Purification and Analytics of a Monoclonal Antibody from Chinese Hamster Ovary Cells Using an Automated Microbioreactor System
Summary
A detailed protocol for the purification and subsequent analysis of a monoclonal antibody from harvested cell culture fluid (HCCF) of automated microbioreactors has been described. Use of analytics to determine critical quality attributes (CQAs) and maximizing limited sample volume to extract vital information is also presented.
Abstract
Monoclonal antibodies (mAbs) are one of the most popular and well-characterized biological products manufactured today. Most commonly produced using Chinese hamster ovary (CHO) cells, culture and process conditions must be optimized to maximize antibody titers and achieve target quality profiles. Typically, this optimization uses automated microscale bioreactors (15 mL) to screen multiple process conditions in parallel. Optimization criteria include culture performance and the critical quality attributes (CQAs) of the monoclonal antibody (mAb) product, which may impact its efficacy and safety. Culture performance metrics include cell growth and nutrient consumption, while the CQAs include the mAb’s N-glycosylation and aggregation profiles, charge variants, and molecular weight. This detailed protocol describes how to purify and subsequently analyze HCCF samples produced by an automated microbioreactor system to gain valuable performance metrics and outputs. First, an automated protein A fast protein liquid chromatography (FPLC) method is used to purify the mAb from harvested cell culture samples. Once concentrated, the glycan profiles are analyzed by mass spectrometry using a specific platform (refer to the Table of Materials). Antibody molecular weights and aggregation profiles are determined using size exclusion chromatography-multiple angle light scattering (SEC-MALS), while charge variants are analyzed using microchip capillary zone electrophoresis (mCZE). In addition to the culture performance metrics captured during the bioreactor process (i.e., culture viability, cell counts, and common metabolites including glutamine, glucose, lactate, and ammonia), spent media is analyzed to identify limiting nutrients to improve the feeding strategies and overall process design. Therefore, a detailed protocol for the absolute quantification of amino acids by liquid chromatography-mass spectrometry (LC-MS) of spent media is also described. The methods used in this protocol take advantage of high-throughput platforms that are compatible for large numbers of small-volume samples.
Introduction
Protein therapeutics are being used to treat a growing variety of medical conditions including tissue transplant complications, autoimmune disorders, and cancers1. Since 2004, the United States Food and Drug Administration (USFDA) has documented an increasing proportion of biologic license applications (BLAs) of all approvals regulated by the Center for Drug Evaluation and Research (CDER), with BLAs accounting for over 25% in 2014 and 20152.
Considering this expanding market, biopharmaceutical manufacturers are challenged with quickly delivering more product with consistent quality. Efforts to augment product yield have focused on CHO cell engineering and production line screening, though the most significant improvements are due to advances in media/feed strategy optimization and cell culture environmental controls1,3,4,5 during the manufacturing process.
Since mAbs are produced in a biological system, there can be inherent protein variability. Antibody composition can be altered post-translationally, such as glycosylation or impacted by degradation or enzymatic reactions. These structural variations may provoke dangerous immune reactions or alter antibody binding, which in turn can reduce or eliminate the intended therapeutic function5. Thus, critical quality attributes (CQAs) of monoclonal antibodies – N-glycan profile, charge variant distribution, and the percentage of antibody in monomeric form – are regularly monitored and controlled as part of a Quality by Design (QbD) approach during manufacturing processes1,6. In a regulated production environment, therapeutic proteins must meet acceptance criteria to be licensed as an approved commercial drug product7. The methods presented herein would typically be part of the quality characterization process for an antibody7,8, and any protein scientist will be familiar with their usage.
In prior work9, the application and operation of microbioreactors for high throughput screening of cell culture conditions in upstream bioprocessing has been described. The purified product obtained from the varying media conditions is subjected to N-glycan analysis using LC-MS. Glycosylation patterns of therapeutic proteins can be detected and characterized using LC-MS techniques10,11, and the presence of various glycan species has been linked to bioprocess parameters such as feed strategy, pH, and temperature12. The effect of the varying media conditions on product quality, indicated by the percentage of the resulting IgG in monomeric form, is also evaluated with Size Exclusion Chromatography- Multi-Angle Light Scattering (SEC-MALS)13,14,15. The charge variant profile represents a number of modifications16 that could impact the function of a product. Microcapillary zone electrophoresis (mCZE) is a technique that offers a considerably faster analysis time compared to traditional cation exchange (CEX) chromatography and capillary isoelectric focusing (cIEF) methods used for charge variant analysis17,18. Spent bioreactor media was analyzed to track amino acid consumption during protein production as it relates to changes in the antibody's identifying attributes19,20,21,22,23.
Protein analytics allow us to identify critical process parameters (CPPs) based on the relationships between process inputs and changes in CQAs. During bioprocess development, identifying and measuring CPPs fundamentally demonstrates process control and ensures that the product has not changed, which is essential in highly regulated manufacturing environments. In this paper, analytical techniques to measure some of the biochemical characteristics of the protein most pertinent to product CQAs (N-glycan profile, charge variants, and size homogeneity) are presented.
Protocol
1. Purification of antibody
NOTE: The equilibration buffer for the in-house antibody is 25 mM Tris, 100 mM NaCl, pH 7.5. The elution buffer used is 0.1 M acetic acid. The buffers and resin (Protein A) are dependent on the specific antibody purified. Column volume is equivalent to the bed height of the resin. The amount of mobile phase used is determined in terms of column volume.
- Initializing the purification system
- Open the software attached to the purification system. Using manual instructions, equilibrate the column with the equilibration buffer at a flow rate of 2 mL/min for 40 min. Stop the manual run after equilibration.
- In the fraction collector, place 15 mL conical tubes to collect purified antibody eluate and 50 mL conical tubes to collect flow-through during high salt wash. Ensure that the fraction collector is reset to the start position by opening and closing the fraction collector before the beginning the run. The fraction collector is maintained at 7 °C.
NOTE: The fraction collector can be reset manually under the Fraction Collector tab in Settings, for both 15 mL and 50 mL tubes.
- Sample injection
NOTE: The harvested cell culture fluid used in the following procedures has been obtained from chinese hamster ovary cells cultured in automated micro-bioreactors9.- Add the 0.22 µm filtered harvest cell culture fluid to an empty 12 mL syringe whose nozzle end is capped.
- Holding the syringe with the nozzle facing down, insert the syringe plunger till a small portion of the plunger is in. Making sure that the fluid is not leaking, turn the syringe with the nozzle facing up and remove the cap.
- Still holding the syringe with the nozzle facing up, push the cylinder to dispel any air until the cell culture fluid is at the tip of the nozzle. Insert the syringe nozzle into the manual injection port on the purification system and twist to tighten.
- Push down on the plunger until all the sample is injected and is visible in the attached 10 mL large volume sample loop.
- Open the saved method file. Save the result file in the required location and specify file name when prompted. Hit run after the sample has been injected into the large volume sample loop.
- Running the purification method
- Select the saved method and click run when prompted by instrument software (step 1.2.5).
NOTE: The system is set up to run the following steps. The user needs not do anything while the instrument is running. - Equilibrate the column with three column volumes (CVs) of equilibration buffer at a flow rate of 2 mL/min. Once the column is equilibrated, the system, using the large volume sample loop, will inject the sample onto the column at a flow rate of 1 mL/min.
- A drop in UV signal at 280 nm indicates that the sample is finished loading. Wash the column with equilibration buffer at a flow rate of 2 mL/min until the UV signal drops below 25 mAU.
- Use four CVs of 25 mM Tris with 1 M NaCl at pH 7.5 to perform a secondary high salt wash at a flow rate of 2 mL/min. The system fraction collector will collect any protein/DNA that comes off the column during the salt wash in 50 mL tubes.
- Apply five CVs of elution buffer at a flow rate of 1 mL/min to elute the antibody off the column. Collect the eluate in 15 mL tubes based on UV signal; when the UV 280 signal is above 35 mAU, collection starts; collection ends when the signal drops below 50 mAU; this is called peak cutting.
NOTE: Peak cutting ensures normalization of elution profiles and to avoid elution peak tailing which may contain protein aggregates24. - Wash the column using three CVs of equilibration buffer. The run ends after the wash step.
- After elution immediately neutralize the purified protein using 1 M Tris base to a pH of ~5.5. Measure the protein concentration using a microvolume UV-Vis spectrophotometer at 280 nm and 260 nm and store at 4 °C.
- Concentrate the purified antibody using centrifugal units (step 2). Then subject the purified antibody to glycan analysis using LC-MS and after aggregation profile analyze using SEC-MALS (steps 3 & 4).
NOTE: The purified antibody should not be frozen without further buffer exchange as frequent freeze-thaw cycles can cause aggregation and precipitation.
- Select the saved method and click run when prompted by instrument software (step 1.2.5).
2. Concentration of purified antibody
NOTE: The Tris-acetate buffer is 0.1 M acetic acid neutralized with 1 M Tris Base to a pH of ~5.5.
- Insert 100 kDa filters into centrifuge tubes.
- Wash the filters with 500 µL of double distilled water. Centrifuge for 10 min at 14,000 x g at room temperature (RT). Repeat this step twice. Discard filtrate.
- Transfer the rinsed filters to fresh centrifuge tubes and add 500 µL of sample to each filter. Centrifuge for 10 min at 14,000 x g.
- Invert the filter into a fresh spin tube. Centrifuge for 2 min at 1,000 x g to collect the concentrated sample.
- Determine the sample concentrations using a UV-Vis spectrophotometer. Blank the spectrophotometer using a solution of Tris-acetate buffer. Use a protein extinction coefficient of 1.37 mL•(mg•cm)-1 at 280 nm for a 1% (% m/v) IgG solution.
- Use the concentrated sample to prepare 12.5 µL of 2 mg/mL working solution for glycan analysis and 30 µL of 3.5 mg/mL working solution for SEC-MALS.
NOTE: The protocol can be paused here. Samples should be refrigerated at 4 °C. At 2 mg/mL concentration, these samples should be stable for at least three months at 4 °C while higher concentrations might precipitate.
3. Analysis of N-glycans using mass spectroscopy
- N-Glycan labeling and isolation
- Start with antibody concentrations of 2 mg/mL in an appropriate buffer such as neutral sodium phosphate, citrate or HEPES buffer. Prepare an intact mAb standard (such as NIST mAb) at 2 mg/mL to process alongside the experimental samples to serve as a positive control.
NOTE: The antibodies should be in a final buffer containing no SDS and less than 0.1 mM nucleophiles (such as Tris, DTT, glycine or histidine). SDS in the sample buffer must be removed. If nucleophiles are in the buffer, then dilute them down or perform a buffer exchange since they will interfere with the kit. The general protocol is provided with the glycan kit. - Dilute 7.5 µL of the antibodies with 15.3 µL of LC-MS-grade water in 1 mL tubes provided with the kit and then denature using 6 µL of 5% solution of an enzyme-friendly and MS-friendly surfactant at 90 °C for 3 min.
- Cool the samples for 3 min to room temperature (RT). Then, add 1.2 µL of PNGase F and incubate for 5 min at 50 °C.
- After cooling 3 min to RT, label the cleaved N-glycans by adding 12 µL of fluorescent tagging reagent dissolved in anhydrous dimethylformamide (DMF) and wait for 5 min. Dilute the labeled N-glycan mixture with 358 µL of acetonitrile (ACN).
- Place a hydrophilic interaction chromatography (HILIC) plate in a vacuum manifold with shims and waste tray. Use a multichannel pipette for large numbers of samples.
- Condition the wells with 200 µL of water, where the vacuum is adjusted so the liquid will take 15-30 s to pass through the HILIC resin. Equilibrate with 200 µL of 85% ACN prior to loading the ACN-diluted labeled glycan mixture (400 µL), applying the vacuum after each new liquid is added to the wells. Wash the resin with 600 µL of 1% formic acid (FA)/90% ACN twice.
- Replace the waste tray with 600 µL collection tubes. Elute the labeled N-glycans with SPE elution buffer (3 elutions of 30 µL each) into the collection tubes. Dilute the pooled elutions with 310 µL of DMF/ACN sample diluent. Pipette the samples into auto sampler vials to be ready for fluorescence (FLR)-MS analysis.
NOTE: These samples are stable when stored at -80 °C for at least 1 month. Store the HILIC plate in its original packaging, taped shut and inside a desiccator for future use.
- Start with antibody concentrations of 2 mg/mL in an appropriate buffer such as neutral sodium phosphate, citrate or HEPES buffer. Prepare an intact mAb standard (such as NIST mAb) at 2 mg/mL to process alongside the experimental samples to serve as a positive control.
- LC-MS analysis of labeled N-glycans
- Analyze the labeled N-glycan elution samples on an Ultra Performance Liquid Chromatography (UPLC) system coupled to a fluorescence detector and quadrupole time-of-flight (Q-ToF) mass spectrometer. Use a column approved for chromatographic separation of the labeled glycans and heat to 60 °C during separations.
NOTE: The column must be flushed with 60% acetonitrile and 40% H2O before use: 50 CVs before the first use or 20 CVs if the column has been used before.- Use 50 mM ammonium formate (AmF) (made with mobile phase concentrate) and 100% LC-MS-grade ACN for the mobile phases. The AmF is sensitive to pH changes and is usable for 1 month after mixing. Set the initial flow rate to 0.4 mL/min, with the LC gradient providing increasing AmF during the elution phase.
- Set the FLR detector to measure at EX 265/EM 425 nm with a sampling rate of 2 Hz. Set the Q-ToF to MS1 positive ion sensitivity mode, with a mass range of 100-2,000 daltons (Da), a scan time of 0.25 seconds and continuum data acquisition. Use leucine enkephalin (2 ng/µL in 50% ACN/0.1% FA) for the internal mass reference, in the "Do NOT apply correction" mode.
NOTE: The internal mass reference correction will be applied later during data processing. - Re-suspend the dextran ladder sequentially in 22.5 µL of H2O, 25 µL of DMF and 52.5 µL of ACN. Prepare 10 µL aliquots for storage at -80 °C, as the ladder is not stable for more than 24 h at higher temperatures (room temperature, 4 °C). The dextran ladder degrades after more than one freeze-thaw cycle.
- Place the samples in the auto sampler set to 10 °C. Load a vial of dextran ladder along with samples, as the retention time information of the ladder will be used for assignments while the mass information used to validate identifications. Use 10 µL injections for the samples and 7.5 µL injections for the ladder. Inject samples in triplicate. Run the loaded method.
- Analyze the labeled N-glycan elution samples on an Ultra Performance Liquid Chromatography (UPLC) system coupled to a fluorescence detector and quadrupole time-of-flight (Q-ToF) mass spectrometer. Use a column approved for chromatographic separation of the labeled glycans and heat to 60 °C during separations.
- N-Glycan identification for LC-MS data
- Perform data processing with a program optimized for hydrophilic interaction chromatography fluorescence mass spectrometry (HILIC-FLR-MS) data.
- Apply internal mass reference corrections within the program. Designate the dextran ladder injections as "Standard" in Sample Information. In the Analysis Method, set the Separation Compound retention times to those of the ladder compounds that were detected during the run.
- To ensure that Area % will be returned for identified glycans, modify the Analysis Method: Under the Processing tab, click Quantitation Settings - Calibrate and set "Calibration curve fit type" to "Relative response (%)".
4. Analysis of antibody aggregation using SEC-MALS
- Sample preparation
- Transfer the 3.5 mg/mL (step 2) diluted protein to a vial with a 150 µL glass insert. Use a gel loading tip to pipet into the bottom bell of the insert to avoid the introduction of bubbles.
- Cap the vial with a septa cap and analyze immediately. Store at 4 °C if analyzing later.
- SEC-MALS configuration and equilibration
NOTE: Analyze aggregation on SEC-MALS configured with an Ultra High-Pressure Liquid Chromatography (UHPLC) with a MALS Detector and Refractive Index Detector controlled by the MALS software.- Configure a method file in the UHPLC software for control of the UHPLC system, setting the flow rate to 0.4 mL/min with a mobile phase of 1x Phosphate Buffered Saline (PBS) (diluted from 10x), the injection volume to 5 µL, the column temperature to 25 °C, and the Diode Array Detector (DAD) to monitor 280 nm. Set run time to 20 min. Equilibrate the system for at least 4 h prior to any sample analysis.
- The interface between UHPLC and Multi Angle Light Scattering – Refractive Index (MALS-RI) detectors requires the use of the analog output on the DAD. Set the DAD attenuation to 1,000 mAU in the DAD method file and AU/UV setting to 1 (UV instrument>Channels>Channel 1).
- Turn on the DAD lamp 30 min prior to beginning analysis and set the wavelength to 280 nm. At the same time, purge the Refractive Index (RI) reference cell for 15 min or until the baseline is stable and then close the reference cell.
- Configure SEC-MALS software sequence, setting the collection time to 12 min, the injection volume to 5 µL, the dn/dc to 0.185 mL/g, A280 extinction coefficient if previously experimentally determined or to 1.37 mL*(mg*cm)-1, and the concentration of the sample. Click Run and wait for the "waiting to inject" dialog to appear on the screen.
NOTE: The A280 extinction coefficient is specific to the protein of interest and should be determined experimentally. - Configure a sample list in the UHPLC software in the same order as in MALS-RI software and submit.
NOTE: It is important to run a system suitability check before and after a run. Bovine serum albumin typically is used to check for peak broadening, a sign that the SEC column may need cleaning or replacement. The same BSA standard injection can be used to specify signal alignment, peak broadening, and detector normalization.
- Aggregate analysis with MALS software
- Click on the tab marked Procedures. Specify the minimum level of despiking required; none is usually sufficient.
- Verify that baselines have been drawn correctly and adjust if necessary, for LS1, LS2, LS3, RI, and UV channels. Set the peak area of interest.
- Review the molecular mass distribution to confirm that the called peaks contain particles of similar size.
5. Charge variant analysis
- Sample preparation and labeling
- Start with 80 µL of a 3.5 mg/mL antibody solution.Desalt the sample using a 0.5 mL Desalting Column (7k MWCO). Prepare the column by first snapping off the bottom stopper, then loosening the top stopper, and placing it in a 1.7 mL micro-centrifuge tube. Centrifuge the desalting column for 1 min at 1,500 x g.
NOTE: Mark the exterior of the column with a dot so that it can be placed in the original orientation for the next steps. - Transfer the column to a new micro-centrifuge tube. Add the 80 µL of diluted protein to the top of the column. Align the column to the original orientation. Centrifuge for 2 min at 1,500 x g. Remove the sample from the centrifuge, discard the desalting column and mix the sample well.
NOTE: Desalting is only required if the sample matrix contains primary amines, excipients that will perturb the sample electrophoresis, or other incompatible substances. - Dilute the sample to a final concentration of 2 mg/mL in a volume of 25 µL and add 5 µL of the labeling buffer (see Table of Materials: Charge Variant Reagent Kit) in the 96-well plate. Prepare the labeling reagent by diluting the necessary amount of labeling reagent (see Table of Materials: Charge Variant Reagent Kit) 1:30 in dimethylformamide. Incubate the sample for 10 min at room temperature away from light.
NOTE: It is important to thaw and then immediately use this reagent and use it within 10 min of mixing with DMF. - After incubating, add 60 µL of reagent grade water and mix well by pipetting. Cover the plate with a plate seal and centrifuge the plate at 1,000 x g for 1 min.
- Start with 80 µL of a 3.5 mg/mL antibody solution.Desalt the sample using a 0.5 mL Desalting Column (7k MWCO). Prepare the column by first snapping off the bottom stopper, then loosening the top stopper, and placing it in a 1.7 mL micro-centrifuge tube. Centrifuge the desalting column for 1 min at 1,500 x g.
- Preparing the charge variant chip
- Prepare the Charge Variant chip by removing storage solution and washing wells 1, 3, 4, 7, 8, and 10 with water. Then replace the water with pH 7.2 running buffer (see Table of Materials: Charge Variant Reagent Kit).
- Add 750 µL of pH 7.2 running buffer to the buffer tube and place the buffer tube in the indicated spot on the upper left-hand corner of the sample tray. Now remove the plate seal from the 96-well plate, press Unload Plate on the instrument user interface, and insert the plate into the GXII sample tray.
NOTE: pH 7.2 buffers were used for this analysis. pH 5.6-7.2 buffers may be used depending on the protein pI. When using lower pH buffers, longer sample run times may be required. - Press the Unload Chip button on the user interface. Ensure that the electrodes are free of any particles, and if not, clean with a lint free swab. When inserting the chip make sure that the window in the center of the chip is free of particles or smudges. If necessary, clean with a lint-free soft cloth.
NOTE: When working with capillary electrophoresis chips, remove buffer by vacuum aspiration, followed by the immediate addition of the next solution to prevent wells from drying out. To minimize the introduction of bubbles, practice reverse pipetting technique. When handling the chip, be mindful of the fragile capillary extending from the bottom of the chip, making sure that it does not dry out and does not break through rough handling. - Close the lid to the chip chamber and select the HT Protein Charge Variant assay. Click the Run button. Follow the prompts to select the sample wells, plate type, assay time (68, 90, or 100 s), and file name. Click Start at the end of the prompts.
- Chip cleanup requires the washing of each well 2x with water, followed by the addition of Storage Buffer (see Table of Materials: Charge Variant Reagent Kit). Once in storage buffer, replace the chip in the instrument and, when prompted, select the HT Protein Charge assay. On the main screen select Wash on the user interface. Once completed, remove the chip, wipe down the electrodes with water and a lint-free swab, and store the chip at 4 °C.
- Charge variant analysis
- Open the instrument analysis software. Import the run by going to File>Import Data File… and clicking on the desired *.gxd file. Only the name will be carried over to software, so renaming the wells is advantageous (Tools > Sample Name Editor). Select the files to be exported by holding down shift while selecting the files. Click File>Export… and select the Raw Data box and then the AIA Format box.
- Open the Browse Projects tab within the analysis software. Click on Database>Import Data… and select the exported *.CDF files.
- Once imported, navigate to the Injections tab, select the files to be analyzed, right click and go to Process… In the window that pops up, select the check box next to Process and select the "Use specified processing method" radio box and the desired processing method from the drop-down box. In the drop-down box immediately below labeled "How:" select Calibrate and Quantitate. Once processed, navigate to the Results tab and check the integration of the chromatograms.
NOTE: The processing method needs to be verified for each method. As a starting point, the parameters used for the current processing method are included in Supplementary file.
NOTE: Exporting of the data can be done in the form of a report or only the peak quantification may be exported. These can be done the same time as processing or from the results window.
6. Amino acid analysis
- Setting up the standard curve for absolute amino acid quantification by LC-MS
- Prepare the extended amino acid (EAA) mixture by dissolving 59.45 mg of Asn, 59.00 mg of Hyp, 65.77 mg of Gln, and 91.95 mg of Trp in 25 mL of 0.1 N HCl. The final concentration of each amino acid in the EAA mixture is 18 nmol/µL.
- Prepare the internal standard (ISTD) stock solution by dissolving 58.58 mg of Nva and 44.54 mg of Sar in 50 mL of HCl.
- Prepare the complete amino acid standards by combining the amino acid stock solution containing Ala, Asp, Arg, Cys, Glu, Gly, His, Ile, Leu, Met, Phe, Pro, Ser, Thr, Trp, Tyr, Val at 1 nmol/µL each with the EAA mixture for final amino acid concentrations of 900, 225, 90, 22.5, and 9 pmol/µL. Add the prepared ISTD stock to the amino acid standards for a final concentration of either 90 pmol/µL or 900 pmol/µL to create "low" and "high" internal standards to use as positive controls for the method.
- Place the amino acid concentrations in sample vials into the autosampler of the UPLC. Generate a calibration curve (9 to 900 pmol/µL) in the instrument software based on the amino acid standard concentrations using the following instructions.
- Use the Q-ToF in electrospray ionization (ESI) positive sensitivity mode coupled to a UPLC for intact amino acid analysis. For chromatographic separation, use a normal phase column made for amino acid separations. Prepare the following buffers with mass spectrometry grade reagents: A = acetonitrile + 0.1% formic acid and B = 100 mM ammonium formate. Set the LC flow rate to 0.6 mL/min and column temperature to 40 °C.
- Use the following 15-minute gradient conditions for amino acid separations: 14% B (0-3 min), 14-100% B (3-10 min), 100% B (10-13 min), 100-8% B (13-14 min), 8% B (14-15 min).
- Use the "Sample Type" and "Conc A" column listings in the MS acquisition program to create a calibration curve for future analysis of crude bioreactor media in the quantitation program. To make these columns appear in acquisition program, use the Customize Display… command when right-clicking on the top menu bar.
- The Sample Type for the amino acid standards will be "Standard" while the media samples will be "Analyte". Fill in the "Conc A" column with the numerical concentrations of the standards in the required units (ex: pmol/µL).
- Run the prepared amino acid standard concentrations at least twice. Validate that the UPLC instrument and mass spectrometer are working properly by checking the ISTD peaks.
- Use the "Edit Method" option in the quantitation application to create the quantitation method (*.mdb file). Define all the amino acids of interest in the quantitation application, such as the compound name, m/z value and expected retention time. Change the integration parameters for the method here.
- Use the created amino acid method on the standard samples to create the calibration curve. This curve can be exported to a *.cdb file for use with the media samples using the Export>Calibration… command.
- In the quantitation application, save the desired layout to a *.qlt file to apply to future datasets using "Save Layout As…". Name (Injection name), Area and Conc are the most important output columns.
- Amino acid analysis of crude bioreactor media by LC-MS
- Centrifuge crude bioreactor media at 1,962 x g for 5 minutes and pass through a 0.22 µm filter.
- Follow up with a perchloric acid cleanup to remove protein and particulate matter: mix the filtered bioreactor media with 0.4 N HClO4 at a 1:1 ratio and centrifuge at 14,700 x g for 5 min at RT. Collect the clarified media in autosampler vials.
NOTE: Adjust the injection volume as needed to make the amino acid concentrations fall within the calibration range. Depending on the instrument, the injection volume can be adjusted between 0.1 µL and 10 µL. - Run media samples in triplicate by LC-MS. Use "Process Samples" under the quantitation program along with the method (*.mdb) and calibration file (*.cdb). The method and calibration curve will be automatically applied to the crude media samples by the quantitation application once all the injections are completed.
- To export data for analysis in another program (such as a spreadsheet), use the "Print" command and create an *.xps or *.pdf file.
Representative Results
The harvested cell culture fluid from the automated microscale bioreactor is purified using fast protein liquid chromatography (FPLC), as seen in Figure 1 and the purified proteins' critical quality attributes (CQAs) were characterized by various downstream analytical methods. This is a key benefit of the automated microbioreactor system; differences in CQAs can be rapidly assessed across a wide range of conditions. N-glycan data from CHO-produced mAbs that are processed by mass spectrometry should appear like the chromatograms shown in Figure 2. The figure depicts a comparison between two chromatograms showing that the mannose 5 peak (M5) from one sample is considerably lower. If only a noisy baseline is observed instead of peaks, this may mean that the chromatography setup is faulty or that the procedure is not successful. Using controls, troubleshooting can be simplified. First, assess the FLR peaks from the dextran ladder; these peaks indicate that the chromatographic system is working correctly. Next, compare the experimentally obtained peaks with those obtained from a processed intact mAb standard. If peaks from the standard are visible, but no sample peaks are identified, then the mAb samples were not processed correctly. This may be due to SDS or nucleophile presence in the buffer interfering with N-glycan labeling and purification.
SEC-MALS can be used to assess two more CQAs: the aggregation profile and the molecular weight of the antibody. A representative SEC-MALS chromatogram is comparable to the one shown in Figure 3. The molecular mass distribution and the absolute molecular weight were determined using the required software with an extinction coefficient of 1.37 mL*(mg*cm)-1 and a dn/dc of 0.185 mL/g. As peak calling and setting the baseline in the software is performed manually, results may vary slightly from user to user. The absolute molecular weight of monomeric IgG1 from Figure 3 is 1.504 x 105 Da ± 0.38% (blue) and the higher order complex is 7.799 x 105 Da ± 3.0% (red). The polydispersity of the aggregates is much greater than that of the monomer, as indicated by the red molar mass distribution of Peak 1 (Figure 3). The small quantity of sample and importance of aggregation as a CQA make this technique a highly valuable complementary analytical tool to the automated microbioreactor system.
The result of mCZE is an electropherogram, such as in Figure 4, which shows the charge variant profile for a monoclonal antibody. The profile is a unique signature for the protein being investigated and is highly sensitive to the operating pH. Also visible is a free-dye peak to the left of the charge variant profile. When establishing an operating pH, there is some discretion to the operator to balance resolution and signal; in addition, the operator must ensure good separation from the free-dye peak which migrates at ~30 s. The sample can be desalted after labelling to remove this peak, though this leads to a significant loss in signal. Once an operating pH is established, the sample profiles can be compared. While generally consistent, changes in labeling efficiency or differences in excipients can lead to minor differences in the migration of a sample and the charge variant profile making electropherograms hard to directly compare. Instead, the method of comparison is usually based on the percentages of basic, main, and acidic species. In this case, relative differences as small as 1-2% can be identified using mCZE.
Amino acid consumption can be monitored to determine if depletion is causing changes in CQAs. Chromatogram readouts from the mass spectrometer can be used to evaluate the successful creation of a calibration curve for the absolute quantification of amino acids in crude bioreactor media samples. Figure 5 depicts two total ion chromatograms (TIC) and one extracted ion chromatogram (XIC) as representative results during this process. In Figure 5A, the TIC shown depicts the background signal from the buffer system as only a water blank was injected. Figure 5B depicts a representative TIC of the amino acid standard where, when compared to the water blank, small peaks that correspond to the individual amino acid species can be observed (such as lysine at 7.96 minutes). To integrate the peak and facilitate the quantification of peak area (and therefore the concentration), the XIC is used where only the signal from a defined "chromatogram mass window" is displayed. Depending on the sensitivity of the instrument and the quality of the chromatographic separation, the optimal mass window will have to be determined by the user. In this example (Figure 5C), the XIC of lysine (m/z = 147.1144) with a mass window of 10 ppm is shown where lysine in the amino acid standard elutes off the column at 8.03 minutes.
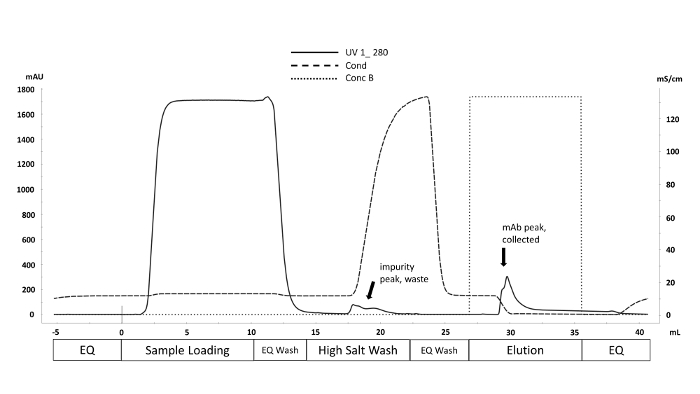
Figure 1. Representative chromatogram of the purification scheme using the Fast Protein Liquid Chromatography (FPLC) technique. Purification method phases corresponding to volume (mL) are labeled along the x-axis. UV absorbance at 280 nm (mAU y-axis, solid line) is monitored throughout the purification cycle. Non-specifically bound impurities are displaced by increasing conductivity (mS/cm y-axis, dashed line) during the High Salt Wash. Antibody is eluted from Protein A column with the introduction of elution buffer (Conc B, dotted line) when the pH decreases to 4 (not shown). Please click here to view a larger version of this figure.
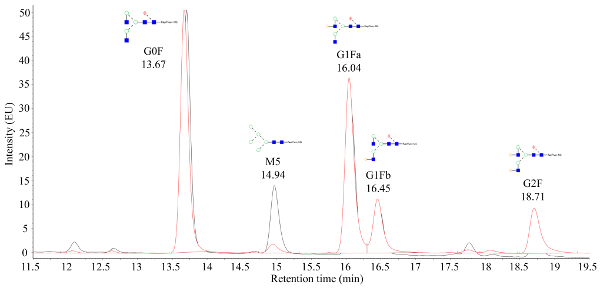
Figure 2. A representative fluorescence chromatogram obtained from tagged glycans that are mass verified. The x-axis is retention time (minutes) while the y-axis is signal intensity. The peak at 14.94 min represents the Mannose 5 (M5) glycan, where a large difference between the M5 signal strength can be observed between the two samples that are overlaid. Please click here to view a larger version of this figure.
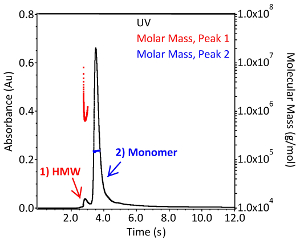
Figure 3. Molecular weight distribution of IgG1 monoclonal antibody. Chromatogram of an intact IgG1 monoclonal antibody separated by size exclusion chromatography in 1x PBS (pH7.4). Absorbance is monitored at 280 nm (black; left axis) and light scattering and refractive index detectors were used to calculate the absolute molecular weight of each peak (red and blue; right axis). High Molecular Weight species are indicated with the peak labeled "HMW". Please click here to view a larger version of this figure.
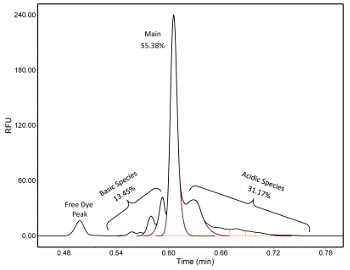
Figure 4. Charge variant profile of a IgG1 monoclonal antibody. This electropherogram is generated on a mCZE platform. A free-dye peak migrates at ~30 s and is well separated from the IgG1. For quantification, peaks were split into basic, main, and acidic species using instrument data analysis software. The red line outlines the integrated peak areas. Please click here to view a larger version of this figure.
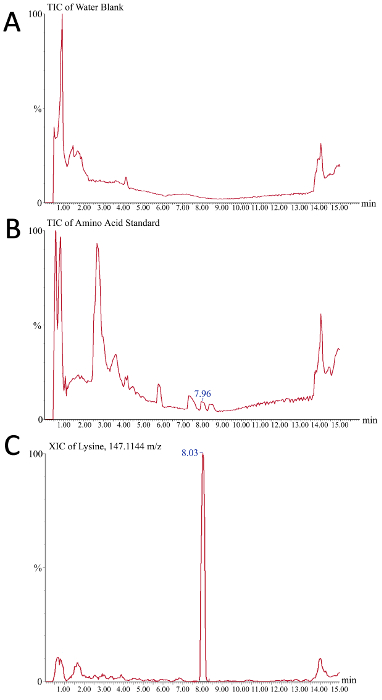
Figure 5. Representative results of the ion chromatograms for mass spectrometry-based amino acid analysis of crude bioreactor media. The x-axis is time (minutes) while the y-axis is signal intensity (A) A water blank serves as the negative control and reveals the background signal observed over the course of the liquid chromatography gradient (B) The 225 pmol/µL amino acid standard is used here as a positive control, as the individual peaks observed in this total ion chromatogram represent the different amino acids of the standard mix being resolved chromatographically (C) A representative extracted ion chromatogram for m/z 147.1144, which is lysine. The 7.96 min peak in B corresponds to the 8.03 peak in C of lysine. Please click here to view a larger version of this figure.
Discussion
HCCF contains debris and large particles that can clog and destroy costly instrumentation, thus culture clarification is needed before further downstream processing. Centrifugation is generally the first approach to separate cells and other insoluble particles from proteins followed by filtration. This filtered HCCF is then subjected to Fast Protein Liquid Chromatography (FPLC) for purification. Purification of HCCF from automated microbioreactors to obtain the product is an important step in downstream processing. Here, a benchtop FPLC system with a protein A column is used to obtain monoclonal antibodies from the HCCF. Analytics for upstream processes can provide useful insight into cell behavior and guide bioprocess design, helping to obtain a consistent and reliable quality product. Analytics also allows us to link Critical Quality Attributes (CQAs) to upstream and downstream processes. Presented here are four assays that are commonly used in the characterization of monoclonal antibodies. These techniques are robust, reliable, and readily deployable for process and product analysis from a variety of upstream sources which are only partially purified and may still contain residual levels of DNA and HCP.
When cleaning up samples for analytics, an important balance must be struck between creating a sample that is sufficiently clean for analysis while preserving the variability present in the bioreactor. The two most common contaminants impacting product are DNA and HCP, which can be checked by measuring the ratio absorbance at 260/280 nm and through SDS-PAGE or µCE-SDS. The assays presented here are not sensitive to low levels of DNA content. The purity of the product is >95% pure, as determined by µCE-SDS.
Charge variant analysis with a microcapillary electrophoresis system provides a high-throughput method to identify charge variants, with chips and reagents that are relatively easy to implement. The nature of the technique and the chemistry of the labeling reagent are both sensitive to excipients and other primary amines, thus requiring a desalting step for most sample matrixes. From experience, low levels of DNA co-migrate with the free-dye from the labeling reaction and do not impact the quality of results. While variability of basic, main, and acidic peak quantification is typically <1%, higher levels of DNA and other contaminants can increase the variability of the assay. It is extremely important to be consistent with the protein labeling and ensure the prompt use of DMF after removal from the bottle and being mixed with the dye. Lysine and/or histidine standards are recommended as labeling controls. Over time and depending on sample quality, chips can foul or lose the coating on the microfluidics channels, leading to greater noise, the presence of ghost peaks, and greater sample-to-sample variation. To identify this occurrence, blanks and a system suitability standard (i.e. NISTmAb) were concurrently analyzed with the samples at regular intervals. When chip issues arise, the chips can be washed with the storage solution or replaced.
The methods used for glycan analysis of therapeutic glycoproteins primarily involves liquid chromatography (LC) and/or mass spectrometry (MS), with lectin microarray analysis gaining popularity as a third option25. The method described in this paper uses both LC and MS, which has benefits and disadvantages. Mass spectrometric methods have the advantage of mass verification of the analyzed glycans, which is not possible with LC-based methods using a fluorescent detection output or lectin microarrays. This method uses LC and fluorescence detection to assign glycan identities using retention time comparison to a dextran ladder standard. Fluorescence monitoring allows for increased sensitivity and quantification due to the ease of its detection, where MS alone might not be able to quantify low abundance species due to the poor ionization efficiency of oligosaccharides. The mass information from MS is used to confirm glycan identities, but the processing software does not use mass information as the primary assignment criteria. Hence, without reproducible chromatography and easily resolvable peaks, this method can suffer regarding glycan assignments. Fortunately, the mass information can help with glycan assignments even in situations when the chromatography is subpar, such as shifts in retention time that hinder reproducible glycan assignments. If this method is used without MS, the chromatography must be at the highest level since mass information cannot be used to correct for residence time drift.
The amino acid analysis method described here utilizes LC-MS for rapid quantitation of underivatized amino acids in crude cell culture media. Alternative amino acid analysis methods require amino acid derivatization agents to enable UV detection26. The LC-MS method offers important advantages over the LC-UV method: it allows for identification based on both retention time and ion mass as opposed to the LC-UV method, which is limited by a lack of mass characterization. Furthermore, the LC-MS method offers time and reproducibility advantages, as the LC-UV method requires a time-consuming derivatization reaction, which may impart sample variability27. However, the injection of crude cell culture media in the LC-MS method can cause detrimental effects on MS signal due to ion skimmer fouling. A calibration ladder is injected frequently as a system suitability check, and sample order is randomized to prevent bias in the data.
The cell culture process for antibody production using microbioreactors is previously described9. In this study, detailed protocols for monoclonal antibody characterization methods that maximize data acquired from limited sample volumes are well-defined. Limited amounts of harvested cell culture fluid can sometimes restrict the product information acquired and selection of the right analytical procedures to obtain product quality data is essential. Analytics are important to link together upstream process parameters to the changes in product quality. Here, a guideline is provided for users to characterize mAbs when working with microbioreactors.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank Scott Lute for the analytical support he provided. Partial internal funding and support for this work is provided by the CDER Critical Path Program (CA #1-13). This project is supported in part by an appointment to the Internship/Research Participation Program at the Office of Biotechnology Products, U.S. Food and Drug Administration, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and FDA.
Materials
| CHO DG44 Cell Line | Invitrogen | A1100001 | |
| Akta Avant 25 | General Electric Life Sciences | 28930842 | |
| Pro Sep vA Ultra Chromatography Resin | Millipore Sigma | 115115830 | Purification Stationary Phase |
| Omnifit 10cm Column | Diba Fluid Intelligence | 006EZ-06-10-AA | Housing for Stationary Phase |
| Tris Base | Fisher Scientific | BP154-1 | |
| Superloop 10 mL | GE Healthcare | 18-1113-81 | |
| µDawn Multi Angle Light Scattering Detector | Wyatt | WUDAWN-01 | |
| 0.22 µm Millex GV Filter Unit PVDF Membrane | Merck Millipore | SLGV033RB | |
| 10X Phosphate Buffered Saline | Corning | 46-013-CM | |
| 12 mL Syringe | Covidien | 8881512878 | |
| 1290 Infinity Binary Pump | Agilent Technologies | G4220A | |
| 1290 Infinity DAD | Agilent Technologies | G4212A | |
| 1290 Infinity Sampler | Agilent Technologies | G4226A | |
| 1290 Infinity Thermostat | Agilent Technologies | G1330B | |
| 1290 Infinity Thermostatted Column Compartment | Agilent Technologies | G1316C | |
| 15 mL Falcon tube | Corning Inc. | 352097 | |
| 150 uL Glass Inserts with Polymer Feet | Agilent Technologies | 5183-2088 | |
| 50 mL Falcon tube | Corning Inc. | 352070 | |
| 96-Well Plate | Bio-Rad | 127737 | |
| Acetic Acid | Sigma-Aldrich | 695072 | |
| Acetonitrile | Fisher Chemical | BPA996-4 | |
| ACQUITY I-Class UPLC BSM | Waters Corporation | 18601504612 | |
| ACQUITY I-Class UPLC Sample Manager | Waters Corporation | 186015000 | |
| ACQUITY UPLC FLR Detector | Waters Corporation | 176015029 | |
| Amicon Ultra-4 100 kDa centrifugal filters | Merck Millipore | UFC810096 | |
| Amino Acid Standard, 1 nmol/µL | Agilent Technologies | 5061-3330 | |
| Amino Acid Supplement | Agilent Technologies | 5062-2478 | |
| Ammonium Formate Solution – Glycan Analysis | Waters Corporation | 186007081 | |
| Blue Screw Caps with Septa | Agilent Technologies | 5182-0717 | |
| CD OptiCHO AGT Medium | Thermo Fisher Scientific | A1122205 | |
| Centrifuge Tubes | Eppendorf | 22363352 | |
| Charge Variant Chip | Perkin Elmer | 760435 | |
| Charge Variant Reagent Kit | Perkin Elmer | CLS760670 | |
| Chromatography Water (MS Grade) | Fisher Chemical | W6-4 | |
| Dimethylformamide | Thermo Scientific | 20673 | |
| Extraction Plate Manifold for Oasis 96-Well Plates | Waters Corporation | 186001831 | |
| Formic Acid | Fisher Chemical | A117-50 | |
| GlycoWorks RapiFlour-MS N-Glycan Starter Kit – 24 Sample | Waters Corporation | 176003712 | |
| GXII Buffer Tubes | E&K Scientific | 697075- NC | |
| GXII Detection Window Cleaning Cloth | VWR | 21912-046 | |
| GXII HT Touch | Perkin Elmer | CLS138160 | |
| GXII Ladder Tubes | Genemate | C-3258-1 | |
| GXII Lint-Free Swab | ITW Texwipe | TX758B | |
| Hydrochloric Acid | Fisher Scientific | A144-500 | |
| Intact mAb Mass Check Standard | Waters Corporation | 186006552 | |
| Intrada Amino Acid Column 150 x 2 mm | Imtakt | WAA25 | |
| NanoDrop One Microvolume UV-Vis Spectrophotometer | Thermo Fisher Scientific | 840274100 | |
| Optilab UT-rEX Differential Refractive Index Detector | Wyatt | WTREX-11 | |
| Perchloric acid | Aldrich Chemistry | 311421 | |
| Pipet Tips with Microcapillary for Loading Gels | Labcon | 1034-960-008 | |
| Polypropylene 96-Well Microplate, F-bottom, Chimney-style, Black | Greiner Bio-One | 655209 | |
| RapiFlour-MS Dextran Calibration Ladder | Waters Corporation | 186007982 | |
| Screw Top Clear Vial 2mL | Agilent Technologies | 5182-0715 | |
| Sodium Chloride | Fisher Scientific | S271-1 | |
| Sodium Iodide | Sigma Aldrich | 383112 | |
| TSKgel UP-SW3000 4.6mm ID x 30 cm L | Tosoh Biosciences | 003449 | |
| UNIFI Scientific Information System | Waters Corporation | 667005138 | |
| Vacuum Manifold Shims | Waters Corporation | 186007986 | |
| Vacuum Pump | Waters Corporation | 725000604 | |
| Xevo G2 Q-ToF | Waters Corporation | 186005597 | |
| Zeba Spin Desalting Column, 0.5 mL | Thermo Scientific | 89883 |
References
- . . Pharmaceutical cGMPs for the 21st Century: A Risk-Based Approach. , (2004).
- New Molecular Entity (NME) Drug and New Biologic Approvals. FDA Available from: https://www.dfa.gov/Drugs/DevelopmentApprovalProcess/HowDrugsareDevelopedandApproved/DrugandBiologicAprrovalReports/NDAandBLAApprovalReports/ucm373420.htm (2015)
- Foltz, I. N., Karow, M., Wasserman, S. M. Evolution and Emergence of Therapeutic Monoclonal Antibodies. Circulation. 127, 2222-2230 (2013).
- Kondragunta, B., Drew, J. L., Brorson, K. A., Moreira, A. R., Rao, G. Advances in clone selection using high-throughput bioreactors. Biotechnology Progress. 26 (4), 1095-1103 (2010).
- Hmiel, L., Brorson, K., Boyne, M. Post-translational structural modifications of immunoglobulin G and their effect on biological activity. Analytical & Bioanalytical Chemistry. 407 (1), 79-94 (2015).
- Rathore, A. S. Roadmap for implementation of quality by design (QbD) for biotechnology products. Trends in Biotechnology. 27 (9), 546-553 (2009).
- . International Council for Harminisation of Techinical Requirements for Pharmaceuticals for Human Use. ICH. , (1999).
- Berkowitz, S. A., Engen, J. R., Mazzeo, J. R., Jones, G. B. Analytical tools for characterizing biopharmaceuticals and the implications for biosimilars. Nature Reviews Drug Discovery. 11 (7), 527-540 (2012).
- Velugula-Yellela, S. R., et al. Use of high-throughput automated microbioreactor system for production of model IgG1 in CHO cells. Journal of Visualized Experiments. , (2018).
- Largy, E., Cantais, F., Van Vyncht, G., Beck, A., Delobel, A. Orthogonal liquid chromatography-mass spectrometry methods for the comprehensive characterization of therapeutic glycoproteins, from released glycans to intact protein level. Journal of Chromatography A. 1498, 128-146 (2017).
- Yang, J. -. M., et al. Investigation of the correlation between charge and glycosylation of IgG1 variants by liquid chromatography-mass spectrometry. Analytical Biochemistry. 448, 82-91 (2014).
- Agarabi, C. D., et al. Bioreactor Process Parameter Screening Utilizing a Plackett-Burman Design for a Model Monoclonal Antibody. Journal of Pharmaceutical Sciences. 104 (6), 1919-1928 (2015).
- Wen, J., Arakawa, T., Philo, J. S. Size-Exclusion Chromatography with On-Line Light-Scattering, Absorbance, and Refractive Index Detectors for Studying Proteins and Their Interactions. Analytical Biochemistry. 240 (2), 155-166 (1996).
- Veurink, M., Stella, C., Tabatabay, C., Pournaras, C. J., Gurny, R. Association of ranibizumab (Lucentis) or bevacizumab (Avastin) with dexamethasone and triamcinolone acetonide: An in vitro stability assessment. European Journal of Pharmaceutics and Biopharmaceutics. 78 (2), 271-277 (2011).
- Li, Y., Weiss, W. F., Roberts, C. J. Characterization of high-molecular-weight nonnative aggregates and aggregation kinetics by size exclusion chromatography with inline multi-angle laser light scattering. Journal of Pharmaceutical Sciences. 98 (11), 3997-4016 (2009).
- Espinosa-de la Garza, C. E., et al. Analysis of recombinant monoclonal antibodies by capillary zone electrophoresis. Electrophoresis. 34 (8), 1133-1140 (2013).
- Han, H., Livingston, E., Chen, X. High throughput profiling of charge heterogeneity in antibodies by microchip electrophoresis. Analytical Chemistry. 83 (21), 8184-8191 (2011).
- Wheeler, T. D., et al. Microchip zone electrophoresis for high-throughput analysis of monoclonal antibody charge variants. Analytical Chemistry. 86 (11), 5416-5424 (2014).
- Carrillo-Cocom, L., et al. Amino acid consumption in naive and recombinant CHO cell cultures: producers of a monoclonal antibody. Cytotechnology. 67 (5), 809-820 (2015).
- Chen, P., Harcum, S. W. Effects of amino acid additions on ammonium stressed CHO cells. Journal of Biotechnology. 117 (3), 277-286 (2005).
- Xing, Z., et al. Optimizing amino acid composition of CHO cell culture media for a fusion protein production. Process Biochemistry. 46 (7), 1423-1429 (2011).
- Fan, Y., et al. Amino acid and glucose metabolism in fed-batch CHO cell culture affects antibody production and glycosylation. Biotechnology and Bioengineering. 112 (3), 521-535 (2015).
- Read, E. K., et al. Fermentanomics informed amino acid supplementation of an antibody producing mammalian cell culture. Biotechnology Progress. 29 (3), 745-753 (2013).
- Mazzer, A. R., Perraud, X., Halley, J., O’Hara, J., Bracewell, D. G. Protein A chromatography increases monoclonal antibody aggregation rate during subsequent low pH virus inactivation hold. Journal of Chromatography. A. 1415, 83-90 (2015).
- Zhang, L., Luo, S., Zhang, B. Glycan analysis of therapeutic glycoproteins. MAbs. 8 (2), 205-215 (2016).
- Wahl, O., Holzgrabe, U. Amino acid analysis for pharmacopoeial purposes. Talanta. 154, 150-163 (2016).
- Le, A., Ng, A., Kwan, T., Cusmano-Ozog, K., Cowan, T. M. A rapid, sensitive method for quantitative analysis of underivatized amino acids by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Journal of Chromatography B. 944, 166-174 (2014).

