Synthesis, Functionalization, and Characterization of Fusogenic Porous Silicon Nanoparticles for Oligonucleotide Delivery
Summary
We demonstrate the synthesis of fusogenic porous silicon nanoparticles for effective in vitro and in vivo oligonucleotide delivery. Porous silicon nanoparticles are loaded with siRNA to form the core, which is coated by fusogenic lipids through extrusion to form the shell. Targeting moiety functionalization and particle characterization are included.
Abstract
With the advent of gene therapy, the development of an effective in vivo nucleotide-payload delivery system has become of parallel import. Fusogenic porous silicon nanoparticles (F-pSiNPs) have recently demonstrated high in vivo gene silencing efficacy due to its high oligonucleotide loading capacity and unique cellular uptake pathway that avoids endocytosis. The synthesis of F-pSiNPs is a multi-step process that includes: (1) loading and sealing of oligonucleotide payloads in the silicon pores; (2) simultaneous coating and sizing of fusogenic lipids around the porous silicon cores; and (3) conjugation of targeting peptides and washing to remove excess oligonucleotide, silicon debris, and peptide. The particle’s size uniformity is characterized by dynamic light scattering, and its core-shell structure may be verified by transmission electron microscopy. The fusogenic uptake is validated by loading a lipophilic dye, 1,1'-dioctadecyl-3,3,3',3'-tetramethylindocarbocyanine perchlorate (DiI), into the fusogenic lipid bilayer and treating it to cells in vitro to observe for plasma membrane staining versus endocytic localizations. The targeting and in vivo gene silencing efficacies were previously quantified in a mouse model of Staphylococcus aureus pneumonia, in which the targeting peptide is expected to help the F-pSiNPs to home to the site of infection. Beyond its application in S. aureus infection, the F-pSiNP system may be used to deliver any oligonucleotide for gene therapy of a wide range of diseases, including viral infections, cancer, and autoimmune diseases.
Introduction
Gene therapy modulates specific gene expression to obtain a therapeutic outcome. Numerous tools for gene modulation have been discovered and studied, including ribonucleic acid interference (RNAi) using oligonucleotides (e.g., short interfering RNA (siRNA)1,2, microRNA (miRNA)3,4), DNA plasmids5,6, nucleases (e.g., zinc finger, TALENS)7,8, and CRISPR/Cas9 systems9,10. While each tool’s mechanism of action differs, all of the tools must reach the cell’s cytoplasm or the nucleus to be active. As such, while these tools have proven to induce significant effect in modulating gene expression in vitro, the in vivo efficacy suffers from extracellular and intracellular obstacles. Due to the fact that the tools are of biological origin, many enzymes and clearance systems exist in our body that have the ability to degrade or remove the foreign molecules11. Even in the case that the tools reach the target cell, they suffer from endocytosis; a mode of cellular uptake that encapsulates and traps the tools in acidic stomach-like vesicles that degrade or expel the tools out of the cell. In fact, studies have shown that lipid nanoparticles are endocytosed via macropinocytosis, from which approximately 70% of the siRNA are exocytosed from the cells within 24h of uptake12,13. The majority of the remaining siRNA are degraded through the lysosomal pathway, and ultimately only 1-2% of the siRNA that initially enters the cell with the nanoparticles achieve endosomal escape to potentially undergo RNAi13,14.
We have recently developed fusogenic porous silicon nanoparticles (F-pSiNPs) that have an siRNA-loaded core composed of porous silicon nanoparticles, and a fusogenic lipid shell15. The F-pSiNPs present three major advantages over other conventional oligonucleotide delivery systems: (1) a fusogenic lipid coating which enables the particles to bypass endocytosis and deliver the entire payload directly in the cell cytoplasm (versus the 1-2% achieved by endocytosed particles13,14) (Figure 1); (2) high mass loading of siRNA in the pSiNPs (>20 wt% compared to 1-15 wt% by conventional systems)15, which rapidly degrade in the cytoplasm (once the core particles shed the lipid coating via fusogenic uptake) to release the siRNA; and (3) targeting peptide conjugation for selective homing to desired cell types in vivo.
The F-pSiNP system has demonstrated significant gene silencing efficacy (>95% in vitro; >80% in vivo) and subsequent therapeutic effect in a fatal mouse model of S. aureus pneumonia; the results of which were published previously15. However, the complex structure of the F-pSiNP system requires delicate handling and fine-tuned optimization to generate uniform and stable nanoparticles. Thus, the purpose of this work is to present a thorough protocol, as well as optimization strategies for the synthesis, functionalization, and characterization of F-pSiNPs to be used in targeted delivery of siRNAs for potent gene silencing effect.
Protocol
1. Synthesis of porous silicon nanoparticles (pSINPs)
CAUTION: Always use caution when working with hydrofluoric acid (HF). Follow all safety guides according to its safety data sheet (SDS), handle any HF-containing chemicals in a fume hood, and wear appropriate personal protective equipment (PPE; double gloves with butyl gloves on the outside, butyl apron with lab coat underneath, face shield with safety goggles underneath). All universities and R&D labs require specific training on HF safety prior to usage. Do not attempt to work with HF without pre-approval of your local lab safety coordinator, as additional safety measures not described here are required.
- Preparation of the etching solutions
- To make the 3:1 HF solution for etching (used in Steps 1.3 and 1.4), fill a plastic graduated cylinder with 30 mL of aqueous 48% HF and 10 mL of absolute ethanol (EtOH). The solution must be contained in high density plastics (e.g., HDPE), as the HF dissolves glass.
- To make a 1:29 HF solution for lift-off (used in Step 1.5), fill a plastic graduated cylinder with 1 mL of aqueous 48% HF and 29 mL of absolute ethanol. The solution must be contained in high density plastics (e.g., HDPE), as the HF dissolves glass.
- Make the 1 M KOH solution in 10% EtOH for excess pSi dissolution (used in Steps 1.3 and 1.5).
- Setting up the etch cell
- In a Teflon etch cell with a 8.6 cm2 etch well (area is calculated by measuring the diameter of the silicon surface available for etching within the O-ring, and calculating the area by A = πr2), place in descending order (Figure 2a): (1) the Teflon cell top; (2) an O-ring; (3) a quarter piece of the single crystal (1 0 0)-oriented p++-type silicon wafer cut into quarters (using a diamond cutter); (4) aluminum foil; and (5) the Teflon cell base with screws gently tightened to prevent leakage.
- Fill the well with EtOH. Take a wipe and insert into the crevice between the Teflon cell top and base. If the wipe is dry upon removal, the etch cell is sealed. If wet, the cell is leaking, and tighten the screws further until sealed.
- Electropolishing the silicon wafer
- Bring the assembled etch cell into a fume hood.
- Fill the well with 10 mL of 3:1 HF solution.
- Connect the positive lead to the aluminum foil (electrode) from the etch cell and connect the negative lead to the platinum coil (counter-electrode) immersed in the 3:1 HF solution of the etch cell to complete the circuit (Figure 2b).
- Run a constant current at 50 mA cm-2 for 60 s. Once etch is finished, remove the cell from the circuit, and rinse out the 3:1 HF carefully using a syringe. Rinse with EtOH three times.
- Dissolve away the etched layer by filling the well slowly with 10 mL of 1 M KOH. Wait until the bubbling subsides.
- Rinse out the KOH with water three times, and then with EtOH three times.
- Electrochemically etching porous layers into silicon wafer
- Run an alternating current of a square waveform, with lower current density of 50 mA/cm2 for 0.6 s and high current density of 400 mA/cm2 for 0.36 s repeated for 500 cycles.
- Once etch is finished, remove the cell from the circuit, and rinse out the 3:1 HF carefully using a syringe. Rinse with EtOH three times.
- Lifting-off the porous layer from the silicon wafer
- Fill the well with 10 mL of 1:29 HF solution.
- Run a constant of 3.7 mA/cm2 for 250 s. Once etch is finished, remove the cell from the circuit. The pSi layer may have visible ripples indicating detachment from the crystalline silicon wafer. Gently wash out the 1:29 HF solution and rinse with EtOH three times, then with water three times.
- Using a pipette tip, firmly crack the circumference of the pSi layer for complete detachment. Using EtOH, collect the pSi fragments (chips) from the Teflon etch cell into a weighing boat. Transfer the chips to a glass vial. The pSi chips may be kept at room temperature in EtOH for over 6 months for storage.
- Dissolve away any remaining porous silicon on the wafer by filling the well slowly with 10 mL of 1 M KOH. Wait until the bubbling subsides.
- Rinse out the KOH with water three times, and then with EtOH three times.
- Repeat Steps 1.3-1.5 until the entire wafer thickness has been etched.
- Sonicating porous layers for nanoparticle formation
- Replace the solvent of the glass vial containing pSi chips from EtOH to 2 mL of DI water.
- Firmly close the cap, and seal using parafilm.
- Place the glass vial in a sonicator bath and suspended such that the volume of pSi chips is completely submerged below the surface.
- Sonicate for 12 h at 35 kHz and RF power of 48W. To prevent significant water loss during sonication, place a volumetric flask filled with water, inverted so that the opening of the flask touches the surface of the water bath.
- Place the glass vial on a flat surface for 1 h to allow larger particles to settle at the bottom. Collect the supernatant using a pipette; the suspension contains sub-100 nm pSiNPs.
2. Preparation of fusogenic lipid film
- Preparation of the lipid stock solutions
- In a fume hood, make 10 mg/mL stocks of lipids by hydrating DMPC, DSPE-PEG-MAL and DOTAP lipids in chloroform. These stock solutions may be kept at -20 °C for up to 6 months under tight parafilm seal.
- Making the fusogenic lipid film
NOTE: The fusogenic composition is made of the lipids, DMPC, DSPE-PEG, and DOTAP, at the molar ratio of 76.2:3.8:20 and 96.2:3.8:0, respectively.- In a glass vial, mix 72.55 μL of DMPC, 15.16 μL of DSPE-PEG, and 19.63 μL of DOTAP by pipetting.
- Optional: For fluorescent labelling of the lipid coating, additionally add 20 µL of DiI dissolved in EtOH at a concentration of 1 mg/mL.
- Place the vial in a fume hood with a loose cap to allow the chloroform to evaporate overnight. The dried film will be a cloudy hard gel-like substance at the bottom of the vial.
3. Loading and sealing of siRNA in pSiNPs
- Preparation of the calcium chloride loading stock
- Dissolve 1.11 g of CaCl2 in 10 mL of RNAse-free water to make a 2 M CaCl2 solution.
- Centrifuge the solution at > 10,000 x g for 1 min to settle the aggregates and collect the supernatant using a pipette. Alternatively, filter the solution through a 0.22 µm filter.
- Preparation of the siRNA loading stock
- Hydrate or dilute the siRNA to 150 µM in RNAse-free water. This stock solution may be aliquoted and kept frozen at -20 °C for at least 30 days given that it does not undergo frequent freeze-thaw cycles.
- Loading the siRNA in pSiNPs with calcium chloride
- Prepare an iced sonication bath.
- Under 15 min ultrasonication, gently pipette 150 µL of siRNA and 700 µL of 2 M CaCl2 into 150 µL of pSiNP. Make sure to leave the lid of the microcentrifuge tube open for gas generation.
- Remove the tube containing 1 mL of siRNA-loaded calcium coated pSiNPs from the sonicator.
4. Coating siRNA-loaded pSiNPs with fusogenic lipids
- Preparation of the liposome extrusion kit
- Fill a wide beaker with DI water. Soak 1 polycarbonate membrane (200 nm pores), and 4 filter supports by floating them on water surface. Assemble the liposome extrusion kit by following manufacturer’s instructions
- Hydrating the lipid film with siRNA-loaded pSiNps
- Hydrate the dried lipid film in the glass vial with 1 mL of of siRNA-loaded calcium coated pSiNPs obtained from step 3.3.3. Pipette until all lipids film have lifted from the bottom of the vial and have mixed into a cloudy homogenous solution.
- Place a magnetic stirring bar in the glass vial, and place the vial onto a hot plate to heat the particles to 40 °C (or high enough above the lipids’ phase transformation temperature to maintain the lipids in the more fluidic liquid crystal phase) while magnetically stirring the solution for 20 min.
- Extruding the lipid-coated pSiNPs for size control
- Aspirate the 1 mL of lipid-coated pSiNP-siRNA solution in the extrusion syringe. Insert an empty syringe on one side of the extruder, and the filled syringe on the other end.
- Begin extrusion by pushing the piston in slowly to push the particles from one syringe, through the polycarbonate membrane, and into the other. Repeat 20 times. If the piston is stuck, the filter and membrane may be clogged. Disassemble the extruder and replace the membrane and filter supports. If the problem persists, dilute the particles until clogging is manageable.
- Collect the extruded particles into a microcentrifuge tube.
5. Conjugation of targeting peptides
- Conjugating targeting peptide to lipid coating
- Prepare the peptide stock with a 1 mg/mL peptide concentration in RNAse-free water.
- In the microcentrifuge tube containing fusogenic lipid-coated pSiNPs, add 100 µL of the 1 mg/mL peptide stock and pipet gently. Keep the tube static at room temperature for 20 min (alternatively, > 2 h at 4 °C).
- To remove excess peptide or siRNA and other excipients, wash in a 30 kDa centrifugal filter by spinning at 5,000 x g at 25 °C for 1 h. Centrifuge twice more with 1 mL of PBS under the same setting.
- Resuspend the final peptide-conjugated fusogenic lipid-coated pSiNPs in PBS at the desired concentration. The particles can now be aliquoted and frozen at -80 °C for storage of at least 30 days.
Representative Results
A successful synthesis of fusogenic pSiNPs should produce a homogenous, slightly opaque solution (Figure 3a). Failure to optimize the ratio and concentration of pSiNPs : siRNA : CaCl2 may lead to aggregation upon loading (Figure 3b). As the particles are extruded through 200 nm membranes, the average hydrodynamic diameter of the fusogenic pSiNPs measured by DLS should be approximately 200 nm, and the average zeta-potential approximately +7 mV as shown in Figure 4. After surface modification with targeting peptides, the overall diameter should be increased to be under 230 nm, and the average zeta-potential decreased down to -3.4 mV15. Any extensive deviation from the extrusion size is indicative of failed extrusion (dparticle >> dextrusion), or failed loading (dparticle << dextrusion). Aggregations may also be quantified using DLS. Moreover, the frozen aliquots must be thawed only once, as repeated freeze-thaw cycles disrupt the lipid membrane and intraparticular fusion and aggregation (Figure 4). As Figure 4a shows, fusogenic pSiNPs may be stored for 30 days and thawed without causing structural changes. However, repeated freeze-thaw cycles of a single aliquot cause severe aggregation (d >> 1,000 nm) and within 4 days of the daily cycle (Figure 4b), thus it is advised that the particles be aliquoted to single-use volumes.
Fusion may be confirmed by labelling the fusogenic lipids with the lipophilic DiI (Step 2.2.2), and observing the in vitro localization using confocal microscopy. Figure 1d shows successful fusion, where the fusogenic pSiNP’s lipids transfer the DiI to the plasma membrane and are localized independent of lysosomes. Unsuccessful fusion will show the DiI localization within the cell’s cytoplasm and colocalization with lysosomes (Figure 1c).
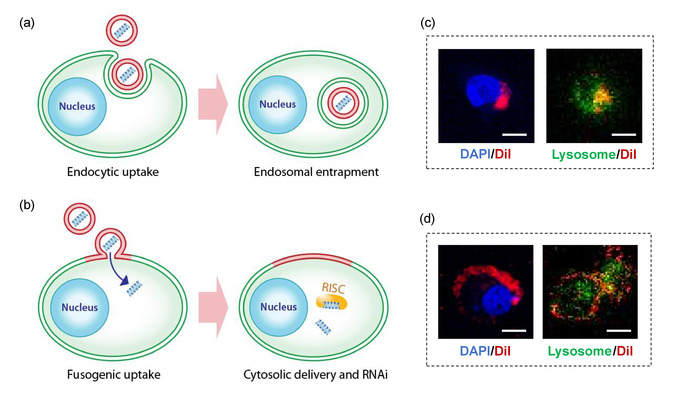
Figure 1: Fusogenic porous silicon nanoparticle system (F-pSiNP). (a) Schematic showing endocytic uptake of conventional nanoparticles and subsequent endosomal entrapment. (b) Schematic showing fusogenic uptake of the F-pSiNPs and subsequent cytosolic delivery of the siRNA payload. (c) Confocal microscopic image of a CAOV-3 cell that has endocytosed non-fusogenic pSiNPs that were loaded with DiI lipophilic dye. CAOV-3 cells were grown to 80% confluence in a 6 well-plate, and treated with 10 µL of nanoparticles, and incubated at 37 °C in 5% CO2 for 15 min. The cells were fixed in 1% paraformaldehyde to be mounted on glass slides with DAPI, dried and kept in the dark until examined by confocal microscopy. (D) Confocal microscopic image of a CAOV-3 cell that has taken up fusogenic pSiNPs that were loaded with DiI lipophilic dye. Cells were pre-stained with LysoTracker Green for 1 h at 37 °C in 5% CO2 according to manufacturer’s instructions. The cells were then washed PBS three times, and were treated with 10 μL of DiI-loaded particles for 15 min. The cells were washed with PBS three times to remove any particles that were not taken up, and the wells were filled with 1 mL of PBS and immediately taken for live-cell imaging by confocal microscopy; DAPI represents nuclear stain and Lysosome represents lysosomal staining by LysoTracker Green; scale bar represents 10 µm. Please click here to view a larger version of this figure.
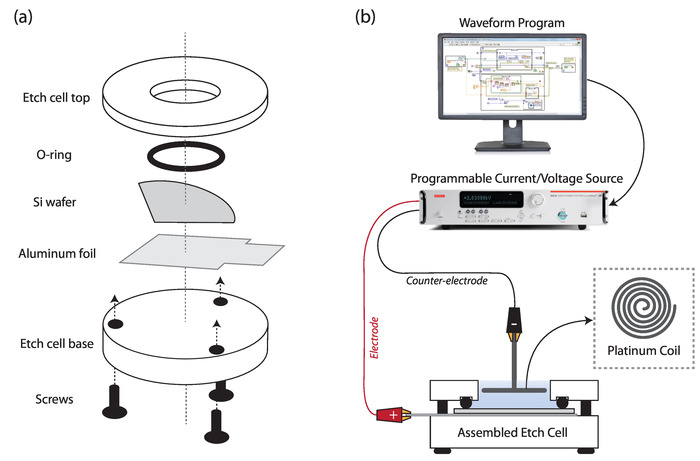
Figure 2: Etch cell setup. (a) Schematic showing etch cell components and assembly order; and (b) diagram of setup for electrochemical etching of silicon. Please click here to view a larger version of this figure.
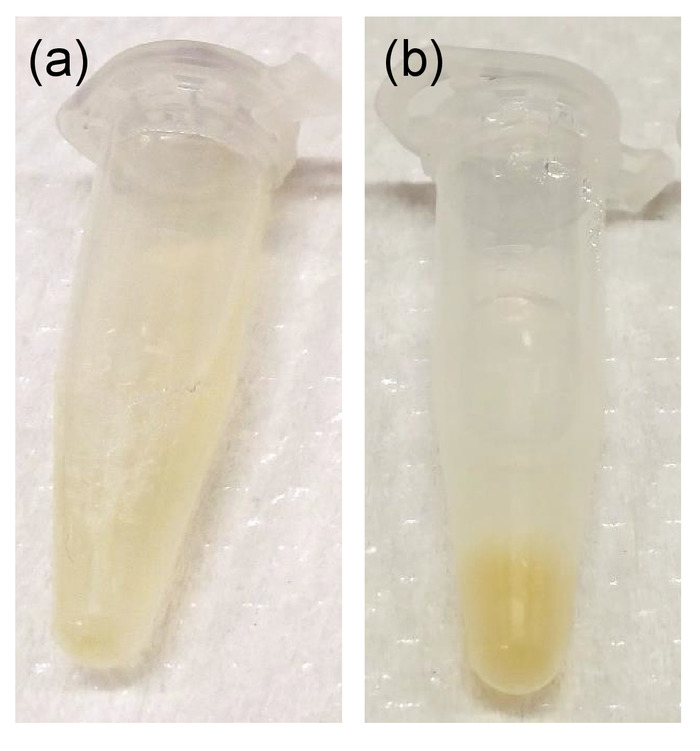
Figure 3: Photograph of final F-pSiNP product. (a) Successful synthesis showing homogenous and cloudy solution; (b) unsuccessful synthesis showing aggregation of particles (yellow). Please click here to view a larger version of this figure.
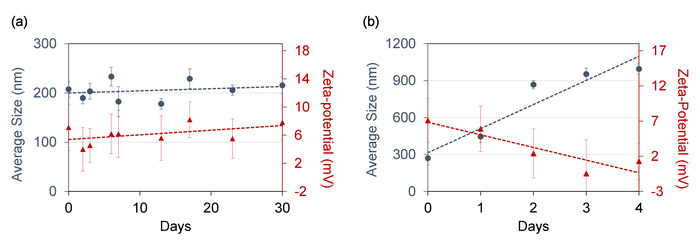
Figure 4: Repeated freeze-thaw cycle of fusogenic pSiNPs cause aggregation. (a) Average size and zeta-potential of fusogenic pSiNPs remains steady for 30 days when thawed once post-freezing; (b) Average size and zeta-potential of the fusogenic pSiNPs shows signs of aggregation and loss of structural integrity within 4 days after undergoing daily freeze-thaw cycles. Error bars represent standard deviation (n = 5). Please click here to view a larger version of this figure.
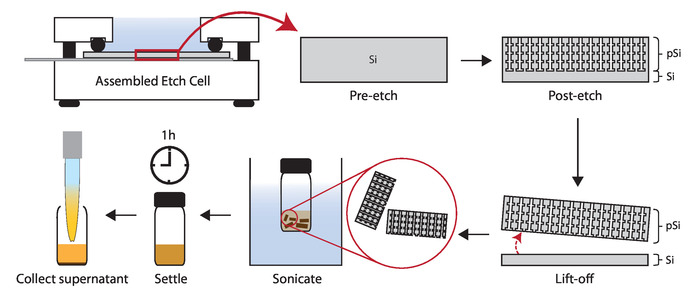
Figure 5: Diagram of porous silicon nanoparticle synthesis. Schematic showing electrochemical etching of silicon wafer (Step 1.4), lift-off of the porous layer (Step 1.5), sonication of the porous layer into particles, and collection of porous silicon nanoparticles (Step 1.6). Please click here to view a larger version of this figure.
Discussion
Synthesis of porous silicon nanoparticles is shown in Figure 5. The critical step in the synthesis of fusogenic pSiNPs is in the loading step (step 3). If the fusogenic nanoparticles are aggregating post-synthesis (Figure 3), the reason may be due to the following: (1) calcium chloride stock was not homogenously prepared, thus step 3.1.2 must be carefully followed or refined; or (2) the ratio of pSiNP : siRNA : CaCl2 or the concentration of one or more of the three components may not be optimal. Re-optimization starting from the CaCl2 concentration is suggested (e.g., altering from 2 M to 1 M or 3 M). Moreover, it is important to make sure that the CaCl2 is from the same vendor, as we found that the same chemical from different vendors resulted in lower pH at the loading step, and subsequent failure to load the siRNA. The pSiNPs may also be concentrated, diluted, or further degraded prior to the loading process by leaving the particles suspended in the RNAse-free water for 2 days after step 1.6.6.
Post-loading, the lipid coating often leads to difficulty in extrusion due to the concentration or density of the particles (step 4). If the extrusion membrane is clogged, forcing the extrusion may rip the membrane. Upon clogging, disassemble the extruder, and replace the membrane and the supports with a new set if the loss from clogging is small. If the loss is great, then dilute the particle suspension, and sonicate for 30 s prior to extrusion. If the problem persists, re-optimization of pSiNP size and loading ratio to minimize aggregates is advised. Lastly, we suggest filtering the particle suspension through a 0.22 µm-pore filter to eliminate any contaminants or aggregates prior to cell or animal treatment. Filtering is especially advised if the particles were synthesized in a non-sterile environment, or after thawing a frozen aliquot of the particles.
The fusogenicity of the particles may be validated by confocal microscopy (as shown in Figure 1c, d), and by transmission electron microscopy of the cells treated with the particles to observe for lipid-shed porous silicon cores in the cytoplasm15.
The fusogenic pSiNPs and its synthesis protocol have a few limitations. For in vitro gene silencing applications, the presented fusogenic pSiNPs have proven to induce >90% knockdown efficiency in a range of cell lines at the 100 nM dose; including the Neuro-2a mouse neuroblastoma cell line, the CAOV-3 human ovarian cancer cell line, and the notoriously difficult-to-transfect RAW 264.7 mouse macrophage cell line. However, we have observed >50% knockdown efficiency at as low as 25 nM dose, which was comparable to that of Lipofectamine. While cationic lipids must be used minimally to reduce cytotoxic effect, we have previously demonstrated its safety at a lipid dose of as high as 1 mg15. For in vivo gene silencing applications, the fusogenic pSiNPs are limited by selectivity. As the cationic lipids electrostatically attract to any cell membrane, the particles must be used as a local therapeutic, or be surface modified with a targeting moiety (e.g., peptides, antibodies, etc.). The synthesis protocol for fusogenic pSiNPs is currently optimized and limited for siRNA delivery only. The same method has been verified to successfully load miRNA, and is hypothesized to be able to load mRNA, cDNA, and other larger nucleotide payloads, but these have yet to be optimized.
The presented work makes a significant contribution to the field of gene therapy. The standard benchmark for in vitro gene silencing is the Lipofectamine 2000. We have demonstrated that the fusogenic pSiNPs can induce knockdown effect of comparable, if not higher, efficiencies15. The major advantage of the fusogenic pSiNPs over Lipofectamine 2000 is its ability to be used for in vivo applications with systemic injections. While other agents, such as Invivofectamine 3.0, have been commercialized for in vivo uses, they are only suitable for liver delivery, and require chemically modified siRNAs (with or without overhangs, or in locked nucleic acid (LNA) structure) to increase stability and specificity.16,17,18 However, the fusogenic pSiNPs can be modified post-synthesis to conjugate targeting moieties with simple thiol-maleimide chemistry, wherein a thiol group in the targeting peptide (which carries an extra cysteine for this purpose) binds a double-bonded carbon in the maleimide ring at the end of PEGylated lipids in the fusogenic coating. Moreover, the high mass loading and delivery efficacy to the cell cytoplasm minimizes the necessity for intracellular specificity and attains strong gene silencing effect with non-modified siRNAs15. One drawback of the fusogenic pSiNPs is that the synthesis protocol is a multi-day process that is more complex than the commercially available transfection agents. However, while the benchmark products must be freshly prepared prior to transfection to obtain the best results, the fusogenic pSiNPs may be aliquoted and frozen for at least 30 days without diminishing the knockdown efficiency.
Future applications for this method include optimization for loading and delivery of larger nucleic acid payloads, such as mRNAs and cDNAs. Additionally, delivery of the CRISPR/Cas9 protein-sgRNA complex, or a cocktail of the Cas9 mRNA and sgRNA, is also a development option, as the system is optimal for loading anionic payloads. Overall, the F-pSiNP system is a modular nanoplatform for gene therapy to treat diseases beyond infections, including viral infections, cancer, and autoimmune diseases.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work is supported by National Institutes of Health through contract # R01 AI132413-01.
Materials
| 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) | Avanti Polar Lipids | 850345P | Powder |
| 1,2-dioleoyl-3-trimethylammonium-propane (chloride salt) (DOTAP) | Avanti Polar Lipids | 890890P | Powder |
| 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (ammonium salt) (DSPE-PEG(2000) Maleimide) | Avanti Polar Lipids | 880126P | Powder |
| Aluminum foil | VWR International, LLC | 12175-001 | |
| Calcium chloride (CaCl2) | Spectrum | C1977 | Anhydrous, Pellets |
| Chloroform | Fisher Scientific | C6061 | |
| Computer | Dell | Dimension 9200 | Any computer with PCI card slot is acceptable |
| Dil Stain (1,1'-Dioctadecyl-3,3,3',3'-Tetramethylindocarbocyanine Perchlorate ('DiI'; DiIC18(3))) | Life Technologies | D3911 | |
| Ethanol (EtOH) | UCSD Store | 111 | 200 Proof |
| Hydrofluric acid (HF) | VWR International, LLC | MK264008 | Purity: 48% |
| Keithley 2651a Sourcemeter | Keithley | 2651A | |
| LabVIEW | National Instruments | Sample program available at: http://sailorgroup.ucsd.edu/sofware/ | |
| LysoTracker Green DND-26 | Thermo Fisher Scientific | L7526 | |
| Liposome extrusion set with heating block | Avanti Polar Lipids | 610000 | |
| Microcon-30kDa Centrifugal Filter Unit | EMD Millipore | MRCF0R030 | |
| O-ring | ChemGlass | CG-305-220 | |
| Phosphate-buffered saline (PBS) | Thermo Fisher Scientific | 10010-049 | |
| Platinum coil | VWR International, LLC | AA10285-BU | |
| Potassium hydroxide (KOH) | Fisher Scientific | P250-3 | |
| Silicon wafer | Siltronix | Custom order | |
| siRNA | Dharmacon | Custom order | IRF5, sense 5’-dTdT-CUG CAG AGA AUA ACC CUG A-dTdT-3’ and antisense 5’-dTdT UCA GGG UUA UUC UCU GCA G dTdT-3’ |
| Sonicator | VWR International, LLC | 97043-960 | |
| Targeting peptide (CRV) | CPC Scientific | Custom order | sequence CRVLRSGSC; made cyclic by a disulfide bond between the side chains of the two cysteine residues |
| Teflon etch cell | Interface Performance Materials, Inc. | Custom order | |
| UltraPure DNase/RNase-Free Distilled Water | Thermo Fisher Scientific | 10977015 |
References
- Ryther, R. C. C., Flynt, A. S., Phillips Iii, J. A., Patton, J. G. siRNA therapeutics: big potential from small RNAs. Gene Therapy. 12, 5 (2004).
- Scherman, D., Rousseau, A., Bigey, P., Escriou, V. Genetic pharmacology: progresses in siRNA delivery and therapeutic applications. Gene Therapy. 24, 151 (2017).
- Broderick, J. A., Zamore, P. D. MicroRNA therapeutics. Gene Therapy. 18, 1104 (2011).
- Geisler, A., Fechner, H. MicroRNA-regulated viral vectors for gene therapy. World Journal of Experimental Medicine. 6 (2), 37-54 (2016).
- Williams, P. D., Kingston, P. A. Plasmid-mediated gene therapy for cardiovascular disease. Cardiovascular Research. 91 (4), 565-576 (2011).
- Ferreira, G. N. M., Monteiro, G. A., Prazeres, D. M. F., Cabral, J. M. S. Downstream processing of plasmid DNA for gene therapy and DNA vaccine applications. Trends in Biotechnology. 18 (9), 380-388 (2000).
- Shim, G., et al. Therapeutic gene editing: delivery and regulatory perspectives. Acta Pharmacologica Sinica. 38, 738 (2017).
- Carlson, D. F., Fahrenkrug, S. C., Hackett, P. B. Targeting DNA With Fingers and TALENs. Molecular Therapy – Nucleic Acids. 1, (2012).
- Dai, W. -. J., et al. CRISPR-Cas9 for in vivo Gene Therapy: Promise and Hurdles. Molecular Therapy – Nucleic Acids. 5, e349 (2016).
- Adli, M. The CRISPR tool kit for genome editing and beyond. Nature Communications. 9 (1), 1911 (2018).
- Houseley, J., Tollervey, D. The Many Pathways of RNA Degradation. Cell. 136 (4), 763-776 (2009).
- Sahay, G., et al. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nature Biotechnology. 31 (7), 653-658 (2013).
- Wang, Y., Huang, L. A window onto siRNA delivery. Nature Biotechnology. 31 (7), 611-612 (2013).
- Gilleron, J., et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nature Biotechnology. 31 (7), 638-646 (2013).
- Kim, B., et al. Immunogene therapy with fusogenic nanoparticles modulates macrophage response to Staphylococcus aureus. Nature Communications. 9 (1), 1969 (2018).
- Zhang, X., et al. Targeted Disruption of G<sub>0</sub>/G<sub>1</sub> Switch Gene 2 Enhances Adipose Lipolysis, Alters Hepatic Energy Balance, and Alleviates High-Fat Diet–Induced Liver Steatosis. Diabetes. 63 (3), 934-946 (2014).
- Schlosser, K., Taha, M., Deng, Y., Stewart, D. J. Systemic delivery of MicroRNA mimics with polyethylenimine elevates pulmonary microRNA levels, but lacks pulmonary selectivity. Pulmonary Circulation. 8 (1), 2045893217750613 (2018).
- Komarov, A. P., et al. Functional genetics-directed identification of novel pharmacological inhibitors of FAS- and TNF-dependent apoptosis that protect mice from acute liver failure. Cell Death & Disease. 7, e2145 (2016).

