High-resolution Confocal Imaging of the Blood-brain Barrier: Imaging, 3D Reconstruction, and Quantification of Transcytosis
Summary
Here we present a microscopy-based protocol for high-resolution imaging and a three-dimensional reconstruction of the mouse neurovascular unit and blood-brain barrier using brain free-floating sections. This method allows for the visualization, analysis, and quantification of intracellular organelles at the BBB.
Abstract
The blood-brain barrier (BBB) is a dynamic multicellular interface that regulates the transport of molecules between the circulation and the brain. Transcytosis across the BBB regulates the delivery of hormones, metabolites, and therapeutic antibodies to the brain parenchyma. Here, we present a protocol that combines immunofluorescence of free-floating sections with laser scanning confocal microscopy and image analysis to visualize subcellular organelles within endothelial cells at the BBB. Combining this data-set with 3D image analysis software allows for the semi-automated segmentation and quantification of capillary volume and surface area, as well as the number and intensity of intracellular organelles at the BBB. The detection of mouse endogenous immunoglobulin (IgG) within intracellular vesicles and their quantification at the BBB is used to illustrate the method. This protocol can potentially be applied to the investigation of the mechanisms controlling BBB transcytosis of different molecules in vivo.
Introduction
The blood-brain barrier (BBB) is a continuous cellular barrier formed by astrocytes, pericytes, neurons, and endothelial cells that separates the central nervous system (CNS) from the blood circulation1. The regulation of transport across the BBB plays a crucial role in maintaining brain homeostasis and is mediated by specialized properties of brain endothelial cells (BECs). The presence of tight intercellular junctions between BECs and a low basal rate of transcytosis limit the paracellular and transcellular transport of blood-borne molecules, respectively2. Recently, the transcytosis pathway in BECs has been harnessed to enhance delivery of therapeutic large molecules to the brain3,4. However, the mechanisms of transcytosis across the BBB have not yet been fully characterized5,6.
Extensive work has been done in vitro to decipher the cellular and molecular mechanisms regulating intracellular transport across BECs7,8,9,10,11, but such systems fail to recapitulate the complex architecture and physiology of the neurovascular unit (NVU). On the other hand, studies in vivo12,13 provide detailed quantitative information on transport rates across the BBB but do not provide insights into the intracellular mechanisms of transport. Therefore, investigating the cellular and intracellular components of the NVU in vivo and ex vivo remains very challenging14. Only a limited number of techniques are amenable to analyze subcellular structures within cells of the NVU. Most studies use electron microscopy but this technique is limited by the complex protocols required for proper tissue preparation and sample handling. Therefore, we established a methodology based on high resolution confocal microscopy that would facilitate the processing of brain samples, the analysis, and the quantification of subcellular compartments within cells of the NVU.
Here, we describe a protocol which utilizes mouse brain free-floating sections to perform quantitative imaging of the BBB and NVU at the cellular and subcellular levels. We tested and validated a number of antibodies to image and reconstruct the NVU in three dimensions. Furthermore, this protocol allows imaging at the maximal optical diffraction-limited resolution of organelles within brain capillaries. Together with image analysis, this protocol can be used to investigate the intracellular transport of macromolecules across the BBB under different experimental conditions, for example in mouse disease models of neurodegeneration.
Protocol
Ethical approval for this study was provided by the Federal Food Safety and Veterinary Office of Switzerland. All animal experiments were conducted in strict adherence to the Swiss federal ordinance on animal protection and welfare as well as according to the rules of the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC).
1. Generation of Brain Free-floating Sections
- To ensure optimal sample quality, prepare a fresh solution of 2% paraformaldehyde (PFA) in phosphate buffer saline (PBS) on the day of the perfusion.
CAUTION: PFA is moderately toxic by skin contact and a probable carcinogen. Use nitrile gloves to handle PFA and prepare the solution under a chemical fume hood.- Prepare 60 mL of PFA solution per animal.
- Bring PFA into solution by increasing the pH with 180 – 200 µL of a 5 M KOH solution for every 100 mL of PBS and heating the solution up to 60 °C.
NOTE: NaOH should not be used as it negatively impacts tissue preservation upon fixation. - Let the solution cool down to room temperature and bring the pH down to 7.4 using HCl. Filter the solution using filter paper (see Table of Materials).
- Perform a whole mouse fixation via transcardial perfusion as previously described15 with some modifications. First, flush the blood from the vasculature using 20 mL of PBS then perfuse with 40 mL of 2% PFA.
- Remove the brain from the skull as previously described15.
- Immerse a freshly 2% PFA-perfused brain in 20 mL of 2% PFA for 7 h at 4 °C for post-fixation.
NOTE: Increasing the incubation in PFA is not recommended as it can prevent detection of intracellular structures. - Extensively wash the brain with ice-cold PBS.
- Embed fixed brains in agarose.
- Prepare a 3% agarose solution in PBS using a microwave for heating. Gently swirl the solution to cool it down but avoid solidification.
- Dry off the excess of PBS around the brain before immersion into the agarose solution in a plastic container. Rotate the brain inside the agarose to remove air bubbles. Allow the agarose to solidify by cooling it down on ice.
- Carefully remove the agarose block from the plastic container and cut a cube around the brain using a razor blade.Mount the brain onto a vibratome specimen holder (see Table of Materials) using cyanoacrylate glue (see Table of Materials). Allow sufficient time for the glue to solidify before proceeding with the sectioning.
- Transfer the brain in the specimen holder to the vibratome buffer tray filled with PBS.Use the vibratome to section 100 µm brain slices (sagittal or coronal).Collect brain sections in a 6-well plate previously filled with PBS.
- Upon finishing brain sectioning, carefully remove PBS and replace with a 1:1 solution of PBS/glycerol.
- Store sections in PBS/glycerol at -20 °C.
2. Cell or Organelle Labelling by Immunofluorescence Staining
- Carefully transfer a brain section into a well of a 24-well plate previously filled with 500 µL of PBS per well. The section will remain in the same well until the end of the procedure.
- Rinse the section twice with 500 µL of PBS for 5 min under gentle agitation.
- Remove the PBS and perform simultaneous blocking and permeabilization of brain slices.
- Prepare a blocking and permeabilization solution with 0.3% Triton-X and 10% donkey serum in PBS.
NOTE: Donkey serum can be replaced with goat serum to match the host species of the specific secondary antibodies used. - Incubate sections with 250 µL of blocking and permeabilization solution for 1 h at room temperature under gentle agitation.
- Prepare a blocking and permeabilization solution with 0.3% Triton-X and 10% donkey serum in PBS.
- Remove the solution and add a PBS solution containing 5% donkey serum and the primary antibody at an appropriate dilution (e.g., 1:100 to 1:1,000). Incubate overnight at 4 °C under gentle agitation.
NOTE: See the Table of Materials for a list of antibodies that successfully label the different cell types of the NVU with this protocol. The optimal dilution of primary antibodies must be determined empirically. For simultaneous labelling of different antigens, dilute all primary antibodies in the same solution. All primary antibodies must be raised in different species. To improve antibody penetration, samples can be incubated for up to 72 h at 4 °C. - Remove antibody solution and wash sections three times for 10 min in PBS under gentle agitation at room temperature. Remove PBS and add 250 µL PBS solution containing 5% donkey serum and the appropriate species-specific fluorescently labelled secondary antibody. Incubate sections at room temperature for 1 h under gentle agitation.
- See Table of Materials for a list of the secondary antibodies used with this protocol.
- Remove the antibody solution and wash sections three times for 10 min in PBS under gentle agitation at room temperature.
- Remove PBS and add 250 µL of a 4',6-diamidino-2-phenylindole (DAPI) solution with a final concentration of 1 µg/mL. Incubate sections at room temperature for 10 minutes under gentle agitation. Remove DAPI solution and wash sections 3 times for 5 min with PBS.
- Mount the brain section on bonding microscopy slides (e.g., histo-bond glass slides).Carefully remove excess PBS around the section on the slide. Add a drop of mounting medium (see Table of Materials) on top of the brain section and carefully cover it with a 0.17 mm (No. 1.5) borosilicate glass coverslip.
- Store samples at 4 °C protected from light until performing the image acquisition.
3. High-resolution Confocal Imaging of the Blood-brain Barrier
- Perform image acquisition with a suitable laser-scanning confocal microscope (see Table of Materials) with laser lines at 405, 488, 561, and 633 nm for excitation of fluorophores in the blue, green, orange, and red regions of the light spectrum. For image acquisition, use a 63X oil objective with a numerical aperture of 1.4.
- In the acquisition menu of the software that controls the microscope, set the image acquisition parameters. In the drop-down menu of ‘Image size’, select a value of 1024 x 1024 pixels. Modify the pixel size to a value between 200 and 300 nm by adjusting the value of the ‘Zoom’ menu.”
NOTE: For the analysis of intracellular structures, reduce the pixel size to 75 nm. This image size setting substantially increases the acquisition time and can be reduced to 512 x 512 pixels to decrease the acquisition time by imaging a smaller field of view. - In the 'Acquisition Speed' drop-down menu, select a value of 400 Hz, i.e. 400 lines per second. In the 'systems settings' menu, select the 'Pixel depth' drop down menu, and change the value to 12 bit.
- In the microscope software, within the 'acquisition' tab, toggle on the option for sequential frame acquisition. Set the optical section thickness to a value between 0.75 and 1 µm for each channel by modifying the value in the 'Pinhole' menu.
- In the panel for fluorescent excitation, activate the laser required to optimally excite the fluorophores in the sample, for example a 488 nm laser line for a green-emitting fluorophore.
NOTE: Use a fluorophore spectra viewer (see Table of Materials) to select the adequate excitation laser. - In the panel for fluorescent detection, move the slider to select the wavelengths that will be measured in each channel, for example between 510 and 550 nm for a green-emitting fluorophore.
NOTE: Use a fluorophore spectra viewer (see Table of Materials) to select the adequate detection wavelengths. - Add a drop of immersion oil with a refractive index of 1.52 on top of the coverslip with the brain section to match the refractive index of the glass coverslip and objective. Place the sample in the microscope and turn on the epifluorescent lamp to visualize the sample using the microscope binoculars.
- Using the buttons on the microscope stand, change the filter wheel to select a filter appropriate for visualizing DAPI-stained nuclei. Use the coarse focus to bring the signal from DAPI-stained nuclei into focus.
- Using the buttons on the microscope stand, change filters to visualize the signal from vascular markers (for example, CollagenIV or CD31) and center the field of view on an individual capillary segment.
- Press the button to start the live-scan mode. During scanning adjust the gain and laser intensity for each channel to maximize the dynamic range of the images and avoid pixel saturation.
NOTE: Use a look-up table which labels saturated pixels to visually assess over-saturation. To avoid signal over-saturation, adjust the settings with the sample which is expected to have the highest fluorescent signal. - Set the value for line averaging to 2.
NOTE: When using higher acquisition speeds, increase this value to reduce noise. - Establish begin and end sections for performing optical z-stacks that span the whole volume of the capillary. Use a step size of 0.45 µm.
NOTE: If images will be used for subsequent quantification, keep the same acquisition settings for the complete data set. - For quantification, acquire 10 to 20 z-stacks per section from at least three different mice for statistical comparisons.
NOTE: Acquire the full data set in the same session to reduce the variability arising from sample bleaching and laser intensity fluctuations.
4. Image Processing and 3D Reconstruction of the Neurovascular Unit
- (Optional) To increase the axial resolution of the acquired z-stacks, perform deconvolution of the image data set using appropriate software (see Table of Materials).
- In the deconvolution software, change the settings to perform "Blind" deconvolution, (i.e., using an adaptive point-spread function).
- In the deconvolution software settings, toggle on the option for Background removal; this option will determine the smallest intensity value in the image stack and subtract it from the intensity values from all pixels in the image.
- In the deconvolution software settings, toggle off the options for rescaling intensity and resizing to 16 bit depth; these options will keep the original intensity values and dynamic range of the images.
- In the menu of 'Refractive index', set the value to 1.52. In the 'Set Wavelength' menu, select the emission values for each image channel. For example, select 520 for a green-emitting fluorophore.
- Open the image data set using software suitable for visualization and analysis of 3-dimensional data sets (see Table of Materials). In the image analysis software, press the "Surpass" button to render the 3D volume of capillaries.
- In the image analysis software press the "Add new surface" button to segment the capillary surface. Use the bottom arrows to move between steps in the surface creation wizard.
- In the 'Create' tab, set the source channel to the channel with capillary marker, for example CollagenIV, and set the surface area detail to a value of 0.5 µm.
NOTE: The latter option applies a Gaussian filter to the image and determines the smoothness of the surface. - In the 'create' tab, toggle on the option for absolute intensity threshold.
NOTE: This option will create a surface by generating a mask based on the absolute intensity of the selected channel in the image. - On the next step in the surface creation wizard, adjust the lower intensity threshold for the surface by moving the sliding window across the intensity histogram until the rendered mask covers the whole capillary in the image. Determine the value range for this parameter empirically for each data set.
- On the final step of the surface creation wizard, click on the drop-down menu under "Filter Type" and select the option 'Number of Voxels'. Set the minimum number of voxels to a value between 1.0e5 to 2.0e5.
NOTE: This option will exclude surfaces with a small number of voxels, for example, small segments of capillaries at the edge of the frame. - Finish the surface creation wizard and save the creation parameters by clicking "Remember parameters" in the rebuild tab of the surface menu.
- Repeat the procedure for the whole imaging data set by loading the parameter set used for the first image. Adjust the parameters for the intensity, threshold, and minimum number of voxels for each image to account for differences in background intensity and capillary morphology, respectively.
- In the 'Create' tab, set the source channel to the channel with capillary marker, for example CollagenIV, and set the surface area detail to a value of 0.5 µm.
- To segment intracellular vesicular structures (e.g., immunoglobulin-filled endosomes), first create a new channel that only includes fluorescent signal from within the capillary.
NOTE: This process will define a region of interest by creating a mask using the capillary surface created in step 4.3- Select the 'edit' menu in the 'capillary surface' panel. Under 'mask properties', click "Mask All". Select the channel that contains the intracellular vesicle signal as the 'Source Channel' and toggle on the option "Duplicate channel before applying mask".
- In the 'mask settings' column, select "Constant inside/outside" and "Set voxels outside surface to:" and set its value to 0.00.
NOTE: This process will create an additional channel in the image where all voxels outside the capillary mask will have an intensity value of 0. To quantify events outside of the capillary (i.e., in the brain parenchyma), choose "Set voxels inside surface to:" and set its value to 0.00. - To segment vesicular structures, press the button to start the "Add new Spots" wizard. Use the bottom arrows to move between steps in the surface creation wizard.
- In the create tab, use the drop down menu under "Source Channel" to select the intracellular masked channel created in step 4.4.4.
- In the create tab, under the "Spot detection" section, set the estimated XY diameter to a value between 0.5 and 1 µm, depending on the average size of vesicles observed in the experiment. This option determines the smallest size which will be detected by the segmentation algorithm.
- In the create tab, under the "Spot detection" section, toggle on the option for "Background Subtraction".
NOTE: This option will smooth the image with a Gaussian filter of 0.75 spot radius (from the value selected in step 4.4.7) and then subtracting the intensity of the original image Gaussian filtered by 0.88 spot radius. - In the 'create' tab, press the dropdown menu under 'Filter Type' and select "Quality". Note that this will segment the image by applying thresholds based on the intensity at the center of the spots. Adjust the lower intensity threshold by moving the sliding window across the 'Quality' histogram until the majority of identifiable vesicles are labelled.
NOTE: The value range for this parameter needs to be determined empirically for each data set. - Finish the spot creation wizard and save the creation parameters by pressing the button "Remember parameters" in the rebuild tab of the surface menu.
- Repeat the procedure for the whole imaging data set by loading the parameter set used for the first image. Adjust the parameters for the 'Quality intensity threshold' for each image to account for differences in background intensity.
5. Quantification of Intracellular Transport at the BBB
- Quantify the volume (in µm3) and area (in µm2) of the capillary segment. In the analysis software, access the statistics panel of the vascular surface created in step 4.3. Select the 'Detailed' tab, and use the drop down menu to select "All values" to find the values for volume and area.
- Quantify the number of vesicles within the capillary segment. In the analysis software, access the statistics panel of the spots created in step 4.4. Select the 'Overall' tab to find the value of the total number of spots in the image.
- To calculate the number of vesicles per capillary volume, for each image normalize the number of spots by the volume calculated in step 5.1. Multiply this value by 1,000 to get the number of vesicles per 1,000 µm3 of the capillary volume.
- To quantify the total intensity within the capillary create a new surface that covers the entire image following the instructions described in steps 4.3 with the following modifications. Select the channel that contains the pertinent signal (e.g., mIgG) as source channel and set the value of the surface area detail level to 5 µm. To create a surface that covers the whole image, set the value of the lower intensity threshold to 0.
- In the analysis software, access the statistics panel of the surface created in step 5.4. Select the "Detailed" tab, and use the drop down menu to select "All values". Record the values for 'Intensity Sum' for the channels of interest; this parameter corresponds to the sum of individual intensity values over all pixels.
NOTE: For an intracellular signal, this corresponds to the duplicated channel with voxels outside surface set to 0.0. For a signal in the brain parenchyma, this corresponds to the duplicated channel with voxels inside surface set to 0.0. - To calculate the fluorescence intensity per capillary volume, for each image normalize the intensity by the volume calculated in step 5.1. Multiply this value by 1,000 to get the fluorescence intensity value per 1,000 µm3 of the capillary volume.
NOTE: For fluorescent signals in the brain parenchyma, normalize the values by the total image volume minus the capillary volume and multiply by 1,000 to get the total intensity per 1,000 µm3 of brain parenchyma. - Repeat the procedure for the whole data set and use appropriate software to perform statistical analysis of the data.
Representative Results
As representative examples of images obtained from the protocol described here, mouse brain sections were stained with antibodies recognizing different components of the NVU including the basement membrane, astrocytes, pericytes, and endothelial cells (see Table of Materials for specific antibodies used) (Figure 1A, D, and E). At this resolution it is possible to distinguish the individual astrocytic processes and end-feet that are in direct contact with capillaries.
To highlight the suitability of this protocol for detecting intracellular structures, brain sections from animals peripherally injected with the human anti-Tau mAb86 antibody16 were stained with a fluorescently labelled anti-human antibody (Figure 1B). mAb86 is known to specifically target neurons expressing a pathological form of Tau16. Using the protocol described herein, mAb86 was detected with diffraction-limited resolution within individual vesicular structures within neurons (Figure 1B-C). In addition, endogenous mouse IgG was detected in intracellular structures within endothelial cells but not in pericytes (Figure 1D-E and Figure 2).
The acquisition of high-resolution confocal z-stacks of the brain vasculature allows for three-dimensional segmentation of capillaries and intracellular vesicles at the BBB. Figure 2 shows an example of the process of rendering and segmenting a capillary labelled with CollagenIV and mouse IgG-positive intracellular vesicles. By quantifying a full dataset of segmented images, for example by measuring the number of vesicles per capillary volume, it is possible to study changes in intracellular transport processes under different conditions. Figure 3B-C shows the differences in mIgG vesicle number and fluorescence intensity corresponding to mIgG at the brain parenchyma, respectively, upon pericyte depletion in the pdgf-bret/ret mouse model as previously reported17. The same approach was also recently used to analyze changes in intracellular transport at the BBB between different brain regions18.
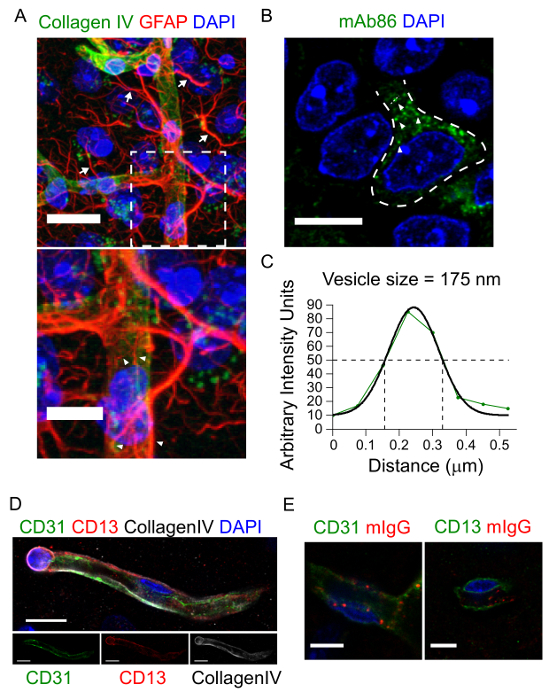
Figure 1: Labelling of multiple cell types and subcellular structures of the neurovascular unit. Representative images of the neurovascular unit (A and D) and intracellular vesicles contained within neurons (B) or endothelial cells (E) obtained with this protocol. The maximum intensity projection image in A (top) shows the distribution of GFAP-positive astrocytes (red) surrounding capillaries labelled by CollagenIV (green). Arrows point to individual astrocytic processes. Scale bar = 20 µm. At this resolution, the individual astrocyte processes and end-feet are clearly visible, as shown in the zoomed image of the boxed region (bottom). Arrowheads point to astrocytic end-feet. Scale bar = 10 µm. The image in B shows the accumulation of a peripherally-injected antibody, mAb86 (green), within a hippocampal neuron. Arrowheads point to individual mAb86-positive vesicles. Scale bar = 10 µm. The graph in C shows the line profile intensity of a single vesicle. The vesicle size was estimated from the full width at half maximum of a Gaussian fit (black solid line) of the intensity curve (green line and circles). The images in D show a three-dimensional reconstruction of an endothelial cell (green) surrounded by a pericyte (red) within the basal lamina (CollagenIV, grey). The lower panels show the individual fluorescence channels. Scale bar = 10 µm. The images in E show the localization of mIgG (red) in intracellular vesicles within endothelial cells (left panel, CD31 in green) but not in pericytes (right panel, CD13 in green). In all images, DAPI stained nuclei are shown in blue. Scale bar = 5 µm. Panels D and E have been modified from reference17. Please click here to view a larger version of this figure.
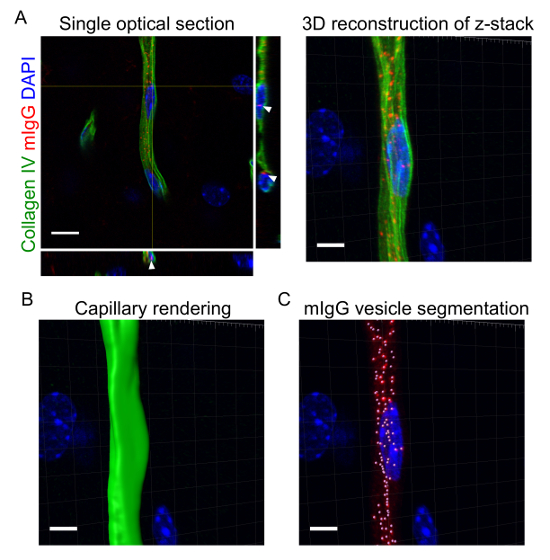
Figure 2: Three-dimensional rendering of capillaries and intracellular vesicles at the blood-brain barrier. With the protocol described, high-resolution confocal images of CollagenIV-positive capillaries (green) and mouse IgG intracellular vesicles (red) were acquired (A). The left panel shows a single optical section with cross-sections. The arrows point to individual mIgG-positive vesicles within brain endothelial cells. The panel on the right shows the 3D reconstruction of the full z-stack using image processing software (see Table of Materials). The capillary volume (B) and individual vesicles (C) were rendered in three dimensions and quantified using image processing software (see Table of Materials). In all images, DAPI stained nuclei are shown in blue. Scale bars = 5 µm. Please click here to view a larger version of this figure.
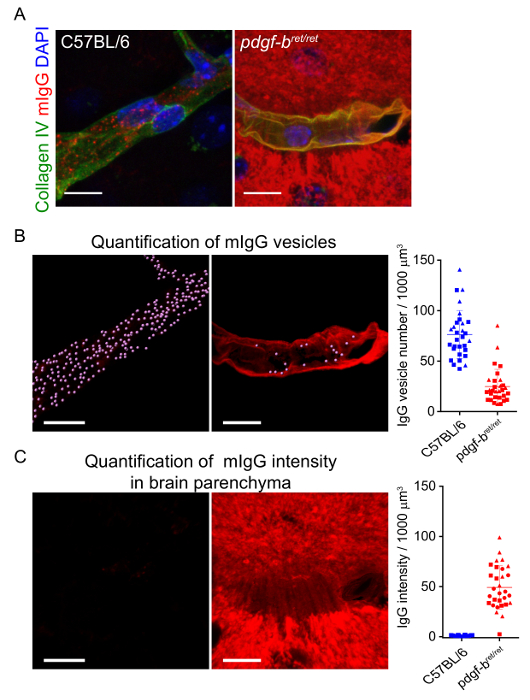
Figure 3: Quantification of mIgG intracellular localization at the BBB. Representative images showing (A) three-dimensional reconstructions of capillaries (labelled with collagenIV, green) and the distribution of mIgG (red) in C57BL/6 mice and in pdgf-bret/ret pericyte depleted mice previously described in19. Scale bars = 10 µm. The images in B show the segmentation of intracellular vesicles within the CollagenIV mask. The graph in B shows the quantification and comparison of mIgG vesicle number per volume of capillary. Each point represents measurements from individual capillary segments. The solid line shows the mean and the error bars represent the standard deviation of the data. The images in C show the mIgG fluorescence signal outside the CollagenIV mask. Similarly, the graph in C shows the quantification and comparison of mIgG fluorescence intensity in the brain parenchyma between C57BL/6 mice and pdgf-bret/ret pericyte depleted mice. Fluorescence intensity units were normalized by the average mIgG parenchyma intensity measured in all C57BL/6 mice. This figure has been modified from reference17. Please click here to view a larger version of this figure.
Discussion
The protocol outlined above describes the preparation of brain free-floating sections, immunofluorescent staining, image acquisition, and analysis parameters for high-resolution microscopy of the BBB. This method has been recently used to investigate the localization of antibody delivery platforms3, the transport of endogenous IgG across the BBB17, and the heterogeneity of the BBB upon pericyte loss18. Different steps in the protocol can be modified to adapt to the specific goal of the experiment. First, the use of thick (100 µm) sections facilitates their handling during the immunostaining and mounting procedures. It also allows for 3D reconstruction of the capillary network, the neurovascular unit, and for the generation of capillary and NVU cross-sections. However, penetration of antibodies within the tissue sections may vary and some antibody staining can be restricted to the superficial layer of the tissue close to the coverslip. The protocol can be modified by increasing the concentration of detergent during the permeabilization step and/or the length of the permeabilization step to improve antibody penetration into the tissue. Second, image quality may be compromised when attempting to acquire images deeper within the tissue (usually 20 to 30 µm below the surface) due to light scattering as well as optical aberrations from refractive index mismatch. To overcome this problem, new methods for tissue clearing and active antibody penetration20 can be combined with this protocol to image larger volumes of tissue. Third, deconvolution is performed after image acquisition to improve the axial resolution of the image. The choice of the blind deconvolution algorithm used in this protocol was based on (i) its ease of use, as no pre-calculation of the point-spread function is required, (ii) its robustness for improving image quality21, and (iii) the lack of artefacts on mIgG intracellular structures after implementation. Depending on the intracellular structures visualized in the sample, other deconvolution algorithms may result in higher image quality. The following references21,22 provide an extensive discussion on the advantages and limitations of additional algorithms for image deconvolution. Finally, the use of an image analysis software package allowed the segmentation of capillaries and intracellular structures in three dimensions. Clearly image analysis is not restricted to the software described in the protocol and alternative packages, for example those discussed in reference23, can be used to segment images. The suitability of different software programs for the analysis of intracellular structures across the BBB should be verified empirically by assessing the accuracy of image segmentation.
Since this method is based on fixed samples, it does not provide direct information about the dynamics of transcytosis across the BBB. However, it can be combined with time-course experiments24, for example by intravenously injecting the molecule of interest and measuring its accumulation within BECs at different time points after injection, to reconstruct the kinetics of intracellular transport. The advantage of this approach is that it allows for the analysis of deep brain regions, as shown in18, which are currently inaccessible to intravital live imaging approaches. A critical step during the protocol is the careful monitoring of tissue fixation. Fixation with 4% PFA dramatically reduces the immunogenicity of intracellular organelles and of endogenous or peripherally administered immunoglobulins17 (Figure 1B). A limitation of this protocol is its requirement of high-quality antibodies (i.e., low non-specific staining, low cross-reactivity) suitable for immunofluorescence. Provided that such reagents are available, the method can be applied to investigate the intracellular localization of any protein of interest. For example, the protocol was used to identify lysosomes in brain endothelial cells17. It should also be noted that since this protocol is based on confocal microscopy, the lateral resolution is limited by diffraction and cannot resolve structures smaller than approximately 175 to 250 nm (Figure 2).
Previous studies have performed detailed analysis of the cellular composition of the neurovascular unit using confocal microscopy25,26. However, investigating intracellular transport at the BBB relies mostly on the use of transmission electron microscopy19,27,28. While this method offers the highest lateral resolution of intracellular structures, electron microscopy remains a challenging technique with low throughput. Moreover, the number of different molecular targets which can be visualized by EM is very limited. This protocol offers an accessible alternative to investigate intracellular transport at the BBB. The complete procedure, from brain collection to image analysis, can be performed in 5 to 6 days. If suitable antibodies are available, immunofluorescence allows simultaneous detection of multiple cell types/molecules within the same sample. Moreover this protocol could be combined with super-resolution microscopy techniques to overcome the limitations in spatial resolution29. Overall, the protocol described above enables the quantification of changes in the intracellular localization of proteins of interest within the neurovascular unit. Its application for different genetic or pharmacological perturbations will allow investigating the intracellular structure and transport functions of the BBB in vivo.
Disclosures
The authors have nothing to disclose.
Acknowledgements
R.V. work was supported by a Roche Postdoctoral Fellowship (2014-2017).
Materials
| Rat monoclonal antibody clone ER-MP12 against CD31/PECAM | Novus Biologicals | MCA2388 | Labels Brain Endothelial Cells Use at dilution 1:100 detect with donkey polyclonal anti-rat IgG (H+L) coupled with suitable AlexaFluor dye, used at dilution 1:400 |
| Rat monoclonal antibody against Podocalyxin | R&D Systems | MAB1556 | Labels Lumen of capillaries Use at dilution 1:100 detect with donkey polyclonal anti-rat IgG (H+L) coupled with suitable AlexaFluor dye, used at dilution 1:400 |
| Rabbit polyclonal antibody against CollagenIV | Biotrend | BT21-5014-70 | Labels Basement membrane of capillaries Use at dilution 1:100 detect with donkey polyclonal anti-rabbit IgG (H+L) coupled with suitable AlexaFluor dye, used at dilution 1:400 |
| Goat polyclonal antibody against CD13 | R&D Systems | AF2335 | Labels pericytes Use at dilution 1:100 detect with donkey polyclonal anti-goat IgG (H+L) coupled with suitable AlexaFluor dye, used at dilution 1:400 |
| Mouse monoclonal antibody clone G-A-5 against GFAP coupled to Cy3 | Abcam | ab49874 | Labels Astrocytes Use at dilution 1:100 |
| Rabbit polyclonal antibody against Iba-1 | Wako | 019-19741 | Labels Microglia Use at dilution 1:100 detect with donkey polyclonal anti-rat IgG (H+L) coupled with suitable AlexaFluor dye, used at dilution 1:400 |
| Rat monoclonal antibody ABL93 gainst LAMP2 | Fitzgerald | 10R-CD107BBMSP | Labels Lysosomes Use at dilution 1:100 detect with donkey polyclonal anti-rat IgG (H+L) coupled with suitable AlexaFluor dye, used at dilution 1:400 |
| donkey polyclonal antibody against mouse IgG (H+L) AlexaFluor594 | LifeTechnologies | A21203 | Use at dilution 1:400 |
| donkey polyclonal antibody against mouse IgG (H+L) AlexaFluor488 | LifeTechnologies | A21202 | Use at dilution 1:400 |
| goat polyclonal antibody against mouse IgG (H+L) AlexaFluor555 | LifeTechnologies | A21422 | Use at dilution 1:400 |
| Filter paper | Sigma-Aldrich | WHA10334347 | Whatman prepleated qualitative filter paper Grade 0858 1/2, grained |
| Cyanoacrylate glue | Roth Carl | 258.1 | Roti Coll 1 |
| Fluorescent Mounting medium | Dako | S3023 | Dako fluorescent mounting medium |
| Adult mice | The Jackson Laboratory | 000664 | C57BL/6J male or female mice between 9 and 19 months of age |
| Vibratome | Leica Biosystems | 14047235612 | Leica VT100S vibrating blade microtome |
| Laser Scanning Confocal microscope | Leica Microsystems | NA | Leica TCS SP8 X with HyD detectors and White light laser |
| Image processing software | Leica Microsystems | NA | Leica Application Suite AF version 3.1.0 build 8587 |
| Image analysis software | Bitplane scientific software | NA | Imaris version 7.6.5 build 31770 for x64 |
| Fluorescence SpectraViewer | ThermoFisher Scientific | NA | https://www.thermofisher.com/ch/en/home/life-science /cell-analysis/labeling-chemistry/fluorescence-spectraviewer.html |
References
- Blanchette, M., Daneman, R. Formation and maintenance of the BBB. Mech Dev. 138, 8-16 (2015).
- Chow, B. W., Gu, C. The Molecular Constituents of the Blood-Brain Barrier. Trends Neurosci. 38 (10), 598-608 (2015).
- Niewoehner, J., et al. Increased Brain Penetration and Potency of a Therapeutic Antibody Using a Monovalent Molecular Shuttle. Neuron. 81 (1), 49-60 (2014).
- Yu, Y. J., et al. Therapeutic bispecific antibodies cross the blood-brain barrier in nonhuman primates. Sci Transl Med. 6 (261), 154 (2014).
- Preston, J. E., Joan Abbott, N., Begley, D. J. Transcytosis of macromolecules at the blood-brain barrier. Adv Pharmacol. 71, 147-163 (2014).
- De Bock, M., et al. Into rather unexplored terrain-transcellular transport across the blood-brain barrier. Glia. 64 (7), 1097-1123 (2016).
- Bien-Ly, N., et al. Transferrin receptor (TfR) trafficking determines brain uptake of TfR antibody affinity variants. J Exp Med. 211 (2), 233-244 (2014).
- Sade, H., et al. A human blood-brain barrier transcytosis assay reveals antibody transcytosis influenced by pH-dependent receptor binding. PLoS One. 9 (4), 96340 (2014).
- Tian, X., et al. LRP-1-mediated intracellular antibody delivery to the Central Nervous System. Sci Rep. 5, 11990 (2015).
- Hsu, J., Rappaport, J., Muro, S. Specific binding, uptake, and transport of ICAM-1-targeted nanocarriers across endothelial and subendothelial cell components of the blood-brain barrier. Pharm Res. 31 (7), 1855-1866 (2014).
- Siupka, P., et al. Bidirectional apical-basal traffic of the cation-independent mannose-6-phosphate receptor in brain endothelial cells. J Cereb Blood Flow Metab. , (2017).
- Zlokovic, B. V., et al. A saturable mechanism for transport of immunoglobulin G across the blood-brain barrier of the guinea pig. Exp Neurol. 107 (3), 263-270 (1990).
- Deane, R., et al. IgG-assisted age-dependent clearance of Alzheimer’s amyloid beta peptide by the blood-brain barrier neonatal Fc receptor. J Neurosci. 25 (50), 11495-11503 (2005).
- Cabezon, I., et al. Serial block-face scanning electron microscopy applied to study the trafficking of 8D3-coated gold nanoparticles at the blood-brain barrier. Histochem Cell Biol. , (2017).
- Gage, G. J., Kipke, D. R., Shain, W. Whole animal perfusion fixation for rodents. J Vis Exp. (65), (2012).
- Collin, L., et al. Neuronal uptake of tau/pS422 antibody and reduced progression of tau pathology in a mouse model of Alzheimer’s disease. Brain. 137 (10), 2834-2846 (2014).
- Villasenor, R., et al. Trafficking of Endogenous Immunoglobulins by Endothelial Cells at the Blood-Brain Barrier. Sci Rep. 6, 25658 (2016).
- Villasenor, R., et al. Region-specific permeability of the blood-brain barrier upon pericyte loss. J Cereb Blood Flow Metab. , (2017).
- Armulik, A., et al. Pericytes regulate the blood-brain barrier. Nature. 468 (7323), 557-561 (2010).
- Richardson, D. S., Lichtman, J. W. Clarifying Tissue Clearing. Cell. 162 (2), 246-257 (2015).
- Sibarita, J. B. . Deconvolution Microscopy. 95, 201-243 (2005).
- Shaw, P. J. . Comparison of Widefield/Deconvolution and Confocal Microscopy for Three-Dimensional Imaging. , 453-467 (2006).
- Chenouard, N., et al. Objective comparison of particle tracking methods. Nat Methods. 11 (3), 281-289 (2014).
- Foret, L., et al. A general theoretical framework to infer endosomal network dynamics from quantitative image analysis. Curr Biol. 22 (15), 1381-1390 (2012).
- Daneman, R., Zhou, L., Kebede, A. A., Barres, B. A. Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature. 468 (7323), 562-566 (2010).
- Bell, R. D., et al. Pericytes control key neurovascular functions and neuronal phenotype in the adult brain and during brain aging. Neuron. 68 (3), 409-427 (2010).
- Ben-Zvi, A., et al. Mfsd2a is critical for the formation and function of the blood-brain barrier. Nature. 509 (7501), 507-511 (2014).
- Stewart, P. A. Endothelial vesicles in the blood-brain barrier: are they related to permeability. Cell Mol Neurobiol. 20 (2), 149-163 (2000).
- Follain, G., Mercier, L., Osmani, N., Harlepp, S., Goetz, J. G. Seeing is believing – multi-scale spatio-temporal imaging towards in vivo cell biology. J Cell Sci. 130 (1), 23-38 (2017).

