Utilizing 18F-FDG PET/CT Imaging and Quantitative Histology to Measure Dynamic Changes in the Glucose Metabolism in Mouse Models of Lung Cancer
Summary
In this protocol, we describe how to utilize [18F]-2-fluoro-2-deoxy-D-glucose positron emission tomography and computed tomography (18F-FDG PET/CT) imaging to measure the tumor metabolic response to the targeted therapy MLN0128 in a Kras/Lkb1 mutant mouse model of lung cancer and coupled imaging with high resolution ex vivo autoradiography and quantitative histology.
Abstract
A hallmark of advanced tumors is a switch to aerobic glycolysis that is readily measured by [18F]-2-fluoro-2-deoxy-D-glucose positron emission tomography (18F-FDG PET) imaging. Co-mutations in the KRAS proto-oncogene and the LKB1 tumor suppressor gene are frequent events in lung cancer that drive hypermetabolic, glycolytic tumor growth. A critical pathway regulating the growth and metabolism of these tumors is the mechanistic target of the rapamycin (mTOR) pathway, which can be effectively targeted using selective catalytic mTOR kinase inhibitors. The mTOR inhibitor MLN0128 suppresses glycolysis in mice bearing tumors with Kras and Lkb1 co-mutations, referred to as KL mice. The therapy response in KL mice is first measured by 18F-FDG PET and computed tomography (CT) imaging before and after the delivery of MLN0128. By utilizing 18F-FDG PET/CT, researchers are able to measure dynamic changes in the glucose metabolism in genetically engineered mouse models (GEMMs) of lung cancer following a therapeutic intervention with targeted therapies. This is followed by ex vivo autoradiography and a quantitative immunohistochemical (qIHC) analysis using morphometric software. The use of qIHC enables the detection and quantification of distinct changes in the biomarker profiles following treatment as well as the characterization of distinct tumor pathologies. The coupling of PET imaging to quantitative histology is an effective strategy to identify metabolic and therapeutic responses in vivo in mouse models of disease.
Introduction
Our research has focused on investigating and targeting cancers with mutations in the Liver kinase B1 (LKB1, also referred to as STK11) mutant cancers1. LKB1 is a master tumor suppressor that represses mTOR complex 1 (mTORC1) through the activation of the AMP kinase (AMPK) leading to the regulation of growth and metabolism. Therefore, the loss of LKB1 leads to an unrestrained mTORC1 activation, the activation of the HIF1-alpha resultant in a glycolytic metabolic phenotype commonly referred to as the Warburg Effect2,3,4. LKB1 inactivating mutations directly lead to the development of a rare familial cancer pre-disposition syndrome known as Peutz-Jeghers Syndrome (PJS) that is characterized by the development of benign gastrointestinal polyps known as hamartomas5,6,7. Moreover, LKB1 frequently co-mutates with oncogenic KRAS resulting in hypermetabolic and aggressive human lung tumors8,9.
Lkb1-related diseases are readily modeled in mice. The heterozygous inactivation of Lkb1 in mice leads to the development of hamartomas accurately modeling PJS10,11,12,13. Additionally, Lkb1 mutations that are readily modeled in mice accurately recapitulate cancer phenotypes of the lung, skin, pancreas, and breast14. The co-mutation of Kras/Lkb1 in the lung tissue of transgenic mice, using a Cre recombinase-mediated activation of the oncogenic KrasG12D allele and biallelic deletion of Lkb1, results in the formation of aggressive and metastatic lung tumors15,16. The characterization of KrasG12D;Lkb1-/- (KL) lung tumors isolated from mice shows these tumors have a high mTORC1 activation and are highly glycolytic, using both direct metabolite measurements of glucose and lactate or measuring the consumption of [18F]-2-fluoro-2-deoxy-D-glucose (18F-FDG) by positron emission tomography (PET) with computed tomography (CT) 17. The mTORC1 hyper-activation in LKB1 mutant tumors provides a clear rationale for testing both allosteric and catalytic kinase inhibitors of mTOR to treat these cancers.
In a previous study, we demonstrated that the allosteric mTORC1 inhibitor rapamycin (RAPA) successfully inhibited the growth of and glycolysis in gastrointestinal (GI) tumors using an Lkb1+/- transgenic mouse model of PJS3. RAPA is currently approved as a single agent therapy for the treatment of renal cell carcinoma but showed limited efficacy in NSCLC18,19,20. RAPA is an allosteric mTORC1 inhibitor and may be improved upon by the development of next-generation mTOR catalytic kinase inhibitors that deliver a more nearly complete inhibition of the mTOR complexes 1 and 2 (mTORC1 and mTORC2, respectively)21. Drugs such as MLN0128 are now being evaluated in preclinical studies and early phase clinical trials22,23. A recent study from our laboratory demonstrated that MLN0128 is a potent mTOR inhibitor in human lung tumor cell lines and in vivo in KL GEMMs of lung cancer15,16. MLN0128 suppressed the lung tumor growth and glucose metabolism in these mice24.
In this study, we take advantage of the well-characterized adenoviral Cre-induced mouse models of lung cancer initiated by a conditionally activated Lox-Stop-Lox-KRASG12D oncogene15,25. These KrasG12D mice were crossed with mice having floxed alleles of Lkb1 (Lkb1L/L) to generate KrasG12D; Lkb1L/L (KL) mice16. Following the intranasal delivery of adeno- or lentivirus expressing Cre recombinase, the KL mice develop early lesions by 4 weeks post-tumor induction. By 6 weeks, the tumors in the KL mice change from adenomatous tumors to a more malignant, aggressive tumor phenotype typical of lung carcinomas, and by 8 – 10 weeks, the mice develop frank carcinomas with a 100% penetrance16,26.
Both PET/CT imaging and quantitative immunohistochemistry may be utilized to determine the molecular and metabolic responses as well as the therapeutic responses in tumors following the delivery of targeted therapies such as MLN012817,26,27. Described here is an experimental protocol that utilizes 18F-FDG PET imaging to measure the metabolic response to an MLN0128-targeted therapy. Coupling PET imaging with quantitative histology enables the measurement of the molecular response to mTOR inhibition as well as the quantification of the tumor burden and tumor histology.
Protocol
All procedures described in the protocol have been approved by the institutional animal care and use committee (IACUC) at the University of California, Los Angeles.
1. 18F-FDG PET and CT Imaging in Mice
CAUTION: Use protective equipment when handling radioactivity. Follow all applicable regulatory procedures when handling radioactivity.
- Place the cage with the mice to be imaged on a warm bed at 37 °C 1 h prior to the 18F-FDG injection to reduce the brown fat consumption of 18F-FDG.
NOTE: Fasting the mice for 4 – 16 h can help reduce the myocardial consumption of 18F-FDG. - Weigh the mouse and record its weight.
- Anesthetize the mouse using 2 – 3% isoflurane in oxygen at 0.5 – 2 L/min for 2 – 3 min using an anesthesia chamber kept at 37 °C. Ensure that mouse has been anesthetized by pinching the toe; no reaction will be observed if the mouse has been anesthetized. Apply ophthalmic ointment to the eyes to prevent any dryness during the anesthesia.
- Dilute 18F-FDG (109 min radioactive half-life) in sterile saline at an adjusted decay-corrected injection concentration of 70 – 75 µCi/100 µL.
NOTE: Follow the PET scanner manufacturer's recommended 18F dose for optimal scanner imaging. - Draw 70 – 75 µCi with an insulin syringe with a 28 G needle, measure the radioactivity dose using a dose calibrator, and record the measurement and time. Place the syringe in a lead syringe holder.
NOTE: The amount of 18F-FDG radioactivity in each dose is measured with a dose calibrator, which is calibrated against a standard reference material, such as cesium-137, according to the manufacturer's protocols. The time of the reading is also recorded to determine the decay correction. - Prick the distal end of the mouse tail and measure the mouse's blood glucose with a glucometer.
- Warm the tail for 1 – 2 min with a gauze soaked in warm water. Wipe the tail with 70% isopropanol to dilate the tail vein just prior to the injection. Administer 100 µL of 18F-FDG (the entire volume in the syringe) with a bolus injection via the lateral tail vein and record the time of injection. Measure the remaining dose in the syringe using the dose calibrator and record the measurement and time.
NOTE: There will be some amount of probe left in the syringe. The use of insulin syringes is preferred over syringes connected to needles via Luer locks due to the decreased amount of the dose trapped in the syringe/needle after the injection has been administered. - Place the injected mouse in the anesthesia chamber kept under 1.5 – 2% isoflurane at 37 °C to allow the probe to be distributed via the mouse's systemic circulation for 1 h prior to the PET scan.
NOTE: It might be beneficial to void the bladder prior to the scanning to allow for an easier 18F-FDG PET visualization of the tumors implanted in the lower flanks of the mouse. - After 1 h, place the mouse in an imaging chamber under nose-cone isoflurane anesthesia and at 37 °C, and secure its limbs in place with medical tape in a supine position.
- Place the imaging chamber in the PET/CT scanner.
- Acquire the PET and CT scans as described in the PET/CT scanner manual28.
NOTE: The PET images are acquired for 600 s with an energy window of 150 – 650 keV, reconstructed using maximum-likelihood expectation maximization with corrections for photon attenuation, detector normalization, and radioisotope decay (a scatter correction was not applied). The CT images are acquired in a continuous mode for 50 s using a 50 kVp, 200 µA X-ray source and a flat-panel detector, and they are reconstructed using the Feldkamp algorithm. - After the PET/CT is completed, remove the mouse from the imaging chamber and allow it to recover in its cage. Monitor the mouse until it has fully regained consciousness and can maintain sternal recumbency.
- Import the reconstructed PET/CT images into AMIDE software by clicking File, then Open, and selecting the appropriate file.
- Convert the PET data to the unit of percent-injected dose per gram (%ID/g) by entering the dose at the time of injection after accounting for any residual dose left in the syringe, or to the unit of standardized uptake value (SUV) by additionally entering the subject's weight. To do this, right-click on the PET data set and locate the %ID/g field on the Basic Info tab. Enter the %ID/g recorded earlier.
- Draw the regions-of-interest (ROIs) on the tumors and normal tissues (liver, muscle, lung, heart, brain, and subcutaneous fat). To do this, click on Edit, select Add ROI, select ROI shape and give the ROI a name. Draw ROIs over tumors and tissues and adjust their dimensions to cover tissue of interest in all 3 axes.
NOTE: To account for differences in PET probe biodistribution between animals, tumor ROI values can be further normalized to ROI values of the liver, a well-perfused organ with minimal glycolytic activity representing 18F-FDG in circulation. ROI analyses of tumor and normal tissues are performed on the same mouse. Lung tumor lesions are generally identified by 18F-FDG PET since the 18F-FDG retention in a normal lung is relatively low. CT is also used to identify lesions, particularly lesions that are 18F-FDG non-avid. An ex vivo analysis of isolated lungs also helps to localize tumor lesions.
2. 18F-FDG Autoradiography
- Prepare the mouse for imaging with 18F-FDG by following steps 1.1 – 1.12, except now, dilute 18F-FDG in sterile saline at an adjusted decay-corrected injection concentration of 1,000 µCi/200 µL.
NOTE: Higher doses of 18F-FDG are used for autoradiography to account for additional sample processing time and an optimal detection by phosphor plates. - Euthanize the mouse via a lethal inhalation of isoflurane at 5% or by CO2 (an IACUC-approved procedure).
NOTE: Cervical dislocation should not be used as this can damage the lung tissue. - Pin down the mouse with the ventral surface exposed and spray it with 70% ethanol to mat down its hair prior to the incision.
- Open the thoracic cavity by applying a midline incision, cutting away the diaphragm and removing the chest walls. Expose the trachea by carefully removing the salivary gland. Place the bulldog clamp on the trachea as close to the jaw as possible, ensuring a tight fit on the trachea. Place a 23 G needle attached to a 3 mL syringe inside the trachea below the bulldog clamp and inject ~2 mL of an OCT:PBS (1:1) (Optimal Cutting Temperature:Phosphate Buffered Saline) solution.
NOTE: OCT is very viscous and is mixed with PBS to allow for an easier injection into the lungs. - Remove the needle from the trachea and use forceps to clamp on the injection point to prevent any leaking of the OCT:PBS solution.
- Carefully remove the lungs from the thoracic cavity and separate the left lobe from the rest of the lungs. Place the left lobe in labeled cryomold filled with a few drops of OCT. Once the lung lobe is inside the mold, fill the cryomold to the top with OCT.
- Repeat the same procedure with the right half of the lungs.
NOTE: If the 18F-FDG signal in the heart is expected to be high or if the lung tumors are located close to the heart, it might be beneficial to remove the heart in order to prevent any tracer leakage. Alternatively, entire lungs can be embedded into a single cryomold. It is important to avoid air bubbles when working with OCT. - Using long forceps, place the prepared cryomold in a closed-cell extruded polystyrene foam container containing a mixture of dry ice and isopentane.
NOTE: This mixture should be at approximately -70 °C before placing the cryomold in it for freezing. After hardening, the OCT compound will become white. If multiple samples are processed at the same time, the frozen samples in OCT cryomolds can be temporarily stored on dry ice. - Remove the frozen block from the cryomold and mount it onto a cryostat for sectioning. Section the block at a 4 µm thickness using microtome blades (34°/80 mm, high profile). Transfer the tissue sections to a glass slide that has been stored at room temperature.
- Place the specimen slides onto a phosphor imaging plate. Place the plate in the cassette and gently close it to prevent slides from shifting. Store the cassette in a -20 °C freezer for the plate exposure, generally overnight.
NOTE: The plates and cassettes need to be pre-chilled to -20 °C before use. Placing the samples at -80 °C is acceptable as well. - After the exposure, remove the slides from the plate and read the plate on the image reader.
- The slides can be wrapped in plastic wrap and stored at -80 °C, or they can be prepared for hematoxylin & eosin staining or immunohistochemistry.
3. Harvesting Lung Tissue for Histology
- Follow steps 2.2 – 2.4, except now, instead of using the OCT:PBS solution, inject 2 – 3 mL of 10% normal buffered formalin to fix the lungs.
- Remove the needle from the trachea and use forceps to clamp on the injection point to prevent any leaking of the formalin. Carefully remove the lungs from the thoracic cavity and place them into a 50 mL conical tube containing ~20 mL of 10% normal buffered formalin for 16 – 24 h to ensure a complete fixation.
NOTE: Fixing the lungs allows for the preservation of the optimal anatomical features. - The next day, transfer the fixed lungs from the formalin to 70% ethanol and prepare the lungs to be placed in a tissue cassette.
- Prepare the lungs for histology by carefully dissecting lobes using scissors to cut at branch points between 5 lobes, numbered 1 – 5 as shown in Figure 1D, and place them in a non-overlapping orientation in the tissue cassette. Place a foam pad gently on the lung tissue to keep the orientation intact.
- Store the dissected lung lobes in 70% ethanol until the paraffin embedding.
- Paraffin-embed the tissue in cassettes and cut 4 µm thick sections for the staining, using standard procedures.
4. Tissue Segmentation and Quantification Using Commercial Software
- Image hematoxylin and eosin (H&E) stained lung sections at a 1.25X magnification using a commercial multispectral imaging system.
- Convert the images to digital image cubes and load (click on File, then on Load Spectral Library) pre-made spectral libraries for H&E.
NOTE: The spectral libraries were developed in advance by acquiring spectral images from singly stained lung sections, one section stained only with eosin and the other only stained with hematoxylin, which were saved in an open-source proprietary spectral library file (.csl). The imaging system has operating software that acquires an image at each wavelength that is needed (as defined by the acquisition protocol). Those images (the "image cube") are stored in an open-source proprietary multispectral file format (.im3). Spectra are extracted from the image cube using the morphometric software and stored in separate spectral library files. - Spectrally unmix the pseudo-colored H&E images of the whole lungs by clicking on the Unmix button.
NOTE: Unmixing takes less than 1 s. The number of pixels in each tissue type were quantified using morphometric image analysis software. - Visualize each image cube by converting the multispectral data into the equivalent 3-color (red, green, blue) image by using the eye's wavelength-dependent color response convolved with the intensity of each image.
NOTE: The resulting 3 images are displayed as a standard 24-bit color image. - Pseudo-color each unmixed image by scaling that image in a user-chosen color (e.g., red, green, purple, etc.) and add that together with the other pseudo-colored unmixed images into one standard 24-bit color image.
- Use the default settings for all analyses.
NOTE: In general, since this kind of segmentation relies on a visual assessment of the results (i.e., an assessment of how well the segmentation on the images outside of the training set works), it is important to properly divide the images up into training, test, and validation sets. - Start with 2 – 3 images as a training set (10 – 15 for human biopsy samples), train on those until the results look good, and then apply that algorithm to another 2 – 3 images. Then, apply the resulting algorithm to the full validation set.
NOTE: Most likely, some retraining will be needed. - Analyze the tumor area in whole lung sections by calculating the total pixel count for the red pseudo-colored tumors in lobes 1 – 5 of each mouse.
NOTE: The normal tissue was pseudo-colored green and blood/blood vessels were pseudo-colored pink as shown in Figure 3. The mean tumor burden for each treatment group was calculated by measuring the total pixel count for each mouse in the treatment group.
Representative Results
18F-FDG PET imaging was performed on KL mice and demonstrated that the tumors in these mice were highly glycolytic as shown by an elevated 18F-FDG consumption (Figure 1A), agreeing with previously published studies26,29. A resection of whole lungs revealed the presence of several tumors (Figure 1B). Mouse lungs can be divided up into 5 separate lobes represented in Figures 1C and 1D. Lobes 1 – 5 were labeled on sectioned lungs that were stained with H&E or glucose transporter 1 (Glut1) (Figure 1D). Glut1 is a primary transporter of both glucose and 18F-FDG and its expression and localization to the plasma membrane of tumor cells directly correlate with 18F-FDG SUV29. A higher resolution analysis of Glut1 staining (40X) in 18F-FDG-avid lung tumors shows an elevated expression and localization of the transporter at the plasma membrane (Figure 1D).
Due to the limited resolution of PET imaging, both PET/CT and tissue autoradiography were performed. The higher resolution of autoradiography might identify smaller tumors and/or heterogeneity of the tumor 18F-FDG distribution. After the tumor induction, 18F-FDG PET/CT imaging was performed on KL mice (Figure 2A) followed by autoradiography on lungs isolated from these mice (Figures 2B and 2C). As seen in Figures 2B and 2C, the autoradiography identified two additional smaller tumors that were positive for 18F-FDG yet were not readily visible by PET. Following autoradiography, the slides with tissue could also be used for the immunohistochemical (IHC) staining of biomarker(s). H&E staining for the tumors confirmed the presence of tumors in the left lobe (Figure 2D).
Next, 18F-FDG PET imaging was performed on MLN0128-treated KrasG12D;Lkb1-/- mice in order to utilize 18F-FDG as a functional biomarker of glucose metabolism in lung tumors (Figure 3). We identified that a treatment with MLN0128 robustly inhibited the mTORC1 signaling and glycolysis as shown by a reduced 18F-FDG consumption (Figures 3A and 3B). These results agree with preclinical studies assessing MLN0128 in KL mice as previously published by our laboratory17,27. Lastly, IHC staining was performed on the tumors (Figure 3C). Tumors were stained either for H&E or with antibodies against phospho-S6, which is a conserved substrate of mTORC1 and is used to indicate an mTORC1 activation (P-S6) vs. inactivation (S6). Figure 3C shows a robust inhibition of P-S6 by MLN0128 in KL tumors compared to those treated with a vehicle, which agrees with previously published work17. In addition to KRAS, oncogenic drivers such as the epidermal growth factor receptor (EGFR) support the glycolytic metabolism in lung tumors as well. Therefore, we tested whether the inhibition of constitutively active mutant EGFR with erlotinib suppressed the 18F-FDG metabolism in mouse xenografts. Figures 3D and 3E show that the human lung tumor line HCC827, which harbors an EGFR del19 mutation, showed a significantly reduced 18F-FDG consumption following five days of erlotinib treatment.
Lastly, morphometric tissue analysis was performed on sectioned lungs and lung tumors to quantify the total tumor burden as well as to differentiate tumor pathologies that included tissue subtype, necrosis, blood vessels from normal lung tissue, and air space. The KL GEMMs developed a complex and pathologically heterogeneous disease that presented itself with lung tumors of varying histopathologies. These include adenocarcinomas (ADC) and squamous cell carcinomas (SCC)-this heterogeneity makes the treatment of this cancer a formidable challenge. Figure 4A shows a single H&E stained lung lobe with two large tumors present. The higher magnification images shown in Figure 4B identify a normal lung, vessels, and airspace, and tumor necrosis as well as adenocarcinoma, characterized by a well-defined papillary structure and a squamous cell carcinoma. Figure 4C represents the pseudo-coloring of the lung lobe and tumor using Inform morphometric software. Figure 4D shows the percentages of normal lung, vessels, and individual pathologies such as tumor necrosis and tumor subtypes that segmented well-differentiated adenocarcinoma from squamous cell carcinoma.
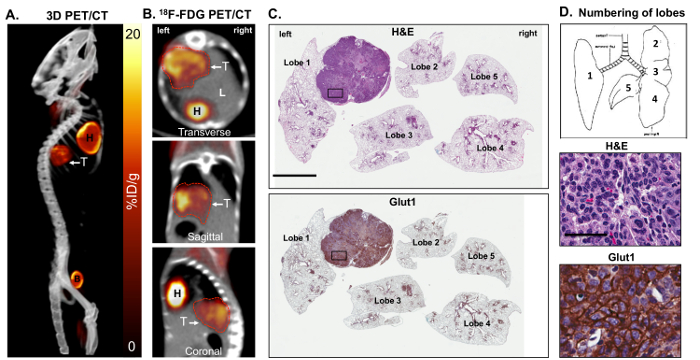
Figure 1: Metabolically active KrasG12D; Lkb1-/- (KL) mutant lung tumors are 18F-FDG positive and express high levels of glucose transporter 1 (Glut1). Panels A and B show a maximal intensity projection [also referred to as a 3-dimensional (3D) image] of the 18F-FDG-PET and CT analysis on some FDG-avid KL mice harboring squamous lung tumors. Shown are (A) a 3D reconstruction and (B) transverse, sagittal and coronal views of lung tumor(s) as (T). (C) This panel shows a whole lung histology of the KL mouse imaged in panels A and B, either stained for H&E (top panel) or with an antibody specific for Glut1 (bottom panel). The lung lobes are numbered. The scale bar = 2 mm. (D) This diagram represents the orientation and numbers of lobes in the mice (top panel) and higher resolution 40X images of the H&E- or Glut1-stained tumors from the slides shown in panel C for H&E (middle panel) or stained with an antibody specific for Glut1 (bottom panel). The scale bar = 25 µm. Please click here to view a larger version of this figure.
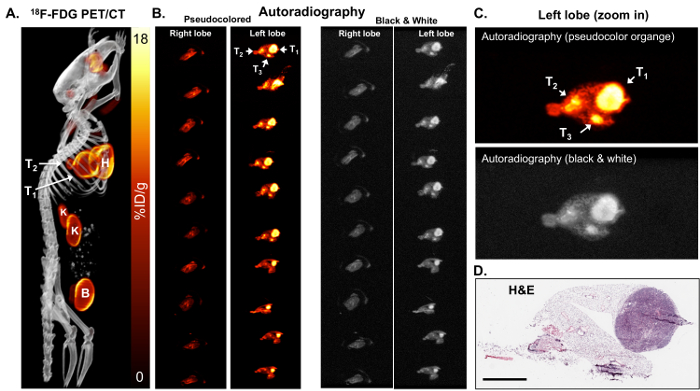
Figure 2: 18F-FDG autoradiography can identify small tumors that are metabolically active. (A) This 18F-FDG PET/CT image shows 18F-FDG-avid tumors in a KL mouse shown as a maximum-intensity projection image. T1 and T2 = tumors, H = the heart, B = the bladder, K = the kidneys. (B) This panel shows the ex vivo autoradiography on serial sections of the right and left lung lobes of the mouse. The lungs in the left and right panels are identical. The lungs in the left panels are pseudo colored orange. The lungs in the right panels are colored in black and white. The tumors (T1, T2, and T3) are indicated with arrows. (C) This is a magnified view of the autoradiography pseudocolored orange (top panel) and black and white (bottom panel). (D) This panel shows the H&E staining of the top slice of the left lobe shown in panel B. The scale bar = 200 µm Please click here to view a larger version of this figure.
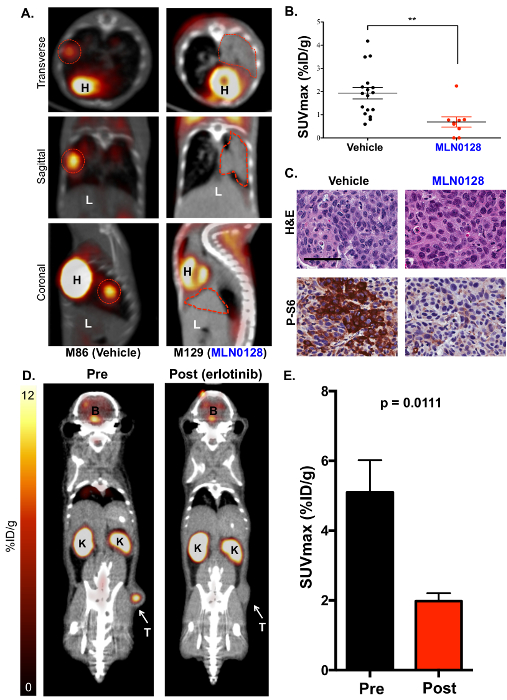
Figure 3: The mTOR inhibitor MLN0128 suppresses glucose consumption in lung tumors of KL mice as detected by 18F-FDG PET. (A) This panel shows representative 18F-FDG PET/CT images of KL mice treated with a vehicle (18F-FDG avid, left) or MLN0128 (18F-FDG non-avid, right). The transverse (top panel), coronal (middle panel), and sagittal (lower panel) views are shown. The tumors are outlined with red lines; H = the heart, L = the liver. (B) This panel shows a quantification of SUVmax (%ID/g) between the vehicle- and MLN0128-treated tumors. (C) This panel shows the H&E and P-S6 staining of whole lung sections from KL mice treated with vehicle or MLN0128. The scale bar = 25 µm. (D) This panel shows representative 18F-FDG-PET and CT images of HCC827 EGFR (del19) xenografts pre- and post-erlotinib treatment. The tumor (T) is indicated with an arrow, K = the kidney, B = the brain. (E) This panel shows a quantification of the SUVmax (%ID/g) for HCC827 xenografts before and after the erlotinib treatment. n = 10 tumors/group. Please click here to view a larger version of this figure.
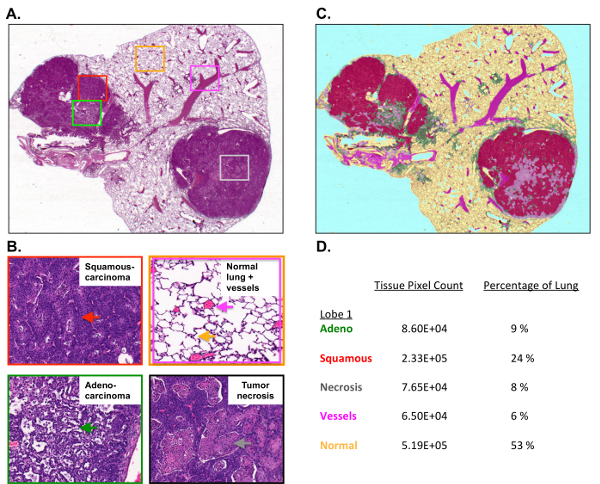
Figure 4: Tumor burden and tumor histology are quantified using morphometric software.
(A) This panel shows the H&E staining of a single mouse lung lobe with a tumor collected from a KL mouse. (B) These higher resolution images show the squamous cell carcinoma (top left), the normal lung, the vessels, and the air space (top right), and the well-differentiated papillary adenocarcinoma (bottom left) and necrosis (bottom right). (C) This panel shows the pseudocoloring of the H&E-stained lung lobe using morphometric software. (D) This panel shows the percentages for the individual lung lobe and tumor pathologies measured by Inform. Please click here to view a larger version of this figure.
Discussion
This article described an imaging-based experimental approach that utilized 18F-FDG PET/CT imaging with qIHC in order to measure both the metabolic and molecular responses in lung tumors following the delivery of the mTOR inhibitor MLN0128. MLN0128 effectively reduced the 18F-FDG consumption, indicating a significant metabolic response in the tumors. By linking PET/CT imaging to immunohistochemistry, we were able to spatially register sectioned tumors to the 3D PET/CT images and perform a detailed examination of whole tumors at a cellular and molecular level. This made it possible to confirm that MLN0128 inhibited the mTOR signaling, thus confirming an on-target molecular response to the drug in the tumors. Lastly, by taking advantage of quantitative histology, we were able to map and separate distinct tumor pathologies, such as overall tumor mass from tumor necrosis, define adenocarcinoma from squamous cell carcinomas, and complement microPET imaging.
MicroPET is currently limited by a spatial resolution of approximately 1 mm. In addition, 18F-FDG retention in certain tissues can be affected by various factors, including plasma glucose levels, the type and duration of anesthetic exposure, the environmental temperature, and the general health of the animal, which may impact 18F-FDG pharmacokinetics30. These parameters have been optimized for this protocol but should be optimized for each animal model. Reproducibility studies of 18F-FDG imaging of subcutaneous tumors in mice demonstrate a coefficient of variation for the mean %ID/g of approximately 15%, suggesting that the tumor therapeutic response of an individual mouse assessed by 18F-FDG PET should be larger than this threshold to be considered reliable and significant31.
The cellular and even subcellular distribution of PET tracers can be assessed by tissue autoradiography with the sections subsequently stained and co-registered with qIHC. Co-registering PET with CT allows a PET image to be put in an anatomical context; this is extremely valuable, even with low soft tissue contrast. The lack of soft tissue contrast by CT can be overcome with magnetic resonance imaging (MRI). In addition, biomarkers for fluorescence imaging can be used to assess glycolysis in vivo, but photon absorption and scatter in the lung cavity may affect the accurate quantitation or detection sensitivity32. In summary, utilizing whole animal PET/CT imaging with quantitative histology provides an accurate and real-time map of tumor biology following therapeutic intervention.
Multispectral imaging (MSI) is applicable in any situation where a color image might be used. At the very least, MSI provides the same information as a color image, and for some applications, MSI can provide more detailed information about the spectral properties of a sample than a simple broad-band three-color (RGB) image. In general, the limitations of MSI are those of color imaging, except that MSI is slower and takes more time to acquire images. The morphometric software was used to obtain reproducible, accurate segmentation results for the images and is described in the Table of Materials. There are additional commercially available products that can be used for tissue segmentation and the quantification of histology.
The complexity of cancer metabolism extends beyond the Warburg Effect and glucose metabolism33,34. It is highly likely that tumors will readily adapt to single agent treatments that inhibit glycolysis. The reliance on amino acid metabolism has been well documented in cancer, and it is expected that tumors rely on a host of amino acids such as glutamine, glycine, and serine, as well as other metabolites such as free fatty acids35,36,37. In addition to 18F-FDG, probes such as 18F- and 11C-labeled glutamine, choline, acetate, 1-(2'-Deoxy-2'-fluoroarabinofuranosyl)cytosine (FAC), and fluorothymidine (FLT) have been successfully used to image amino acid, nucleotide, and lipid metabolism in animal models of cancer38,39,40,41. Automation and microscale tracer radiochemistry technologies coupled with higher resolution, higher sensitivity PET scanners will improve the accessibility of PET for measuring various biological procedures42,43. As the understanding of metabolism increases, it is logical that the repertoire of PET radiotracers will increase as well, enabling researchers and physicians to noninvasively profile tumor metabolism.
The utilization of PET/CT imaging and quantitative histology addresses a clinical need, which is to rapidly translate bench discoveries into clinical use. To accomplish this, researchers must be able to accurately measure the therapeutic response as well as the acquired resistance to drugs, which PET/CT imaging enables. Moreover, the PET/CT and immunohistochemical analysis of lung tumors are used as standard of care for patients, and thus, are directly translatable into clinical practice. Importantly, PET/CT imaging readily identifies therapy-resistant tumors, which researchers can isolate and interrogate at a molecular level in order to better understand the mechanisms of disease. This is an iterative process that has made it possible to better understand the mechanisms of resistance and design more effective therapeutic strategies for the clinical translation.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank the University of California Los Angeles’ Crump Preclinical Imaging Technology Center for their assistance with the PET/CT imaging of the mice, the Translational Pathology Core Laboratory and Statistics Core at the University of California Los Angeles’ David Geffen School of Medicine for their assistance with the tumor sample preparation and analysis. For funding, David B. Shackelford was supported by the CTSI and KL2 Translational Science Award grant numbers KL2TR000122 and UL1TR000124 at the David Geffen School of Medicine at UCLA and by the Department of Defense Lung Cancer Research Program Translational Research Partnership W81XWH-13-1-0459 and ACS RSG-16-234-01-TBG. Sean T. Bailey was supported by an NIH T32 training grant HL072752 through the David Geffen School of Medicine at UCLA. Anthony Jones is supported by the UCLA Tumor Cell Biology Training Program (USHHS Ruth L. Kirschstein Institutional National Research Service Award # T32 CA009056). Gihad Abdelhady is supported by an NIH/NCI Diversity Supplement R01CA208642.
Materials
| G8 PET/CT | Perkin Elmer | CLS139564 | Used for 18F-FDG PET and CT imaging of mice |
| Axio Imager.M2 | Zeiss | 490020-0003-000 | Acquiring images of FFPE lung tumor sections |
| Inform software | Perkin Elmer | CLS135781 | Morphometric used for image analysis of tumor pathologies |
| Glut1 antibody | Alpha Diagnostics | GT12-A | IHC staining of FFPE lung tumor sections |
| Phospho-S6 Ribosomal Protein (Ser235/236) (D57.2.2E) XP™ Rabbit mAb | Cell Signaling Technologies | 4858 | IHC staining of FFPE lung tumor sections |
| MX35 Premier microtome blades | Thermo Fisher Scientific | 3051835 | Microtome blades for sectioning tissue for autoradiography |
References
- Shackelford, D. B., Shaw, R. J. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nature Reviews Cancer. 9 (8), 563-575 (2009).
- Shaw, R. J., et al. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 6 (1), 91-99 (2004).
- Shackelford, D. B., et al. mTOR and HIF-1alpha-mediated tumor metabolism in an LKB1 mouse model of Peutz-Jeghers syndrome. Proceedings of the National Academy of Sciences of the United States of America. 106 (27), 11137-11142 (2009).
- Faubert, B., et al. Loss of the tumor suppressor LKB1 promotes metabolic reprogramming of cancer cells via HIF-1alpha. Proceedings of the National Academy of Sciences of the United States of America. 111 (7), 2554-2559 (2014).
- Hemminki, A. The molecular basis and clinical aspects of Peutz-Jeghers syndrome. Cellular and Molecular Life Sciences. 55, 735-750 (1999).
- Hemminki, A., et al. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 391 (6663), 184-187 (1998).
- Sanchez-Cespedes, M. A role for LKB1 gene in human cancer beyond the Peutz-Jeghers syndrome. Oncogene. 26 (57), 7825-7832 (2007).
- Sanchez-Cespedes, M., et al. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Research. 62 (13), 3659-3662 (2002).
- Ding, L., et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 455 (7216), 1069-1075 (2008).
- Ylikorkala, A., et al. Vascular abnormalities and deregulation of VEGF in Lkb1-deficient mice. Science. 293 (5533), 1323-1326 (2001).
- Bardeesy, N., et al. Loss of the Lkb1 tumour suppressor provokes intestinal polyposis but resistance to transformation. Nature. 419 (6903), 162-167 (2002).
- Miyoshi, H., et al. Gastrointestinal hamartomatous polyposis in Lkb1 heterozygous knockout mice. Cancer Research. 62 (8), 2261-2266 (2002).
- Jishage, K., et al. Role of Lkb1, the causative gene of Peutz-Jegher’s syndrome, in embryogenesis and polyposis. Proceedings of the National Academy of Sciences of the United States of America. 99 (13), 8903-8908 (2002).
- Shackelford, D. B. Unravelling the connection between metabolism and tumorigenesis through studies of the liver kinase B1 tumour suppressor. Journal of Carcinogenesis. 12, 16 (2013).
- Jackson, E. L., et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes & Development. 15 (24), 3243-3248 (2001).
- Ji, H., et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 448 (7155), 807-810 (2007).
- Momcilovic, M., et al. Heightening energetic stress selectively targets LKB1-deficient non-small cell lung cancers. Cancer Research. 75 (22), 4910-4922 (2015).
- Wislez, M., et al. Inhibition of mammalian target of rapamycin reverses alveolar epithelial neoplasia induced by oncogenic K-ras. Cancer Research. 65 (8), 3226-3235 (2005).
- Liang, M. C., et al. TSC1 loss synergizes with KRAS activation in lung cancer development in the mouse and confers rapamycin sensitivity. Oncogene. 29 (11), 1588-1597 (2010).
- Hudes, G., et al. Temsirolimus, interferon alfa, or both for advanced renal-cell carcinoma. The New England Journal of Medicine. 356 (22), 2271-2281 (2007).
- Wander, S. A., Hennessy, B. T., Slingerland, J. M. Next-generation mTOR inhibitors in clinical oncology: how pathway complexity informs therapeutic strategy. The Journal of Clinical Investigation. 121 (4), 1231-1241 (2011).
- Pourdehnad, M., et al. Myc and mTOR converge on a common node in protein synthesis control that confers synthetic lethality in Myc-driven cancers. Proceedings of the National Academy of Sciences of the United States of America. 110 (29), 11988-11993 (2013).
- Hsieh, A. C., et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 485 (7396), 55-61 (2012).
- Momcilovic, M., et al. Heightening energetic stress selectively targets LKB1-deficient non-small cell lung cancers. Cancer Research. 75 (22), 4910-4922 (2015).
- Frese, K. K., Tuveson, D. A. Maximizing mouse cancer models. Nature Reviews Cancer. 7 (9), 645-658 (2007).
- Shackelford, D. B., et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell. 23 (2), 143-158 (2013).
- Momcilovic, M., et al. Targeted Inhibition of EGFR and Glutaminase Induces Metabolic Crisis in EGFR Mutant Lung Cancer. Cell Reports. 18 (3), 601-610 (2017).
- Goodwin, J., et al. The distinct metabolic phenotype of lung squamous cell carcinoma defines selective vulnerability to glycolytic inhibition. Nature Communications. 8, 15503 (2017).
- Fueger, B. J., et al. Impact of animal handling on the results of 18F-FDG PET studies in mice. Journal of Nuclear Medicine. 47 (6), 999-1006 (2006).
- Dandekar, M., Tseng, J. R., Gambhir, S. S. Reproducibility of 18F-FDG microPET studies in mouse tumor xenografts. Journal of Nuclear Medicine. 48 (4), 602-607 (2007).
- Luker, G. D., Luker, K. E. Optical imaging: current applications and future directions. Journal of Nuclear Medicine. 49 (1), 1-4 (2008).
- Vander Heiden, M. G., Cantley, L. C., Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 324 (5930), 1029-1033 (2009).
- Vander Heiden, M. G., DeBerardinis, R. J. Understanding the intersections between metabolism and cancer biology. Cell. 168 (4), 657-669 (2017).
- Zhang, W. C., et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 148 (1-2), 259-272 (2012).
- Possemato, R., et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 476, 346-350 (2011).
- Sullivan, L. B., et al. Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell. 162 (7360), 552-563 (2015).
- Hassanein, M., et al. Preclinical evaluation of 4-[(18)F]fluoroglutamine PET to assess ASCT2 expression in lung cancer. Molecular Imaging and Biology. 18 (1), 18-23 (2016).
- Qu, W., et al. Preparation and characterization of L-[5-11C]-glutamine for metabolic imaging of tumors. Journal of Nuclear Medicine. 53 (1), 98-105 (2012).
- Venneti, S., et al. Glutamine-based PET imaging facilitates enhanced metabolic evaluation of gliomas in vivo. Science Translational Medicine. 7 (274), 217 (2015).
- Gambhir, S. S. Molecular imaging of cancer with positron emission tomography. Nature Reviews Cancer. 2 (9), 683-693 (2002).
- Keng, P. Y., et al. Micro-chemical synthesis of molecular probes on an electronic microfluidic device. Proceedings of the National Academy of Sciences of the United States of America. 109 (3), 690-695 (2012).
- Lazari, M., et al. ELIXYS – a fully automated, three-reactor high-pressure radiosynthesizer for development and routine production of diverse PET tracers. EJNMMI Research. 3 (1), 52 (2013).

