA. Non-polarized CD4+ T cells should not express any RORγt or IL-17A and are shown as a negative control in this figure.
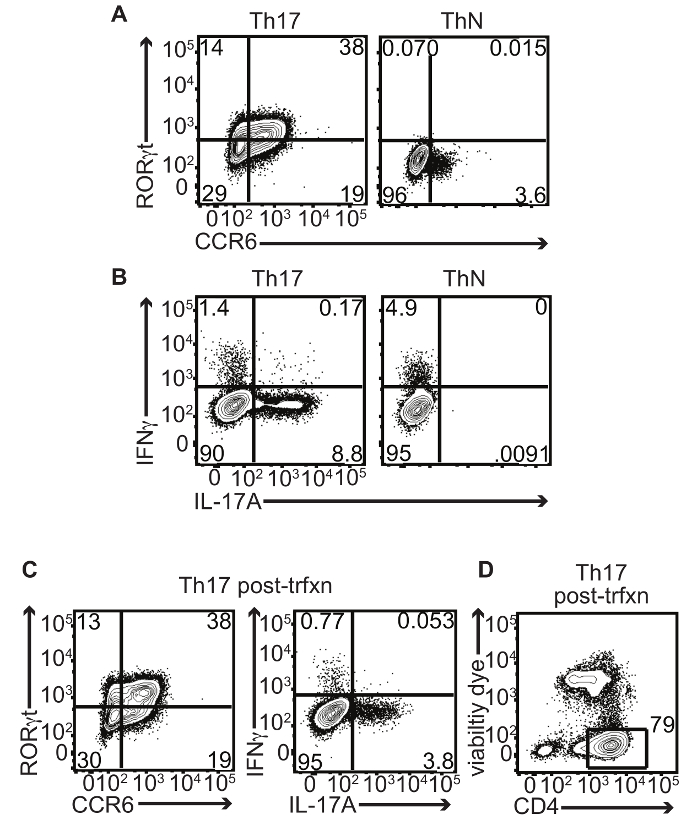
Figure 1: In vitro Differentiated Human Th17 Cells Express CCR6, RORγt, and IL-17A. Human CD4+ T cells isolated from umbilical cord blood were cultured under Th17 conditions (IL-1β, IL-23, IL-6, and TGFβ) or in non-polarizing (ThN) conditions (media only, no polarizing cytokines or blocking antibodies added) in vitro and analyzed on day 4 for Th17 marker expression by flow cytometry: (A) Representative contour plots display surface CCR6 and intracellular RORγt co-staining of CD4+ T cells. Numbers in quadrants indicate percent CCR6 and/or RORγt-positive live singlet CD4+ cells. (B) IL-17A and IFNγ production after restimulation with PMA/ionomycin. Numbers in quadrants indicate percent IL-17A and/or IFNγ-positive live singlet CD4+ cells. (C) Representative contour plot displays surface CCR6 and intracellular RORγt co-staining of CD4+ T cells post-transfection with small RNA control. Numbers in quadrants indicate percent CCR6 and/or RORγt-positive live singlet CD4+ cells (left). Representative contour plot displays IL-17A and IFNγ production after restimulation with PMA/ionomycin post-transfection with small RNA control. Numbers in quadrants indicate percent IL-17A and/or IFNγ-positive live singlet CD4+ cells (right). (D) Representative contour plot displays viability of CD4+ cells post-transfection with small RNA control. Number indicates percent CD4+ live singlet cells. Please click here to view a larger version of this figure.
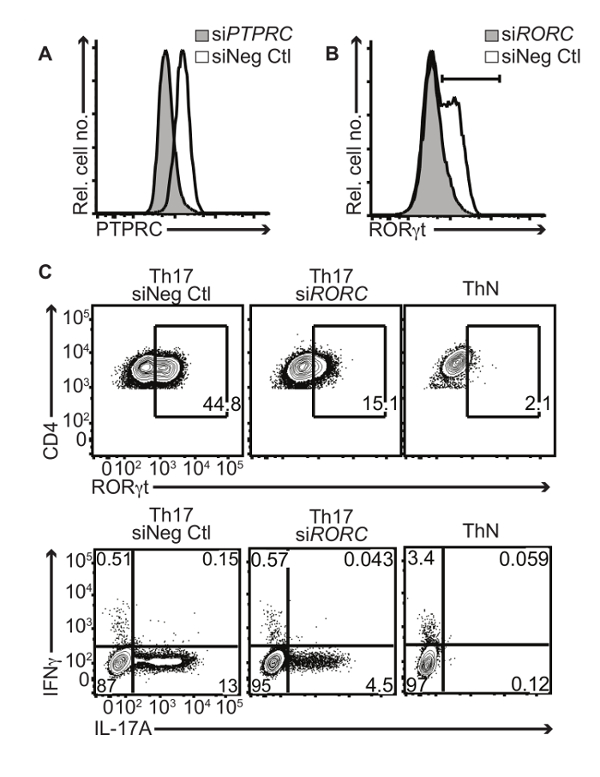
Figure 2: RNA Interference in Primary Human Th17 Cells. Human CD4+ T cells from umbilical cord blood cultured under Th17 conditions were transfected on day 2 with an siRNA nontargeting control (siNeg Ctl) or targeting pool against PTPRC (siPTPRC ) or RORC (siRORC) comprising 4 individual siRNAs. Cells were then analyzed by flow cytometry on day 4: (A) Surface CD45 expression shown in a representative histogram after transfection with siPTPRC (grey tint) or siNeg Ctl (black line). (B) Intracellular RORγt expression is shown in a representative histogram after transfection with siRORC (grey tint) or siNeg Ctl (black line). (C) Representative contour plots display surface CD4 and intracellular RORγt co-staining of CD4+ T cells (top panel) or IL-17A and IFNγ production after restimulation with PMA/ionomycin (bottom panel). Numbers in quadrants indicate percent CD4+RORγt+ or IL-17A and/or IFNγ-positive live singlet cells under ThN or Th17 conditions transfected with siNeg Ctl or siRORC. Please click here to view a larger version of this figure.
Discussion
This protocol provides an improved method for the delivery of small RNAs into human Th17 cells. Although human Th17 cells were used here, this method of electroporation with small RNAs can be used with other primary human T helper subsets, such as Th1, Th2, and Tregs. It has not worked well for naïve CD4+ T cells so the cells must be activated in culture prior to transfection. For this protocol, we first optimized the in vitro culture system for better IL-17A production. The biggest factor was base media. We found that serum-free media was superior to traditional serum-containing media for better induction of IL-17A producing human Th17 cells. Since Th17 cells can be generated with subsets of the cytokines TGFβ, IL-1β, IL-23, and IL-6, we tested different cytokine mixes. In this study, we achieved more robust IL-17A production when all four polarizing cytokines were included. Additionally, we optimized shortening the length of the culture to ensure only one transfection was necessary during the culture period. While this four-day protocol allows results to be generated faster and necessitates only one transfection, culture length could be lengthened to further increase IL-17A production. A longer culture period may even be necessary to achieve optimal differentiation of other primary T cell subsets. Since these transfections are transient, multiple transfections may be required over time for lengthy culture periods. We suggest replenishing small RNA reagents after approximately 3 days by re-electroporating the cells with the reagent to prevent dilution over multiple cell divisions. We have routinely done this for mouse CD4+ T cells. Although it is more technically challenging since the cells in each condition must be collected and counted separately, the transfection is successful and there are minimal effects on cell viability.
In these experiments, human Th17 cells were generated through in vitro culture of human umbilical cord blood. Importantly, an alternative option for this differentiation assay would be to use human peripheral blood mononuclear cells (PBMCs). For both CBMCs and PBMCs, it is necessary to isolate CD4+ T cells prior to the start of the culture. We present here our method for purifying CD4+ T cells by negative selection, which yielded robust transfection efficiency, but we expect other methods of preparing primary human CD4+ T cells would work equally well. We have tried several kits that use either positive or negative selection for the isolation of mouse CD4+ T cells and both methods have resulted in similar transfection efficiency. An additional consideration is the importance of using naïve T cells in order to optimally differentiate into Th17 cells. Using human cord blood is advantageous for this reason because it already has a higher proportion of naïve T cells present and therefore it is not necessary to sort or pre-enrich for naïve T cells prior to the start of culture. However, a clear disadvantage of this approach is the limited availability of umbilical cord blood. Alternatively, sorting of naïve T cells from PBMCs14 can be performed prior to culture since this resource is more readily available, although large quantities would have to be obtained.
While there are multiple methods for transfecting mammalian cells, primary T cells tend to be a more difficult cell type to transfect. Next generation electroporation is a successful physical method of transiently transfecting T cells that is technically easy to perform and is reproducible. Once optimized, it has a very high efficiency of transfection for small RNAs that approaches 100%. This can be seen in Figure 2 where the entire unimodal population of CD45+ cells (Figure 2A) and the population of all the cells expressing RORγt (Figure 2B) shift to a lower expression level. Importantly, optimized transfection with small RNAs does not compromise cell viability. We routinely observe greater than 70% viability post-transfection with small RNAs (Figure 1D). Although viability is not an issue with this protocol, the biology of other T cell characteristics, such as cytokine production, may be affected after transfection with small RNAs. For this reason, it is critically important to use a chemistry-matched small RNA control in every experiment.
We optimized the two electroporation parameters, pulse and voltage, for minimizing cell death and maximizing transfection efficiency. Higher voltages increase cell death during transfection. Some other technical considerations to prevent cell death and ensure successful transfection include minimizing the amount of synthetic reagents used and limiting the time between cell preparation and electroporation. Minimal concentrations necessary for synthetic transfection reagents should always be used to prevent potential T cell toxicity and off-target effects. Reducing cell preparation time prior to transfection to minimize the time of cells in transfection buffer at room temperature is crucial and can be achieved by preparing cells in batches, if necessary.
Currently, there are two commercially available next generation electroporation instruments. Both of these systems can efficiently transfect primary T cells. This protocol was optimized using the Neon transfection system. While this transfection system is more cost-effective, the Amaxa system uniquely offers a 96-well adaptor for electroporation for higher throughput applications. We hope that additional commercial advancements will continue to increase the throughput and efficiency of these transfection systems, while keeping cost at a minimum.
Next generation electroporation of human T cells can efficiently introduce small RNAs including siRNAs against target genes of interest as exemplified in this protocol as well as miRNA mimics or inhibitors. The high efficiency makes it unnecessary to use markers of transfection since nearly every cell is transfected with small RNAs. This is in contrast to the transfection of primary T cells with DNA plasmids where typical plasmid transfections result in only 20-30% expression of the marker gene with greater than 50% toxicity that increases with increasing amounts of plasmid. Introducing synthetic mimics using this method has allowed for dozens of genes to be screened for function in T cell biology in parallel without any need for molecular cloning12,13. It has also enabled testing for the function of all miRNAs expressed by T cells in parallel11,13. With this protocol, these types of moderate throughput screens are applicable to human T cells. Notably, this technique was recently used to introduce small RNA (gRNA)-Cas9 ribonucleoprotein complexes into primary human T cells for genome editing15. These diverse applications highlight the utility, flexibility and power of the protocol presented here as an important tool for immunologists seeking to study T cell biology.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by the US National Institutes of Health grants (R01HL109102, P01HL107202, U19CA179512, F31HL131361), a Leukemia & Lymphoma Society scholar award (K.M.A.), and the National Institute of General Medical Sciences (NIGMS) Medical Scientist Training Program (Grant #T32GM007618) (M.M.).
Materials
| anti-human IL-17A PE | ebioscience | 12-7179-42 | Clone: eBio64DEC17 |
| anti-human IFNg FITC | ebioscience | 11-7319-82 | Clone: 4S.B3 |
| anti-human CD4 eVolve605 | ebioscience | 83-0047-42 | Clone: SK3 |
| mouse anti-human CD196 (CCR6) BV421 | BD biosciences | 562515 | Clone: 11A9 |
| anti-human RORgt AF647 | BD biosciences | 563620 | Clone: Q21-559 |
| anti-human CD45 eFluor450 | ebioscience | 48-9459-42 | Clone: 2D1 |
| Foxp3/Trascription Factor Staining Buffer Set | ebioscience | 00-5523-00 | For intracellular transcription factor flow cytometry staining |
| Hu FcR Binding Inhibitor Purified | ebioscience | 14-9161-71 | |
| ImmunoCult-XF T Cell Expansion Medium | Stemcell Technologies | 10981 | "Serum-Free Base Media" |
| MACS CD28 pure functional grade, human | Miltenyi Biotec | 130-093-375 | Clone: 15E8 |
| anti-human CD3 Purified | UCSF monoclonal antibody core | N/A | Clone: OKT-3 |
| LEAF purified anti-human IL-4 | Biolegend | 500815 | Clone: MP4-25D2 |
| anti-human IFNg, functional grade purified | ebioscience | 16-7318-85 | Clone: NIB42 |
| Recombinant human IL-23 | Peprotech | 200-23 | |
| Recombinant human IL-1β | Peprotech | 200-01B | |
| Recombinant human TGF-β1 | Peprotech | 100-21C | |
| Recombinant human IL-6 | Peprotech | 200-06 | |
| siGENOME Control Pool, Non-targeting #2 | Dharmacon | D-001206-14-05 | |
| siGENOME SMARTpool Human RORC | Dharmacon | M-003442-00 | |
| siGENOME SMARTpool Human PTPRC | Dharmacon | M-008067-01 | |
| Dynabeads Untouched Human CD4 T Cells Kit | ThermoFisher Scientific | 11346D | Human CD4+ T cell Isolation Kit |
| Neon Tranfection System | ThermoFisher Scientific | MPK5000 | Next generation electroporation instrument |
| Neon Tranfection System 10 μl Kit | ThermoFisher Scientific | MPK1096 | |
| Resuspension Buffer T | ThermoFisher Scientific | Provided in kit (MPK1096) | "Transfection Resuspension Buffer" |
| Lymphoprep | Stemcell Technologies | #07801 | Density Gradient Medium |
| costar 6-well tissue culture treated plates | Corning | 3516 | flat bottom plates |
| costar 48-well tissue culture treated plates | Corning | 3548 | flat bottom plates |
| BD Pharm Lyse lysing buffer, 10 X | BD biosciences | 555899 | Must make 1X solution with distilled water prior to use |
References
- Zhu, J., Yamane, H., Paul, W. E. Differentiation of Effector CD4 T Cell Populations*. Ann Rev Immunol. 28 (1), 445-489 (2010).
- Korn, T., Bettelli, E., Oukka, M., Kuchroo, V. K. IL-17 and Th17 Cells. Ann Rev Immunol. 27 (1), 485-517 (2009).
- Gaffen, S. L., Jain, R., Garg, A. V., Cua, D. J. The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nature. 14 (9), 585-600 (2014).
- Romagnani, S., Maggi, E., Liotta, F., Cosmi, L., Annunziato, F. Properties and origin of human Th17 cells. Mol Immunol. 47 (1), 3-7 (2009).
- Acosta-Rodriguez, E. V., et al. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nature Immunol. 8 (6), 639-646 (2007).
- Annunziato, F., et al. Phenotypic and functional features of human Th17 cells. J Exp Med. 204 (8), 1849-1861 (2007).
- Du, C., et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nature Immunol. 10 (12), 1252-1259 (2009).
- Escobar, T. M., et al. miR-155 Activates Cytokine Gene Expression in Th17 Cells by Regulating the DNA-Binding Protein Jarid2 to Relieve Polycomb-Mediated Repression. Immunity. 40 (6), 865-879 (2014).
- Wang, H., et al. Negative regulation of Hif1a expression and TH17 differentiation by the hypoxia-regulated microRNA miR-210. Nature Immunol. 15 (4), 393-401 (2014).
- Kim, T. K., Eberwine, J. H. Mammalian cell transfection: the present and the future. Anal bioanal chem. 397 (8), 3173-3178 (2010).
- Steiner, D. F., et al. MicroRNA-29 Regulates T-Box Transcription Factors and Interferon-γ Production in Helper T Cells. Immunity. 35 (2), 169-181 (2011).
- Simpson, L. J., et al. A microRNA upregulated in asthma airway T cells promotes TH2 cytokine production. Nature Immunol. 15 (12), 1162-1170 (2014).
- Pua, H. H., et al. MicroRNAs 24 and 27 Suppress Allergic Inflammation and Target a Network of Regulators of T Helper 2 Cell-Associated Cytokine Production. Immunity. 44 (4), 821-832 (2016).
- Flaherty, S., Reynolds, J. M. Mouse Naïve CD4+ T Cell Isolation and In vitro Differentiation into T Cell Subsets. J Vis Exp. (98), (2015).
- Schumann, K., et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc Natl Acad Sci USA. 112 (33), 10437-10442 (2015).

