Preparation, Purification, and Use of Fatty Acid-containing Liposomes
Summary
Liposomes containing single-chain amphiphiles, particularly fatty acids, exhibit distinct properties compared to those containing diacylphospholipids due to the unique chemical properties of single chain amphiphiles. Here we describe techniques for the preparation, purification, and use of liposomes comprised in part or whole of these amphiphiles.
Abstract
Liposomes containing single-chain amphiphiles, particularly fatty acids, exhibit distinct properties compared to those containing diacylphospholipids due to the unique chemical properties of these amphiphiles. In particular, fatty acid liposomes enhance dynamic character, due to the relatively high solubility of single-chain amphiphiles. Similarly, liposomes containing free fatty acids are more sensitive to salt and divalent cations, due to the strong interactions between the carboxylic acid head groups and metal ions. Here we illustrate techniques for preparation, purification, and use of liposomes comprised in part or whole of single chain amphiphiles (e.g., oleic acids).
Introduction
Liposomes, or vesicles – compartments bounded by bilayer membranes comprised of amphiphilic lipids – have found use in numerous biomedical applications as delivery vehicles for pharmaceuticals, models of cell membranes, and for the development of synthetic cells. We and others have also employed liposomes as models of primitive cell membranes in early life.1,2,3,4 Typically, in such systems, we employ single-chain amphiphiles containing only one lipid hydrocarbon tail (e.g., oleic acid), as these molecules are simpler to synthesize without the benefit of the coded protein enzymes modern cells employ.
Liposomes comprised of single-chain lipids are similar to those formed from diacylphospholipids (e.g., 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, or POPC) in that the boundary is composed of bilayer membranes. Liposomes formed from either class of lipids can retain a dissolved payload, and can be downsized and purified by different techniques. Several important differences result from the unique chemical characteristics of single-chain lipids. Vesicles formed by phospholipids are stable over a broad pH range, while fatty acid vesicles are only stable at neutral to mildly basic pH (ca. 7 – 9), which requires certain pH buffer for vesicle preparation. Most of the time, this buffer may also contain specific soluble molecules for vesicle encapsulation, which can be either functional materials (e.g., RNA) for compartmentalized biochemical reactions or simple fluorescent dyes (e.g., calcein) for vesicle characterization.
The presence of only a single hydrocarbon chain produces a membrane that is both more permeable to solutes, as well as more dynamic. Additionally, the carboxylic acid head group present in fatty acids results in vesicles that are more sensitive to the presence of salt and divalent cations (e.g., Mg2+). Magnesium is one of the most important divalent cations for catalyzing biochemical reactions in protocells for origin-of-life studies. In early life, prior to the evolution of sophisticated protein enzymes, RNA may have been the dominant polymer, due to its dual capability to self-replicate and perform catalysis. One representative example of a magnesium-requiring RNA related reaction is non-enzymatic RNA copying, first demonstrated in 1960s.5 When chemically activated RNA nucleotides (i.e. 2-methylimidazolide nucleotides) bind to a preexisting primer-template complex, the 3'-hydroxyl group of the primer attacks the 5'-phosphate of an adjacent activated monomer to displace the leaving group (i.e. 2-methylimidazole), and forms a new phosphodiester bond. This RNA copying chemistry requires a high concentration of Mg2+, which needs to be chelated in order to be compatible with fatty acid protocells.6 Another Mg2+-dependent reaction is that catalyzed by the hammerhead ribozyme, which is perhaps the best-characterized catalytic RNA. This ribozyme, which can be reconstituted from two short oligonucleotides, performs a self-cleavage reaction that is convenient to monitor by a gel shift. As such, it is frequently employed as a model ribozyme in origin-of-life studies.7 Due to a requirement by this ribozyme for unliganded magnesium, liposomes are typically constructed by a mixture of fatty acid and fatty acid glycerol esters, which are more stable to magnesium.8,9 In this protocol, we present techniques we have developed for the preparation, manipulation, characterization of these vesicles and demonstrate application of these vesicles as protocells to host non-enzymatic RNA copying and hammerhead ribozyme catalysis.
Protocol
1. Vesicle preparation
- Preparation of thin films
- Use gas tight syringes to transfer certain amount of lipids as described in Table 1.1 to chloroform in a glass vial.
- Evaporate the resulting solution under a stream of nitrogen or argon in the fume hood.
- Subject the resulting thin film to house vacuum for at least 2 h to remove any chloroform residue. The lipid can be left under vacuum overnight at this point.
- Rehydration of vesicles
- Prepare 10 mL of 5 mM calcein 250 mM tris-HCl pH 8.0 hydration buffer by dissolving 31 mg of calcein powder in 2.5 mL of 1 M tris-HCl pH 8.0 buffer and adding another 7.5 mL of deionized (DI) water.
- Pipette 250 µL of above hydration buffer into an empty tube and add 1.875 µL of 10 M NaOH for a final 75 mM base (½ equivalent of total fatty acids). Transfer the solution to the thin film of dry lipid to form vesicles with a total lipid concentration of 0.15 M .
- Use high-speed (>3000 rpm) vortexer to vortex the resulting mixture for 4-5 s.
- Leave the lipid-buffer mixture on a low speed (ca. 30 rpm) rotating shaker to rehydrate, at least overnight.
- Sizing of vesicles (optional)
- Using tweezers, apply one filter support to each inner surface of the syringe ports of the extruder.
- Wet each filter support with 250 mM tris-HCl, pH 8.
- Using tweezers, apply one track-etched 100 nm polycarbonate membrane to one of the extruder O-rings and filter supports. Taking care to not tear or puncture the membrane, gently push the membrane into the O-ring so as to make good contact between the two surfaces.
- Assemble the extruder, taking care not to displace the membrane and filter supports.
- Fill an extruder syringe with ca. 0.5 mL of 250 mM tris-HCl, pH 8. Insert this syringe into one side of the extruder.
- Insert an empty syringe into the other side of the extruder.
- By hand, push the plunger of the syringe containing buffer slowly (≤ 50 µL/s), and verify that resistance is felt, indicating the track-etched membrane is in place and intact. It is helpful to practice this step without a membrane in place in order to gauge the expected level of resistance.
- Remove and empty the two syringes. It is not necessary to clean the two syringes at this stage since they contain buffer of identical composition to the liposome preparation.
- Load the liposome preparation into one of the two syringes.
- Reassemble the extruder with syringe containing vesicle sample on the left side and empty syringe on the right side.
- By hand, push the plunger of the syringe containing vesicle sample very slowly (10-25 µL/s).
- Carefully observe the syringe on the right side of the extruder; clear buffer will initially enter the right side of the extruder (ca. 50 µL, this is due to the inner dead volume of the extruder), followed by a small plume of extruded liposomes. Immediately stop pushing at this point, remove the syringe on the right side of the extruder, and discard this solution.
- Replace the syringe on the right side of the extruder and extrude until the left syringe is empty.
- Reverse the orientation of the extruder and repeat step 13. Continue for the desired number of cycles (typically 7, 9, or 11); an odd number is always used to ensure un-extruded liposomes are not collected from the syringe initially containing the pre-downsizing liposome preparation.
- Gently dispense the contents of the right syringe into an Eppendorf tube and place the tube on a low speed (ca. 30 rpm) rotating shaker for around 0.5 h.
- Proceed with vesicle purification, as described in Section 2. Extruded vesicles should be used within 24 h or re-extruded before next time use.
2. Vesicle purification
- Preparation of vesicle purification mobile phase
- Prepare 5 mL of 250 mM tris-HCl pH 8 hydration buffer by adding 1.25 mL of 1 M tris-HCl pH 8 buffer, 3.75 mL of DI water to a 15 mL Falcon tube. Then add 37.5 µL of 10 M NaOH (½ equivalent of base relative to unesterified fatty acid) to the hydration buffer. Pipette 235 µL of pure oleic acid directly into the Falcon tube resulting a vesicle solution with 0.15 M total lipids.
- Use high-speed (>3000 rpm) vortexer to vortex the mixture for 4-5 s, then tumble on a low speed rotating shaker for at least 2 h. The lipid preparation can be left overnight on the rotating shaker at this point. Filter the mobile phase through 0.22 μm syringe filter unit before use to remove any potential aggregates.
- Purification of vesicles on Sepharose
- Remove ca. 5 mL of ethanolic Sepharose 4B slurry from the bottle using a pipette. Apply this to a disposable 10 mL chromatography column.
- Allow the slurry to settle and ethanol flow-through until ethanol approaches top of the resin bed.
- Apply 5 mL of deionized water to the top of the resin and let flow-through to wash away ethanol residue.
- Apply 250 mM tris-HCl, pH 8 in 1-2 mL portions to the top of the resin, applying a new portion each time the liquid level approaches the top of the resin bed. Repeat for 3-5 times.
- Clamp the column on the retort stand, then connect the tip of column with stopcock connector and tubing to the fraction collector. Add another portion of buffer to flush the tubing, close the stopcock when the liquid level in column approaches the top of resin.
- Apply the extruded vesicles from Section 1 to the top of the resin using a 200 µL pipettor, taking care to apply the vesicle preparation as evenly as possible to the resin without touching the resin bed or column wall.
- Open the stopcock to begin the flow and start collecting fractions into a 96-well plate. Apply vesicle purification mobile phase to the top of the resin bed in 0.5-1 mL portions as the buffer depletes, taking care not to allow the resin bed to dry out. Collect in five-drop fractions, collecting at least 36 wells (three rows of a 96-well plate).
- Fluorescence characterization of purification fractions
- Take the 96-well plate from the previous section and read it on a plate reader (λex=485 nm, λem=515 nm).
- Plot the resulting fluorescence data as fluorescence vs. fraction number. Vesicles elute first, followed by the unencapsulated fraction (Figure 1).
- Repurification of vesicles to monitor vesicle leakage
- Repeat the purification and characterization steps in Sections 2.2 and 2.3. Vesicles that have retained their contents will exhibit no peak of unencapsulated solute (or a very small one of ca. 5-10% of the intensity of the vesicle peak).
3. Use of Vesicles in the Presence of Magnesium
- Use of unliganded magnesium
- Prepare and purify vesicles as described in sections 1, 2.1 and 2.2.
- Prepare 1 mL of 50 mM MgCl2 solution in 250 mM tris-HCl pH 8.0 buffer by adding 250 µL of 1 M tris-HCl pH 8.0 buffer and 50 µL of 1 M MgCl2 to 700 µL of DI water. Vortex to mix well.
- To give the desired magnesium concentration of 5 mM, mix 0.9 mL of purified vesicles and 100 µL of magnesium solution and stir rapidly to minimize vesicle disruption by transient exposure of vesicles to high local concentration of magnesium.
NOTE: Rapid mixing of magnesium solution is critical to vesicle stability. If magnesium and vesicles are not quickly mixed by immediate vortexing, some vesicles will be exposed to higher concentrations of magnesium, resulting in inconsistent results from sample to sample. - Leave the vesicle sample on a tumbler for at least 30 min before repurification as described in 2.4 to check for contents leakage. Add the same concentration of magnesium as in vesicle sample to the repurification mobile phase.
- Use of liganded magnesium
- Prepare and purify vesicles as described in sections 1, 2.1 and 2.2.
NOTE: Use ½ equivalent KOH instead of NaOH to deprotonate fatty acids; it has been found that this produces more stable liposomes in chelated MgCl2 systems. - Prepare 2M potassium citrate solution and adjust pH to 8.0 with KOH.
- Premix MgCl2 and potassium citrate at the specified ratio (ca. 1:4 for stable oleic acid vesicles) in 250 mM tris-HCl pH 8.0 buffer.
- Add premixed magnesium citrate solution to the vesicle sample, briefly vortex.
NOTE: Always premix magnesium and ligand solution. Never expose vesicles to unchelated magnesium solution alone. - Leave the vesicle sample on a tumbler for at least 30 min before repurification as described in 2.4. Add the same concentration of magnesium and citrate as in the vesicle sample to the repurification mobile phase.
- Prepare and purify vesicles as described in sections 1, 2.1 and 2.2.
4. Non-enzymatic RNA Copying in Vesicles
- Preparation of monodisperse RNA encapsulated vesicles
- Prepare a dry oleic acid film as described in section 1.1.
- Prepare vesicle rehydration buffer with 50 µM fluorescent labeled RNA primer, 150 µM RNA template and 250 mM tris-HCl pH 8.0 containing ½ equivalent KOH relative to oleic acid.
- Add 250 µL of rehydration buffer to the lipid film and follow step 1.2.2 and 1.2.3 to make vesicles with total 0.15 M lipid concentration.
- In order to make small unilamellar monodisperse vesicles, follow section 1.3 for vesicle extrusion.
- Magnesium-citrate addition and vesicle purification
- Premix magnesium and citrate stocks, and then mix this with the vesicle sample to a final lipid concentration of 0.1 M.
- Purify vesicles on Sepharose 4B size exclusion column, with 250 mM tris-HCl pH 8.0, 0.1 M lipids and given magnesium and citrate as mobile phase.
- Collect vesicle fractions by following section 2.3.
- Primer extension reaction
- Prepare 200 mM 2-MeImpG (guanosine 5'-monophosphate 2-methylimidazolide) stock solution with 250 mM tris-HCl pH 8.0. (NOTE: Follow previous published protocol10 for 2-MeImpG synthesis.)
- To initiate primer extension reaction, transfer 150 µL of 2-MeImpG stock solution to 450 µL vesicle sample to reach a final concentration of 75 mM lipid and 50 mM activated monomer. Start the timer and keep the reaction sample tumbling all the time.
- (Optional) For continuous fresh monomer feeding, transfer 300 µL of reaction solution to one chamber of a lab constructed small-volume liposome dialyzer11 and put 300 µL feeding solution in the other chamber. The feeding solution should contain all the ingredients at the same concentration as in the reaction solution, except for replacing RNA containing vesicles with empty vesicles. Each round of dialysis takes 1-24 h, depending on the analyte and membrane used.
- For kinetic studies, take a 100 µL aliquot at each time point, and repurify the aliquot through a Sepharose 4B size exclusion column with 250 mM tris-HCl pH 8.0 as the mobile phase.
- Collect the vesicle fractions in a 1.5 mL eppendorf tube.
- Pipette Triton to the vesicle fractions to a final around 0.1% v/v.
- Add 0.5 mL of cold ethanol to the tube and incubate at -20 °C for at least 2 h.
- Centrifuge all the samples at 16.1×1000 rcf (16.1 × g) for 10 min and gently pipette out the liquids. Wash the RNA pellets with 70% cold ethanol and centrifuge again for 5 min. Discard the liquid and put the tube with RNA pellets on an 80 °C heat block for 2 min to evaporate residual ethanol. Dissolve the pellets in 50 µL of 8 M Urea with 1xTBE loading buffer.
- PAGE analysis
- Prepare 20% PAGE gel by mixing 200 mL of UreaGel system concentrate, 25 mL of UreaGel system diluent, 25 mL of 10xTBE buffer, 80 µL of TEMED and 250 mg of ammonium persulfate. Quickly pour the gel mixture between clamped gel plates (35 cm × 45 cm) and insert the comb. Wait for at least 30 min till complete polymerization.
- Assemble the gel plate onto the gel stand and fill the gel boxes with 1xTBE gel running buffer. Take off the comb and flush the wells with syringe. Pre-run the gel with 60 W for 30 min.
- Heat the samples on 80 °C heat block for 2 min and load 5 µL of each sample per well.
- Turn on gel power box and set gel running with constant watts at 60 W for 2.5 h.
- Disassemble the gel plate from the gel stand, wipe clean the glass and put the whole plate in gel scanner to start the gel scanning.
- Quantify the gel with ImageQuant TL 1D analysis. Plot the natural logarithm of the ratio of the amount of primer remaining at a given time point to the initial amount of primer vs. time, fit to a line, and calculate the pseudo-first order reaction rate (Figure 2).
5. Hammerhead RNA Self Cleavage in Vesicles
- Preparation and purification of ribozyme encapsulated in vesicles
- Prepare a lipid thin film (OA: GMO=9:1) as in section 1.1 with components as in Table 1.2. NOTE: warm GMO to at least 60 °C for complete melting before using.
- Prepare vesicle rehydration buffer with 5 µM of each hammerhead ribozyme strand and 250 mM tris-HCl pH 8.0.
- Add 250 µL of rehydration buffer to the lipid film and follow steps 1.2.2 and 1.2.3 to make vesicles with 0.15 M total lipid concentration.
- In order to make small unilamellar monodisperse vesicles, follow section 1.3 for vesicle extrusion.
- Purify vesicles on a Sepharose 4B size exclusion column, with 250 mM tris-HCl pH 8.0, 0.15 M mixed lipids as mobile phase.
- Hammerhead ribozyme self-cleavage reaction
- Prepare magnesium solution and mix with purified vesicles as described in section 3.1 to initiate the self-cleavage reaction.
- For kinetic studies, take 100 µL of the mixture at each time point and directly repurify this aliquot through a Sepharose 4B size exclusion column with 250 mM tris-HCl pH 8.0 as mobile phase. Collect the vesicle fraction.
- PAGE Analysis
- Follow steps 4.3.6 to 4.3.8 to prepare RNA loading sample.
- Place commercially casted 15% TBE-Urea gel into gel box and fill the gel box with 1xTBE gel running buffer.
- Heat the samples on 80 °C heat block for 1 min and load 5 µL of each sample per well.
- Run the gel with constant 200 V for around 1 h.
- Scan the gel.
- Quantify the gel with an analysis software, like ImageQuant TL 1D, to measure the extent of cleavage by comparing the intensity of remaining substrate RNA band and cleaved RNA fragment band (Figure 3).
6. Giant Fatty Acid Vesicles for Microscopy
- Preparation of giant fatty acid vesicles
- Follow section 1.1 and Table 1.3 to prepare an oleic acid thin film with 0.2 mol% of fluorescently labeled lipid for better membrane observation.
- Rehydrate lipid film with 500 µL 250 mM tris-HCl pH 8.0 to make the vesicle sample 10 mM lipids in total. The buffer may contain fluorescent dyes or RNA molecules if desired. Leave vesicle sample tumbling overnight.
- Prepare 150 mL of pure fatty acid dialysis buffer with 10 mM total lipids by mixing 470 µL of oleic acid and 150 mL of 250 mM tris-HCl pH 8.0 containing ½ equivalent NaOH.
- Extrude the vesicle sample through a 10 µm polycarbonate membrane to remove large aggregates.
- Transfer the vesicle sample into 1 µm large-pore dialysis cassette, as previously described.12
- Place the dialysis cassette into a 100-mL beaker containing 30 mL dialysis buffer. Agitate the beaker gently on a table-top shaker at 80-100 rpm.
- Change the dialysis buffer every 30 min for 5 times to remove free dyes or RNA and small vesicles.
- Carefully remove the vesicle sample from the dialysis cassette with a syringe and transfer to an Eppendorf tube.
- Microscopy Observation
- Pipette 10 µL of vesicles onto a clean standard microscope slide and place a glass cover slip over the sample. Seal cover glass with transparent nail polish.
- Observe the vesicles with a 60X oil-dispersion or similar objective by confocal microscopy using the appropriate laser source for the fluorescently-labeled membrane, RNA or encapsulated dye (Figure 4).
Representative Results
We typically perform liposome purifications on size-exclusion columns. Typical liposome preparations contain a fluorophore of some kind. When liposomes are generated and extruded, the species to be encapsulated are present both inside and outside of the liposomes. By purifying liposomes on a size-exclusion resin (Sepharose 4B), unencapsulated solutes are retained within the pores of the resin, while the larger liposomes are not and elute first (Figure 1A). Collecting fractions and plotting fluorescence vs. fraction number (Figure 1B) typically yields a two-peak trace, with the early-eluting fractions corresponding to the liposomes, which are then collected and used in subsequent applications.
We frequently examine nonenzymatic primer extension reactions, which were a likely means of RNA replication prior to the emergence of ribozyme and protein-based RNA polymerases. These reactions typically employ a fluorescently labeled primer (Figure 2A), which is extended by activated monomers. These reactions can be monitored by gel electrophoresis (Figure 2B) and the resulting electropherograms integrated to obtain rate constants for a given reaction condition (Figure 2C).
To demonstrate that RNA could function inside protocells, we employ hammerhead ribozyme self-cleavage (Figure 3A) as a model RNA catalytic reaction. This reaction requires free Mg2+ to facilitate catalysis, and therefore we used OA/GMO vesicles since they are stable in the presence of 5 mM Mg2+. Similar to primer extension reactions, the hammerhead ribozyme self-cleavage reaction can also be monitored by gel electrophoresis (Figure 3B) and later analyzed to acquire the rate constant under specific conditions (Figure 3C).
We image liposomes employing both fluorescence and transmitted light. Liposomes can be labeled using fluorescent lipids, which give a membrane label (Figure 4A), or using a fluorescent solute within their lumen (Figure 4B). Transmitted light can also be used to observe vesicles (also shown in Figure 4B).
| Table 1.1 pure oleic acid in chloroform | |||
| Component | Stock | Amount | |
| Oleic acid | >99% | 11.7 µL | |
| Chloroform | 1 mL | ||
| Table 1.2 oleic acid and glycerol monooleate (9:1) in chloroform | |||
| Component | Stock | Amount | |
| Oleic acid | >99% | 10.5 µL | |
| Glycerol monooleate | >99% | 1.4 µL | |
| Chloroform | 1 mL | ||
| Table 1.3 oleic acid with 0.2mol% Rhodamine-PE in chloroform | |||
| Component | Stock | Amount | |
| Oleic acid | >99% | 1.6 µL | |
| Rhodamine-PE in chloroform | 10 mM | 20 µL | |
| Chloroform | 1 mL | ||
Table 1. Fatty acid chloroform solutions.
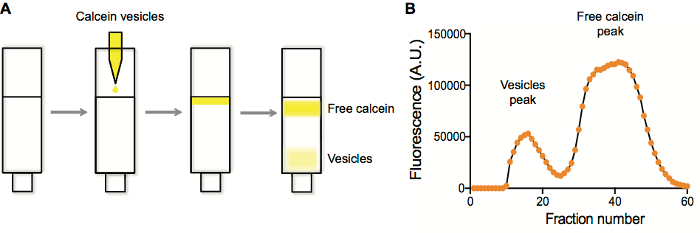
Figure 1. Vesicle purification and fluorescence characterization of purification fraction. A. Separation of vesicles containing calcein from free calcein on a Sepharose 4B column. B. Vesicle and free calcein peak detection by plotting the fluorescence in each well vs. well number after fraction collection. Please click here to view a larger version of this figure.
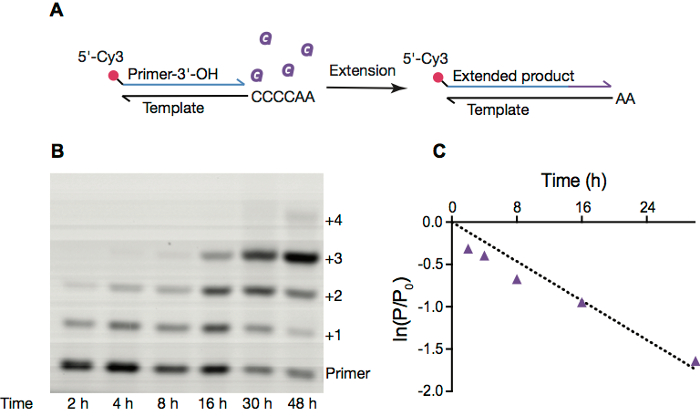
Figure 2. Non-enzymatic RNA replication inside OA vesicles. A. Scheme of non-enzymatic RNA primer extension. B. PAGE image of a primer extension reaction inside pure oleic acid vesicles, with conditions as in section 4. C. Linear fit of the natural logarithm of ratio of amount of primer remaining at given time point to the initial amount of primer vs. time over 30 h. Reaction rate, calculated from the slope of ln(P/P0) vs time, is 0.058 h-1. Please click here to view a larger version of this figure.
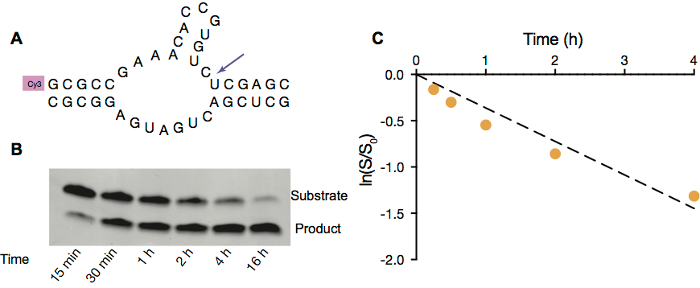
Figure 3. Hammerhead ribozyme cleavage in OA/GMO vesicles. A. Scheme of hammerhead ribozyme cleavage of fluorescently labeled substrate strand (top). B. PAGE image of hammerhead ribozyme cleavage inside OA/GMO vesicles with 5 mM Mg2+. C. Ribozyme activity inside vesicles. Linear fit of natural logarithm of ratio of amount of substrate remaining at given time point to the initial amount of substrate vs. time in first 4 h. Reaction rate, calculated from the slope of ln(S/S0) vs time, is 0.36 h-1. Please click here to view a larger version of this figure.
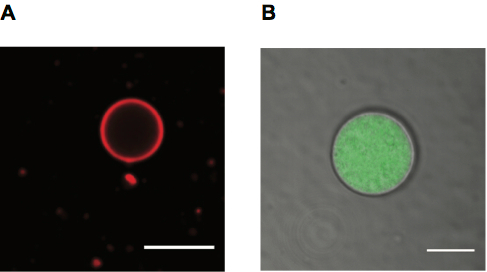
Figure 4. Giant Fatty Acid Vesicles for Microscopy. A. Confocal microscopy image of a Rhodamine PE labeled oleic acid vesicle, scale bar 10 μm. B. Confocal microscopy image of oleic acid vesicle containing Alexa488 labeled RNA with membrane shown in transmitted detector (TD) channel, scale bar 5 μm.
Discussion
Liposomes formed from fatty acids have been suggested by many as potential models for primitive cells due to their high permeability and dynamic properties. The carboxylic head group of single chain fatty acids only allows self-assembly into membranes in a restricted pH range, and the resulting membranes are quite sensitive to the presence of salts. As a result, fatty acid vesicles require different preparation and handling methods compared with phospholipid vesicles.
In this protocol, although we use oleic acid as an example for liposome formation, other long chain unsaturated fatty acids (C14) and their derivatives (ca. myristoleic acid, palmitoleic acid, and the corresponding alcohols and glycerol esters) also form vesicles following the thin film rehydration method as long as the total lipid concentration is above the cmc and the hydration buffer pH is close to the pKa of the fatty acid within the membrane. Other than tris-HCl buffer used in this protocol, other buffer systems (ca. bicine, phosphate, borate) were reported to support fatty acid vesicle formation, though vesicles formed in phosphate or borate buffer are usually quite leaky13. The resulting fatty acid vesicles after rehydration are polydisperse and multilamellar, but are easily converted into small monodisperse unilamellar vesicles by extrusion as described. Compared with sonication as an alternative method for generating small vesicles, extrusion provides more options for the control of vesicle size by applying different pore size membranes. Vesicles after extrusion are usually slightly bigger than the membrane pore size, but by increasing the number of extrusion cycles, vesicles with a narrower size distribution and an average size close to the membrane pore size can be obtained.
In order to synthesize functional protocells, fatty acid vesicles need to host specific biochemical reactions resulting from the encapsulation of RNA or other building blocks. The thin film rehydration method provides an easy way to form vesicles containing desired encapsulated materials. However, the encapsulation efficiency is relatively low and a large fraction of precious materials such as RNA are typically lost during the purification process. In some cases the encapsulation efficiency can be modestly improved by repeated freeze-thaw cycles before extrusion. Microfluidic methods for the high yield preparation of phospholipid liposomes allow for almost 100% encapsulation efficiency, however analogous methods have not yet been developed for fatty acid vesicles.
When handling protocells with either chelated or free Mg2+, purification after magnesium solution addition and repurification before each time point ensures the removal of leaked encapsulated material that might affect the accuracy of reaction rate measurements inside vesicles. Since each purification takes at least 10 min to achieve good separation and to collect vesicle fractions, the analysis of fast reactions is difficult, and the reaction must be stopped prior to column repurification.
The protocol we present here is well suited for the construction of fatty acid liposomes that host reactions mimicking those that might occur in primitive cells. Our protocols also enable potential applications in the development of biomedical delivery systems and bioreactors for other biochemical reactions.
Disclosures
The authors have nothing to disclose.
Acknowledgements
J.W.S. is an Investigator of the Howard Hughes Medical Institute. This work was supported in part by a grant (290363) from the Simons Foundation to J.W.S. Both A.E.E. and K.P.A. acknowledge support from University of Minnesota startup funds.
Materials
| sephorose 4B | SIGMA-ALDRICH INC | 4B200 | |
| calcein | SIGMA-ALDRICH INC | C0875-10G | |
| tris-HCl pH8.0 1M | LIFE TECHNOLOGIES CORP | AM9851 | |
| citric acid | SIGMA-ALDRICH INC | 251275-500G | |
| sodium hydroxide | SIGMA-ALDRICH INC | 71690-250G | |
| potassium hydroxide | Sigma | 30614 | |
| oleic acid | Nu-Chek | U-46-A | |
| glycerol monooleate | Nu-Chek | M-239 | |
| Liss Rhodamine-PE | LIFE TECHNOLOGIES CORP | L1392 | |
| magnesium chloride | Fisher/Thermo Fisher Scientific | AM9530G | |
| sequagel concentrate | National Diagnostics | EC-830 | |
| sequagel DILUENT | National Diagnostics | EC-840 | |
| 15% TBE-UREA GEL | Thermo Fisher Scientific | EC68852BOX | |
| urea | Sigma Aldrich | U6504-500G | |
| titon-100x | SIGMA-ALDRICH INC | T9284-100ML | |
| RNA primer | IDT | 5'Cy3-GCG UAG ACU GAC UGG | |
| RNA template | IDT | 5'-AAC CCC CCA GUC AGU CUA CGC | |
| hammerhead substrate strand | IDT | 5'Cy3-GCG CCG AAA CAC CGU GUC UCG AGC | |
| hammerhead ribozyme strand | IDT | 5'GGC UCG ACU GAU GAG GCG CG | |
| vesicle extruder set | AVANTI POLAR LIPIDS | 610000 | |
| fraction collector | Gilson, Inc. | 171041 | |
| 96-well plates | Fisher | NC9995941/675 | |
| plate reader | Molecular Devices | SpectraMax i3 | |
| confocal microscope | Nikon | Nikon A1R MP Confocal | |
| gel scanner | GE Healthcare Life Sciences | Typhoon 9410 scanner |
References
- Hanczyc, M. M., Fujikawa, S. M., Szostak, J. W. Experimental Models of Primitive Cellular Compartments: Encapsulation, Growth, and Division. Science. 302, 618-622 (2003).
- Mansy, S. S., Schrum, J. P., Krishnamurthy, M., Tobé, S., Da Treco, ., Szostak, J. W. Template-directed synthesis of a genetic polymer in a model protocell. Nature. 454 (7200), 122-125 (2008).
- Apel, C. L., Deamer, D. W., Mautner, M. N. Self-assembled vesicles of monocarboxylic acids and alcohols: conditions for stability and for the encapsulation of biopolymers. Biochim. Biophys. Acta. 1559 (1), 1-9 (2002).
- Walde, P., Wick, R., Fresta, M., Mangone, A., Luisi, P. L. Autopoietic Self-Reproduction of Fatty Acid Vesicles. J. Am. Chem. Soc. 116 (26), 11649-11654 (1994).
- Sulston, J., Lohrmann, R., Orgel, L. E., Todd Miles, H. Nonenzymatic Synthesis of Oligoadenylates on a Polyuridylic Acid Template. Proc. Natl. Acad. Sci. U. S. A. 59 (3), 726-733 (1967).
- Adamala, K., Szostak, J. W. Nonenzymatic template-directed RNA synthesis inside model protocells. Science. 342 (6162), 1098-1100 (2013).
- Uhlenbeck, O. C. A small catalytic oligoribonucleotide. Nature. 328 (6131), 596-600 (1987).
- Chen, I. A., Salehi-Ashtiani, K., Szostak, J. W. RNA catalysis in model protocell vesicles. J. Am. Chem. Soc. 127 (38), 13213-13219 (2005).
- Adamala, K. P., Engelhart, A. E., Szostak, J. W. Collaboration between primitive cell membranes and soluble catalysts. Nat. Commun. 7, 1-7 (2016).
- Joyce, G. F., Inoue, T., Orgel, L. E. RNA Template-directed Synthesis on Random Copolymers. J. Mol. Biol. 176, 279-306 (1984).
- Adamala, K., Engelhart, A. E., Kamat, N. P., Jin, L., Szostak, J. W. Construction of a liposome dialyzer for the preparation of high-value, small-volume liposome formulations. Nat. Protoc. 10 (6), 927-938 (2015).
- Zhu, T. F., Szostak, J. W. Preparation of large monodisperse vesicles. PloS one. 4 (4), e5009 (2009).
- Zhu, T. F., Budin, I., Szostak, J. W. Vesicle extrusion through polycarbonate track-etched membranes using a hand-held mini-extruder. Methods Enzym. 533, (2013).

