Retroviral Scanning: Mapping MLV Integration Sites to Define Cell-specific Regulatory Regions
Summary
Here, we describe a protocol for genome-wide mapping of the integration sites of Moloney murine leukemia virus-based retroviral vectors in human cells.
Abstract
Moloney murine leukemia (MLV) virus-based retroviral vectors integrate predominantly in acetylated enhancers and promoters. For this reason, mLV integration sites can be used as functional markers of active regulatory elements. Here, we present a retroviral scanning tool, which allows the genome-wide identification of cell-specific enhancers and promoters. Briefly, the target cell population is transduced with an mLV-derived vector and genomic DNA is digested with a frequently cutting restriction enzyme. After ligation of genomic fragments with a compatible DNA linker, linker-mediated polymerase chain reaction (LM-PCR) allows the amplification of the virus-host genome junctions. Massive sequencing of the amplicons is used to define the mLV integration profile genome-wide. Finally, clusters of recurrent integrations are defined to identify cell-specific regulatory regions, responsible for the activation of cell-type specific transcriptional programs.
The retroviral scanning tool allows the genome-wide identification of cell-specific promoters and enhancers in prospectively isolated target cell populations. Notably, retroviral scanning represents an instrumental technique for the retrospective identification of rare populations (e.g. somatic stem cells) that lack robust markers for prospective isolation.
Introduction
Cell identity is determined by the expression of specific sets of genes. The role of cis-regulatory elements, such as promoters and enhancers, is crucial for the activation of cell-type specific transcriptional programs. These regulatory regions are characterized by specific chromatin features, such as peculiar histone modifications, transcription factors and co-factors binding, and chromatin accessibility, which have been widely used for their genome-wide identification in several cell types1,2,3. In particular, the genome-wide profile of acetylation of histone H3 lysine 27 (H3K27ac) is commonly used to define active promoters, enhancers and super-enhancers4,5,6.
Moloney murine leukemia virus (MLV) is a gamma-retrovirus that is widely used for gene transfer in mammalian cells. After infecting a target cell, the retroviral RNA genome is retro-transcribed in a double-stranded DNA molecule that binds viral and cellular proteins to assemble the pre-integration complex (PIC). The PIC enters the nucleus and binds the host cell chromatin. Here, the viral integrase, a key PIC component, mediates the integration of the proviral DNA into the host cell genome. mLV integration in the genomic DNA is not random, but occurs in active cis-regulatory elements, such as promoters and enhancers, in a cell-specific fashion7,8,9,10. This peculiar integration profile is mediated by a direct interaction between the mLV integrase and the cellular bromodomain and extraterminal domain (BET) proteins11,12,13. BET proteins (BRD2, BRD3, and BRD4) act as a bridge between host chromatin and mLV PIC: through their bromodomains they recognize highly acetylated cis-regulatory regions, while the extraterminal domain interacts with the mLV integrase11,12,13.
Here, we describe the retroviral scanning, a novel tool to map active cis-regulatory regions based on the integration properties of mLV. Briefly, cells are transduced with mLV-derived retroviral vector expressing the enhanced green fluorescent protein (eGFP) reporter gene. After genomic DNA extraction, the junctions between the 3' long terminal repeat (LTR) of the mLV vector and the genomic DNA are amplified by linker-mediated PCR (LM-PCR) and massively sequenced. mLV integration sites are mapped to the human genome and genomic regions highly targeted by mLV are defined as clusters of mLV integration sites.
Retroviral scanning was used to define cell-specific active regulatory elements in several human primary cells14,15. mLV clusters co-mapped with epigenetically defined promoters and enhancers, most of which harbored active histone marks, such as H3K27ac, and were cell-specific. Retroviral scanning allows the genome-wide identification of DNA regulatory elements in prospectively purified cell populations7,14, as well as in retrospectively defined cell populations, such as keratinocyte stem cells, that lack effective markers for prospective isolation15.
Protocol
1. MLV Transduction of Human Cells
- Isolate target cells and transduce them with an mLV-derived retroviral vector harboring the eGFP reporter gene and pseudotyped with Vesicular Stomatitis Virus G (VSV-G) or the amphotropic envelope glycoprotein16.
- Keep mock-transduced cells as a negative control for the following analyses. Since mLV-based retroviral vectors can transduce efficiently dividing cells, culture the target cell population in conditions that stimulate cell division. Transduction conditions need to be specifically optimized for each cell type under study. Cell growth and transduction conditions for human hematopoietic progenitors, T cells and epidermal cells are described in References 7, 14, 15, and 17.
- 48 h after transduction, resuspend 100,000 cells in 300 μL of phosphate-buffered saline (PBS) containing 2% fetal bovine serum and measure eGFP expression by flow cytometric analysis (488-nm excitation laser). Use mock-transduced cells as negative control. For optimal integration site retrieval, purify GFP+ cells by Fluorescence Activated Cell Sorting (FACS).
- Collect 0.5 to 5 million cells for genomic DNA preparation. A longer culture period (>7 days) is preferred to dilute the unintegrated provirus and necessary when analyzing the long-term progeny of bona fide stem cells (see Discussion Section and reference15). The cell pellets can be snap-frozen and stored at -80 °C until use.
2. Amplification of mLV integration sites by linker-mediated-PCR (LM-PCR)
- Preparation of genomic DNA (gDNA)
- Extract gDNA using a column-based DNA extraction kit and follow the protocol for cultured cells, according to the manufacturer's instructions.
- Restriction enzyme digestion
- Set up 4 restriction enzyme digestions per sample in 1.5 mL tubes. Digest 0.1 to 1 μg gDNA in each tube by adding 1 µL of Tru9I (10U) and 1 µL of Buffer M in a final volume of 10 µL. Incubate the reactions at 65 °C for 6 h or overnight.
- Add to each reaction 1 µL of PstI (10U), 1 µL of Buffer H and 8 µL of water. Incubate the reactions at 37 °C for 6 h or overnight. Samples can be stored at -20 °C until use.
- Linker ligation
- Prepare a 100 μM Tru9I linker stock solution in a 1.5 mL tube by mixing the linker plus strand and linker minus strand oligonucleotides at a 100 μM concentration. Put the tube in a water set at 100 °C and let it cool down at room temperature. Tru9I linker stock solution can be stored at -20 °C until use.
- Set up 8 linker ligation reactions in 1.5 mL tubes. For each reaction, add the following components to 10 µL of restriction enzyme digestion: 1.4 µL of 10X T4 DNA Ligase Reaction buffer, 1 µL of 10 μM linker Tru9I, 1 µL (2,000U) of T4 DNA ligase and 0.6 µL of water. Incubate at 16 °C for 3 to 6 h. Samples can be stored at -20 °C until use.
- First PCR
- Set up 48 PCR reactions (6 reactions from each ligation tube) in 0.2 mL tubes. For each PCR, add the following reagents to 2 µL of ligation reaction: 5 µL of 10X PCR buffer, 2 µL of 50 mM Magnesium Sulfate, 1 µL of 10 mM deoxynucleotide (dNTP) Mix, 1 µL of 10 μM linker primer, 1 µL of 10 μM mLV-3' LTR primer, 0.3 µL (1.5U) of Taq DNA Polymerase and 37.7 µL of water. Primer sequences are provided in Table 1.
- Perform PCR reaction in a thermal cycler with heated lid, as follows: 95 °C for 2 min; 25 cycles of 95 °C for 15 s, 55 °C for 30 s, 72 °C for 1 min; 72 °C for 5 min; hold at 4 °C. Samples can be stored at -20 °C until use.
- Second PCR
- Set up 48 PCR reactions (1 from each "first PCR" tube) in 0.2 mL tubes. For each PCR, add the following components to 2 µL of the first PCR reaction: 5 µL of 10X PCR buffer, 2 µL of 50 mM Magnesium Sulfate, 1 µL of 10 mM dNTP Mix, 1 µL of 10 μM linker nested primer, 1 µL of 10 μM mLV-3' LTR nested primer, 0.3 µL of Taq DNA Polymerase (1.5 U) and 37.7 µL of water. Use nested primers designed for the specific massive sequencing strategy chosen: (i) linker nested primer and mLV-3' LTR nested primer compatible with Illumina platform; (ii) linker nested primer and mLV-3' LTR nested primer compatible with Roche platform. Primer sequences are listed in Table 1.
- Perform PCR reaction in a thermal cycler with heated lid, as follows: 95 °C for 2 min; 25 cycles of 95 °C for 15 s, 58 °C for 30 s, 72 °C for 1 min; 72 °C for 5 min; hold at 4 °C. Samples can be stored at -20 °C until use.
- Pool the 48 nested PCR reactions in a 15 mL tube (final volume of ~2.4 mL). Samples can be stored at -20 °C until use.
- Determine the presence and the size of the LM-PCR products by agarose gel electrophoresis.
- Add 4 µL of Loading Buffer to 20 µL of LM-PCR products and load the sample onto a 1% agarose gel, together with a 100 bp DNA ladder. Run the gel at 5 V/cm for 30 to 60 min and visualize the PCR products by ethidium bromide staining.
- Run an LM-PCR reaction from the mock-transduced sample as negative control.
- Precipitate the amplicons by adding 0.1 volumes of sodium acetate solution (3 M; pH 5.2) and 2.5 volumes of 100% ethanol. Mix and freeze at -80 °C for 20 min.
- Spin at full speed in a standard microcentrifuge at 4 °C for 20 min. Discard the supernatant and wash the pellet with 70% ethanol. Spin at full speed in a standard microcentrifuge at 4 °C for 5 min.
- Discard the supernatant and air dry the pellet. Add 200 µL of PCR-grade water and resuspend the DNA. Samples can be stored at -20 °C until use.
- Add 20 μL of Loading Buffer to 100 µL of the LM-PCR products and load the sample onto a 1% agarose gel, together with a 100 bp DNA ladder.
- Run the gel at 5 V/cm for 30 to 60 min and cut the portion of the gel containing 150 to 500 bp-long amplicons.
- Purify the LM-PCR products with a column-based gel extraction kit and measure the concentration using an UV spectrophotometer.
- Use 1 µL of library to evaluate the length of the LM-PCR products using a bioanalyzer instrument, according to manufacturer's instructions.
- Shotgun cloning of amplicons
- Use 20 ng of the purified LM-PCR products and clone them in the pCR2.1-TOPO vector, according to the manufacturer's instructions.
- Perform sequencing reactions using the M13 Universal primer, followed by genomic mapping of the resulting sequences, to reveal the presence of the viral-genome junctions (including the mLV-3' LTR nested primer) in >50% of the clones to identify samples suitable for massive sequencing. Example of a viral-genome junction:

MLV-3' LTR nested primer 454 and linker nested primer 454 (Table 1) are indicated in bold and the 3' end of the viral LTR in italic. The human genomic sequence is highlighted in red text (chr10:6439408-6439509, hg19).
3. Massive Sequencing of mLV Integration Sites
NOTE: LM-PCR products can be sequenced using commercial platforms (choosing the proper nested primer pair in the second PCR reaction, see subsection 2.5.1). For sequencing by Roche GS-FLX pyrosequencing platform, refer to previous papers7,14,15. In this section, a newly-optimized protocol for Illumina sequencing platform is described.
- Library preparation
- Set up 1 indexing PCR reaction per sample in 0.2 mL tubes. For each PCR, add the following reagents to 5 µL (150-170 ng) of purified LM-PCR product: 5 µL of Index Primer 1, 5 µL of Index Primer 2, 25 µL of 2X Master Mix and 10 µL of PCR-grade water. Use a different index combination for each sample.
- Perform PCR reactions in a thermal cycler with heated lid, as follows: 95 °C for 3 min; 8 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s; 72 °C for 5 min; hold at 4 °C.
- Purify the PCR products using a solid-phase reversible immobilization (SPRI) bead isolation protocol: in new 1.5 mL tubes, add 56 µL of beads to each sample and proceed following manufacturer's instructions. Elute in 25 µL of Tris-HCl 10 mM. Libraries can be stored at -20 °C until use.
- Library check
- Use 1 µL of sample to assess library size using a bioanalyzer instrument.
- Use 1 µL of sample to quantify library molarity using a fluorescence-based Real time PCR assay, according to manufacturer's instructions.
- Library dilution and sequencing
- Dilute libraries to 10 nM using Tris-HCl 10 mM. For pooling libraries, transfer 5 µL of each diluted library to a new 1.5 mL tube and then dilute the pool to 4 nM in Tris-HCl 10 mM.
- Mix 5 µL of diluted pool with 5 µL of 0.2 N NaOH in a new 1.5 mL tube, vortex briefly, spin-down and incubate for 5 min at room temperature to denature the libraries.
- Put tubes on ice and add 990 µL of pre-chilled hybridization buffer (HT1). Aliquot 300 µL of denatured pool in a new 1.5 mL tube and add 300 µL of pre-chilled HT1 to obtain a 10 pM final library pool.
- In parallel, mix 2 µL of PhiX control library (10 nM) with 3 µL of Tris-HCl 10 mM in a 1.5 mL tube. Add 5 µL of NaOH 0.2 N, vortex briefly, spin-down and incubate for 5 min at room temperature to denature the diluted PhiX. Put tube on ice and add 990 µL of pre-chilled HT1.
- Aliquot 300 µL of denatured PhiX in a new 1.5 mL tube and add 300 µL of pre-chilled HT1 to obtain a 10 pM final PhiX.
- In a new 1.5 mL tube, mix 510 µL of denatured library pool with 90 µL of denatured PhiX library, thus obtaining a final 10 pM pool with 15% of PhiX control.
- Pipette these 600 µL of sample volume into the Load Sample reservoir of the thawed sequencing reagent cartridge and proceed immediately to perform a single-read 150-cycle run.
NOTE: The critical reagents and primer sequences required for this protocol are listed in Table 1.
Representative Results
Workflow of the retroviral scanning procedure
The workflow of retroviral scanning procedure is schematized in Figure 1. The target cell population is purified and transduced with a mLV-derived retroviral vector expressing an eGFP reporter gene. The transgene is flanked by the two identical long terminal repeats (5' and 3' LTR), ensuring synthesis, reverse transcription and integration of the viral genome into host DNA. The transduction efficiency is assessed by FACS analysis of eGFP expression. The cell population containing a high proportion (>30%) of mLV-transduced cells is amplified and subsequently lysed to extract genomic DNA containing the integrated mLV viral cassettes. Genomic DNA is digested and ligated with a compatible linker and the junctions between the viral 3' LTRs and the host genome are amplified by LM-PCR. Virus-host genome junctions are then massively sequenced using Roche or Illumina platforms. Finally, mLV integration sites are mapped to the human genome to define genomic clusters of recurrent insertion sites.
Amplification of mLV integration sites by LM-PCR
The LM-PCR is schematized in Figure 2. Genomic DNA is extracted from mLV-transduced cells and digested with the Tru9I restriction enzyme, which cuts frequently the human genome, generating fragments with a median length of 70 bp. A second restriction enzyme (PstI) is used to prevent amplification of integrated and non-integrated internal 5' LTR fragments.A Tru9I double-stranded linker is then ligated to the genomic fragments and LM-PCR is performed with primers specific for the linker and the 3' LTR to amplify the virus-host genome junctions. Nested PCR can be performed using primers compatible with Roche or Illumina sequencing platforms.
Analysis of virus-host genome junction amplicons
In the experiment represented in Figure 3, we purified CD34–CD13+ myeloid progenitor/precursors (MPP) and transduced them with an mLV-derived retroviral expressing the eGFP reporter gene. More than 60% of MPP cells expressed eGFP 48 h after transduction (data not shown). 15 days after transduction, we collected the cells, extracted gDNA and amplified the virus-host genome junctions, as described above. An aliquot of the pooled LM-PCR products was loaded on an 1% agarose gel to verify the presence and the size of the amplicons. We successfully visualized a DNA smear corresponding to the LM-PCR products of different sizes, ranging from 150 to 500 bp (Figure 3A). Amplicons were then concentrated by DNA precipitation and loaded on a 1% agarose gel. LM-PCR products were gel-purified and run on a bioanalyzer system, confirming the expected amplicon sizes (between 150 and 500 bp; Figure 3B).
Mapping of mLV integrations into active and cell-specific regulatory regions
In the experiment reported in Figure 4, hematopoietic stem/progenitor cells (HSPC), erythroid progenitor/precursors (EPP) and myeloid progenitor/precursors (MPP) were transduced with an mLV-derived retroviral vector. These results were obtained processing and sequencing samples derived from different cell types separately, to avoid the potential contamination/collisions18,19. Raw sequence reads generated by massive sequencing were processed by an automated bioinformatics pipeline to eliminate viral and linker sequences. Then, unique sequences of at least 20 bp were mapped on the human genome using Blat17. Raw alignments were filtered requiring the match to start within the first 3 nucleotides, univocal matches and a minimum of 95% identity. Clusters of recurrent mLV integrations were defined by a statistical comparison with a dataset of random genomic sequences, generated randomly extracting genomic positions from the human genome with a Tru91 restriction motif at a distance compatible with the sequencing platform. Control sequences were then processed through the same mapping and filtering pipeline used for integration sequences, to generate a random set of unique sites. To define mLV clusters, we applied the DBSCAN clustering algorithm19, comparing the distribution of consecutive mLV integrations with that of an equal number of random sites to identify regions of highly clustered integrations, which define cell-specific regulatory elements7,14,15. In order to avoid the generation of false clusters, multiple extractions from the random control dataset were performed. We mapped by LM-PCR and pyrosequencing 32,574, 27,546 and 36,358 mLV integration sites in HSPC, EPP and MPP, respectively. Clusters of recurrent mLV integrations co-mapped with acetylated enhancers and promoters (Figure 4A). Most of the mLV-targeted regulatory regions were cell-specific, such as: (i) the promoter of HSPC-specific SPINK2 gene (Figure 4B); (ii) the Locus Control Region containing potent enhancers of the erythroid-specific β-like globin genes (Figure 4C); (iii) enhancers located upstream of the MPP-specific LYZ gene (Figure 4D). Finally, we used luciferase assays to validate a subset of putative cell-specific mLV-targeted enhancers in EPP and MPP (Figure 4E).
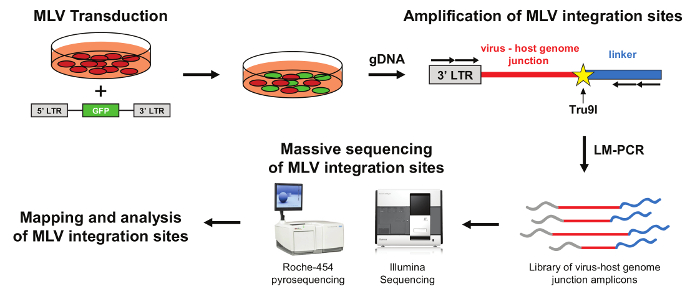
Figure 1: A general scheme of the retroviral integration site mapping procedure. Target cells are transduced with a mLV-based retroviral vector containing a eGFP cassette. Genomic DNA obtained from transduced cells is digested with Tru9I and ligated with a compatible Tru9I double-strand linker. mLV integration sites were amplified by nested LM-PCR and the library of virus-host genome junctions can be massively sequenced using Illumina or Roche platforms. The resulting reads were mapped to the human genome to define clusters of recurrent mLV integrations. Please click here to view a larger version of this figure.
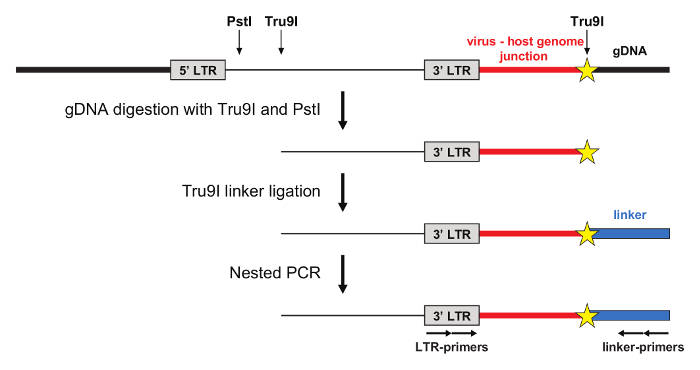
Figure 2: Amplification of virus-host genome junctions by LM-PCR. Genomic DNA (gDNA) containing the integrated mLV provirus is digested with Tru9I and PstI restriction enzymes, and ligated with a compatible Tru9I linker. Nested PCR is performed using primers specific for the LTR and the linker. Tru9I and PstI restriction sites in the viral and in the human genome are indicated. Please click here to view a larger version of this figure.
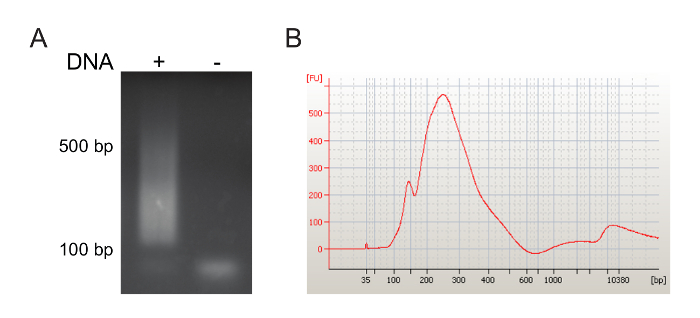
Figure 3: Analysis of LM-PCR amplicons. (A) LM-PCR products (lane +) were run on a 1% agarose gel and visualized by ethidium bromide staining. A no template sample served as negative control sample, where only PCR primers were visualized (lane -). (B) After gel purification, LM-PCR product size was checked by microcapillary electrophoresis. Sample size (bp) and fluorescence intensity (FU) are shown on x and y axes of the electropherogram, respectively. Please click here to view a larger version of this figure.
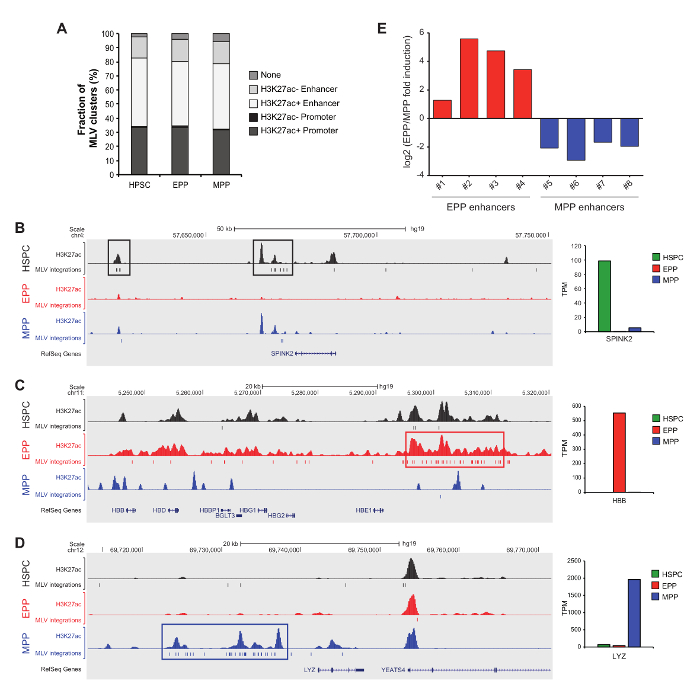
Figure 4: Mapping of mLV integration into epigenetically-defined regulatory regions. (A) We defined 3,498, 2,989 and 4,103 clusters of recurrent mLV integration sites in HSPC, EPP and MPP, respectively. In each cell population, >95% of mLV clusters overlapped with epigenetically defined enhancers and promoters. (B, C, and D) Cell-specific mLV-targeted regions were highly acetylated and associated with cell-specific expression of the targeted gene (SPINK2, HBB and LYZ, almost exclusively expressed in HSPC, EPP and MPP, respectively, as determined by Cap analysis of Gene Expression). mLV single integrations are depicted with small bars. TPM indicates Tag Per Million. (E) Putative cell-specific mLV-targeted enhancers in EPP and MPP. Figure 4 is adapted from reference14. We received the permission to re-use this figure under the creative commons license. Please click here to view a larger version of this figure.
Discussion
Here, we described a protocol for genome-wide mapping of the integration sites of mLV, a retrovirus that targets chromatin regions, epigenetically marked as active promoters and enhancers. Critical steps and/or limitations of the protocol include: (i) mLV transduction of the target cell population; (ii) amplification of virus-host junctions by LM-PCR; (iii) retrieval of a high fraction of integration sites. mLV-based retroviral vectors efficiently transduce dividing cells. The low efficiency of transduction of non-dividing cells (e.g. post-mitotic neuronal cells) is a potential limitation of this technique. However, it can be overcome through cell sorting of the transduced population based on the expression of the reporter gene (e.g. eGFP). The generation of relatively short amplicons by LM-PCR (150 to 500 bp) is mandatory to generate a library of amplicons compatible with the currently used massive sequencing strategies and to allow a comprehensive genome-wide analysis of mLV integration sites. As an example, amplification of >500 bp long LM-PCR products can result from either partial genomic DNA digestion or intra-ligation of Tru9I-digested genomic fragments, as evaluated by shotgun cloning of LM-PCR amplicons (data not shown). In the first case, the issue can be resolved by further optimizing genomic DNA digestion conditions, whereas, in the latter case, the successful massive sequencing of mLV integration sites can be accomplished through a gel purification of amplicons between 150 and 500 bp. Finally, the use of restriction enzymes to cut the genomic DNA can lead to the preferential amplification and detection of integration sites that lie close to a restriction site. The percentage of integration sites that the Tru9I restriction enzyme can retrieve is estimated to be ~50%. Thus, this technique can be further optimized to improve integration site retrieval by using multiple restriction enzymes or performing random DNA shearing by sonication20,21.
Recently, we have optimized the sequencing of viral integration sites using the widely-used Illumina platform, as detailed in this paper. This sequencing approach allows the generation of a higher number of reads per run compared to the Roche platform, greatly increasing the number of integration sites retrieved from a single experiment, thus globally reducing time and costs.
Genome-wide identification of cell-specific regulatory regions requires the mapping of one to three histone modifications by ChIP-seq6 (mono- and tri-methylation of histone H3 lysine 4 to identify enhancers and promoters, and H3K27ac to distinguish between active and inactive regulatory elements)4,5,6 and a systematic comparison of different cell types to define cis-acting elements active exclusively in a determined cell population. We mapped mLV integration sites in prospectively isolated target cell populations, such as multipotent hematopoietic progenitors and their committed erythroid and myeloid progeny14, embryonic stem cells, neuroepithelial-like stem cells and differentiated keratinocytes15. Retroviral scanning allowed the genome-wide definition of cell-specific regulatory elements in each cell population analyzed, making the comparative genome-wide studies not strictly necessary. Importantly, this tool can be used for the identification of active regulatory elements in rare cell populations (e.g. somatic stem cells), that lack robust markers for prospective isolation and cannot be analyzed by ChIP-seq-based analysis of histone modifications15. In these cell populations mLV integration is a permanent genetic marker of active regulatory regions, allowing their retrospective identification in the more abundant cell progeny in vitro and in vivo. As an example, we successfully used mLV integration clusters as surrogate markers of promoters and enhancers in a retrospectively identified keratinocyte stem cell (KSC) population. We transduced an early-passage, foreskin-derived keratinocyte culture containing KSCs with an mLV vector, then we passaged these cells for >35 cell doublings to enrich in the progeny of KSCs, thus defined by their ability to maintain the culture for this number of passages. In this case, mLV integrations permanently marked the regulatory regions active in the original transduced KSC population15. Future studies will aim at identifying in a genome-wide manner cell-specific regulatory elements in a larger number of rare human stem cell populations.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by grants from the European Research Council (ERC-2010-AdG, GT-SKIN), the Italian Ministry of Education, Universities and Research (FIRB-Futuro in Ricerca 2010-RBFR10OS4G, FIRB-Futuro in Ricerca 2012-RBFR126B8I_003, EPIGEN Epigenomics Flagship Project), the Italian Ministry of Health (Young researchers Call 2011 GR-2011-02352026) and the Imagine Institute Foundation (Paris, France).
Materials
| PBS, pH 7.4 | ThermoScientific | 10010031 | or equivalent |
| Fetal Bovine Serum | ThermoScientific | 16000044 | or equivalent |
| 0.2 ml tubes | general lab supplier | ||
| 1.5 ml tubes | general lab supplier | ||
| QIAGEN QIAmp DNA mini Kit | QIAGEN | 51306 | or equivalent |
| T4 DNA ligase | New England BioLabs | M0202T | |
| T4 DNA Ligase Reaction buffer | New England BioLabs | M0202T | |
| Linker Plus Strand oligonucleotide | general lab supplier | 5’-PO4-TAGTCCCTTAAGCGGAG-3’ (Purification grade: SDS-PAGE) | |
| Linker Minus Strand oligonucleotide | general lab supplier | 5’-GTAATACGACTCACTATAGGGCTCCGCTTAAGGGAC-3’ (Purification grade: SDS-PAGE) | |
| Tru9I | Roche-Sigma-Aldrich | 11464825001 | |
| SuRE/Cut Buffer M | Roche-Sigma-Aldrich | 11417983001 | |
| PstI | Roche-Sigma-Aldrich | 10798991001 | |
| SuRE/Cut Buffer H | Roche-Sigma-Aldrich | 11417991001 | |
| Platinum Taq DNA Polimerase High Fidelity | Invitrogen | 11304011 | |
| 10mM dNTP Mix | Invitrogen | 18427013 | or equivalent |
| PCR grade water | general lab supplier | ||
| 96-well thermal cycler (with heated lid) | general lab supplier | ||
| linker primer | general lab supplier | 5’-GTAATACGACTCACTATAGGGC-3’ (Purification grade: PCR grade) | |
| MLV-3’ LTR primer | general lab supplier | 5’-GACTTGTGGTCTCGCTGTTCCTTGG-3’ (Purification grade: PCR grade) | |
| linker nested primer 454 | general lab supplier | 5’-GCCTTGCCAGCCCGCTCAG[AGGGCTCCGCTTAAGGGAC](Purification grade: SDS-PAGE) | |
| MLV-3’ LTR nested primer 454 | general lab supplier | 5’-GCCTCCCTCGCGCCATCAGTAGC[GGTCTCCTCTGAGTGATTGACTACC](Purification grade: SDS-PAGE) | |
| linker nested primer Illumina | general lab supplier | 5'-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-[AGGGCTCCGCTTAAGGGAC](Purification grade: SDS-PAGE) | |
| MLV-3’ LTR nested primer Illumina | general lab supplier | 5'-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-[GGTCTCCTCTGAGTGATTGACTACC](Purification grade: SDS-PAGE) | |
| Sodium Acetate Solution (3M) pH 5.2 | general lab supplier | ||
| Ethanol (absolute) for molecular biology | Sigma-Aldrich | E7023 | or equivalent |
| Topo TA Cloning kit (with pCR2.1-TOPO vector) | Invitrogen | K4500-01 | |
| QIAquick Gel Extraction kit | QIAGEN | 28704 | |
| Agarose | Sigma-Aldrich | A9539 | or equivalent |
| Ethidium bromide | Sigma-Aldrich | E1510 | or equivalent |
| 100 bp DNA ladder | Invitrogen | 15628019 | or equivalent |
| 6X Loading Buffer | ThermoScientific | R0611 | or equivalent |
| NanoDrop 2000 UV-Vis Spectrophotometer | ThermoScientific | ND-2000 | |
| Nextera XT Index kit | Illumina | FC-131-1001 or FC-131-1002 | |
| 2x KAPA HiFi Hot Start Ready Mix | KAPA Biosystems | KK2601 | |
| Dynal magnetic stand for 2 ml tubes | Invitrogen | 12321D | or equivalent |
| Agencourt AMPure XP 60 ml kit | Beckman Coulter Genomics | A63881 | |
| Tris-HCl 10 mM, pH 8.5 | general lab supplier | ||
| Agilent 2200 TapeStation system | Agilent Technologies | G2964AA | or equivalent |
| D1000 ScreenTape | Agilent Technologies | 5067-5582 | or equivalent |
| D1000 Reagents | Agilent Technologies | 5067-5583 | or equivalent |
| KAPA Library Quantification Kit for Illumina platforms (ABI Prism) | KAPA Biosystems | KK4835 | |
| ABI Prism 7900HT Fast Real-Time PCR System | Applied Biosystems | 4329003 | |
| NaOH 1.0 N, molecular biology-grade | general lab supplier | ||
| HT1 (Hybridization Buffer) | Illumina | Provided in the MiSeq Reagent Kit | |
| MiSeq Reagent Kit v3 (150 cycles) | Illumina | MS-102-3001 | |
| MiSeq System | Illumina | SY-410-1003 | |
| PhiX Control v3 | Illumina | FC-110-3001 |
References
- Ernst, J., et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 473 (7345), 43-49 (2011).
- Shlyueva, D., Stampfel, G., Stark, A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 15 (4), 272-286 (2014).
- Roadmap Epigenomics Consortium. Integrative analysis of 111 reference human epigenomes. Nature. 518 (7539), 317-330 (2015).
- Heintzman, N. D., et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 459 (7243), 108-112 (2009).
- Creyghton, M. P., et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 107 (50), 21931-21936 (2010).
- Hnisz, D., et al. Super-enhancers in the control of cell identity and disease. Cell. 155 (4), 934-947 (2013).
- Cattoglio, C., et al. High-definition mapping of retroviral integration sites identifies active regulatory elements in human multipotent hematopoietic progenitors. Blood. 116 (25), 5507-5517 (2010).
- Biasco, L., et al. Integration profile of retroviral vector in gene therapy treated patients is cell-specific according to gene expression and chromatin conformation of target cell. EMBO Mol Med. 3 (2), 89-101 (2011).
- De Ravin, S. S., et al. Enhancers are major targets for murine leukemia virus vector integration. J Virol. 88 (8), 4504-4513 (2014).
- LaFave, M. C., et al. mLV integration site selection is driven by strong enhancers and active promoters. Nucleic Acids Res. 42 (7), 4257-4269 (2014).
- Gupta, S. S., et al. Bromo- and extraterminal domain chromatin regulators serve as cofactors for murine leukemia virus integration. J Virol. 87 (23), 12721-12736 (2013).
- Sharma, A., et al. BET proteins promote efficient murine leukemia virus integration at transcription start sites. Proc Natl Acad Sci U S A. 110 (29), 12036-12041 (2013).
- De Rijck, J., et al. The BET family of proteins targets moloney murine leukemia virus integration near transcription start sites. Cell reports. 5 (4), 886-894 (2013).
- Romano, O., et al. Transcriptional, epigenetic and retroviral signatures identify regulatory regions involved in hematopoietic lineage commitment. Sci Rep. 6, 24724 (2016).
- Cavazza, A., et al. Dynamic Transcriptional and Epigenetic Regulation of Human Epidermal Keratinocyte Differentiation. Stem Cell Reports. 6 (4), 618-632 (2016).
- Miller, A. D., Law, M. F., Verma, I. M. Generation of helper-free amphotropic retroviruses that transduce a dominant-acting, methotrexate-resistant dihydrofolate reductase gene. Mol Cell Biol. 5 (3), 431-437 (1985).
- Cattoglio, C., et al. High-definition mapping of retroviral integration sites defines the fate of allogeneic T cells after donor lymphocyte infusion. PLoS One. 5 (12), e15688 (2010).
- Aiuti, A., et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 341 (6148), 1233151 (2013).
- Biffi, A., et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 341 (6148), 1233158 (2013).
- Gabriel, R., et al. Comprehensive genomic access to vector integration in clinical gene therapy. Nat Med. 15 (12), 1431-1436 (2009).
- Gillet, N. A., et al. The host genomic environment of the provirus determines the abundance of HTLV-1-infected T-cell clones. Blood. 117 (11), 3113-3122 (2011).

