Analysis of Lipid Droplet Content in Fission and Budding Yeasts using Automated Image Processing
Summary
Here, we present a MATLAB implementation of automated detection and quantitative description of lipid droplets in fluorescence microscopy images of fission and budding yeast cells.
Abstract
Lipid metabolism and its regulation are of interest to both basic and applied life sciences and biotechnology. In this regard, various yeast species are used as models in lipid metabolic research or for industrial lipid production. Lipid droplets are highly dynamic storage bodies and their cellular content represents a convenient readout of the lipid metabolic state. Fluorescence microscopy is a method of choice for quantitative analysis of cellular lipid droplets, as it relies on widely available equipment and allows analysis of individual lipid droplets. Furthermore, microscopic image analysis can be automated, greatly increasing overall analysis throughput. Here, we describe an experimental and analytical workflow for automated detection and quantitative description of individual lipid droplets in three different model yeast species: the fission yeasts Schizosaccharomyces pombe and Schizosaccharomyces japonicus, and the budding yeast Saccharomyces cerevisiae. Lipid droplets are visualized with BODIPY 493/503, and cell-impermeable fluorescent dextran is added to the culture media to help identify cell boundaries. Cells are subjected to 3D epifluorescence microscopy in green and blue channels and the resulting z-stack images are processed automatically by a MATLAB pipeline. The procedure outputs rich quantitative data on cellular lipid droplet content and individual lipid droplet characteristics in a tabular format suitable for downstream analyses in major spreadsheet or statistical packages. We provide example analyses of lipid droplet content under various conditions that affect cellular lipid metabolism.
Introduction
Lipids play crucial roles in cellular energy and carbon metabolism, synthesis of membrane components, and production of bioactive substances. Lipid metabolism is fine-tuned according to environmental conditions, nutrient availability and cell-cycle phase1. In humans, lipid metabolism has been connected to diseases, such as obesity, type II diabetes and cancer2. In industry, lipids produced by microorganisms, such as yeasts, represent a promising source of renewable diesel fuels3. Cells store neutral lipids in so-called lipid droplets (LDs). These evolutionarily conserved bodies are composed of triacylglycerols, steryl esters, an outer phospholipid monolayer and associated proteins1. LDs originate in the endoplasmic reticulum, exert cell-cycle or growth-phase dynamics, and are important for cellular lipid homeostasis1. LD number and morphology can be used as a convenient proxy when assaying lipid metabolism under various growth conditions or when screening a panel of mutants. Given their dynamic nature, techniques capable of analyzing the properties of individual LDs are of particular interest in studies of lipid metabolism.
Various yeast species have been used to describe lipid-related metabolic pathways and their regulation, or used in biotechnology to produce interesting compounds or fuels1. Furthermore, for model yeasts, such as the budding yeast Saccharomyces cerevisiae or the distantly related fission yeast Schizosaccharomyces pombe, genome-wide deletion strain libraries are available that can be used for high-throughput screens4,5. Recently LD composition and dynamics have been described in S. pombe6,7,8,9, and mutants related to lipid metabolism have been isolated in the emerging model yeast Schizosaccharomyces japonicus10.
Numerous techniques are available to study LD content and dynamics. Most employ some kind of staining of LDs with lipophilic dyes such as Nile Red or BODIPY 493/503. The latter shows more narrow excitation and emission spectra, and increased specificity towards neutral lipids (LDs) as opposed to phospholipids (membranes)11. Fluorimetric and flow-cytometry methods have been used successfully in various fungal species to uncover genes and growth conditions that affect storage lipid content12,13,14,15. While these methods are suitable for high-throughput applications, they cannot measure the numbers and morphology of individual LDs in cells, which can differ dramatically between growth conditions and genotypes. Coherent Raman scattering or digital holographic microscopy are label-free methods that yield LD-level data, but require specialized expensive equipment16,17,18. Fluorescence microscopy, on the other hand, can provide detailed data on LD content, while utilizing commonly available instruments and image analysis software tools. Several analysis workflows exist that feature various degrees of sophistication and automation in cell/LD detection from image data, and are optimized for different cell types, such as metazoan cells with large LDs19,20,21, or budding yeasts17,22,23. Some of these approaches only work in 2D (e.g., on maximum projection images), which may fail to reliably describe the cellular LD content. To our knowledge, no tools exist for determination of LD content and morphology from fission yeast microscopic data. Development of automated and robust LD-level analyses would bring increased sensitivity and enhanced statistical power, and provide rich information on neutral lipid content, ideally in multiple yeast species.
We have developed a workflow for LD content analysis from 3D fluorescence microscopy images of yeast cells. Live cells are stained with BODIPY 493/503 and Cascade Blue dextran to visualize LDs and determine cell boundaries, respectively. Cells are immobilized on glass slides and subjected to z-stack imaging using a standard epifluorescence microscope. Images are then processed by an automated pipeline implemented in MATLAB, a widely used (commercial) package for statistical analyses. The pipeline performs image preprocessing, segmentation (cells vs. background, removal of dead cells), and LD identification. Rich LD-level data, such as LD size and fluorescence intensity, are then provided in a tabular format compatible with major spreadsheet software tools. The workflow was used successfully to determine the impact of nitrogen source availability on lipid metabolism in S. pombe24. We now demonstrate the functionality of the workflow in S. pombe, S. japonicus and S. cerevisiae, using growth conditions or mutants that affect cellular LD content.
Protocol
1. Preparation of Solutions and Media
- Prepare lipid staining solution.
- To prepare stock lipid staining solution dissolve 10 mg of BODIPY 493/503 in 10 mL of anhydrous DMSO (final concentration 1 mg/mL). Dissolve the whole content of a 10 mg BODIPY 493/503 vial to prevent loss of material during weighing.
CAUTION: DMSO may pass through the skin. Wear appropriate personal protective equipment. - Prepare working lipid staining solution by mixing 100 µL of the 1 mg/mL BODIPY 493/503 stock solution and 900 µL of anhydrous DMSO (final concentration 0.1 mg/mL).
- Aliquot the stock and working solutions, and store at -20 °C.
NOTE: Dissolved BODIPY 493/503 is stable for several years at -20 °C. However, the solution has to be protected from moisture and light.
- To prepare stock lipid staining solution dissolve 10 mg of BODIPY 493/503 in 10 mL of anhydrous DMSO (final concentration 1 mg/mL). Dissolve the whole content of a 10 mg BODIPY 493/503 vial to prevent loss of material during weighing.
- To prepare stock solution for cell boundary visualization, dissolve 25 mg of Cascade Blue dextran (whole vial) in 2.5 mL of deionized water (final concentration 10 mg/mL). Aliquot the stock solution and store at -20 °C protected from light.
- To prepare microscope slide coating solution, dissolve 5 mg of soybean lectin in 5 mL of deionized water (final concentration 1 mg/mL). Aliquot the lectin solution and store at -80 °C.
NOTE: The lectin solution is stable for several years at -80 °C. Aliquots currently at use may be stored at -20 °C. - Prepare cultivation media.
- To prepare 400 mL of complex YES cultivation medium for S. pombe and S. japonicus, dissolve 2 g of yeast extract and 0.1 g of SP supplements (if required for auxotrophic mutants) in 340 mL of deionized water in a 500 mL bottle and autoclave. Add 60 mL of 20% (w/v) of separately autoclaved or filter-sterilized glucose in aseptic conditions.
- To prepare 400 mL of defined EMM cultivation medium for S. pombe and S. japonicus, dissolve 4.9 g of EMM broth without dextrose in 360 mL of deionized water in a 500 mL bottle and autoclave. Add 40 mL of 20% (w/v) of separately autoclaved or filter-sterilized glucose in aseptic conditions.
NOTE: For general guidelines on S. pombe and S. japonicus cultivation see25 and26, respectively. - To prepare 300 mL of complex YPAD cultivation medium for Saccharomyces cerevisiae, dissolve 3 g of yeast extract, 6 g of peptone and 30 mg of adenine sulphate in 270 mL of deionized water in a 500 mL bottle and autoclave. Add 30 mL of 20% (w/v) of separately autoclaved or filter-sterilized glucose in aseptic conditions.
- To prepare 300 mL of defined minimal medium for S. cerevisiae, dissolve 2 g of yeast nitrogen base (without amino acids) in 270 mL of deionized water in a 500 mL bottle and autoclave. Add 30 mL of 20% (w/v) of separately autoclaved or filter-sterilized glucose in aseptic conditions.
NOTE: For general guidelines on S. cerevisiae cultivation see27.
2. Cell Cultivation
- Growing S. pombe or S. japonicus to exponential or early stationary phase.
- In the morning, inoculate 5 mL of YES medium with fresh fission yeast biomass. Incubate at 32 °C with shaking (180 rpm) for several hours.
NOTE: For all cultivations, use Erlenmeyer flasks having 10 times the volume of culture to ensure proper aeration. Some laboratories prefer to grow fission yeasts at 30 °C, but cultivation temperature of 32 °C results in shorter doubling times without detrimental effects to the cells, thus reducing the total time required to perform an experiment25,28. - In late afternoon of the same day (after at least 6 hours of cultivation), dilute the culture with fresh YES medium to a 10 mL final culture volume so that it reaches the desired optical density (OD) (or number of cells/mL) the following morning, and incubate at 32 °C with shaking (180 rpm). It is of advantage to know the doubling time of each used strain to accurately determine the dilution factor (use Equation 1).
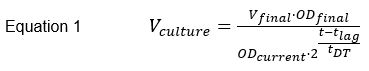
Where Vculture is the preculture volume needed for dilution, Vfinal is the total volume of the new culture (10 mL for standard cultivations), ODfinal is the desired OD to be reached the following morning, ODcurrent is the currently measured OD of the preculture, t is the time of cell growth until harvesting, tlag is duration of the lag phase (depends on laboratory conditions, needs to be empirically defined) and tDT is the doubling time of the strain.
NOTE: When exponential-phase cells are to be analyzed, do not let precultures reach the stationary phase as this dramatically alters cell physiology (including LD content) for several subsequent generations. - In the morning of imaging day, if the culture reached slightly higher OD than required (in case of exponential-phase cells), dilute it with fresh YES and continue incubation for at least two more doubling times before staining of LDs. Otherwise proceed directly to staining (Section 3).
- In the morning, inoculate 5 mL of YES medium with fresh fission yeast biomass. Incubate at 32 °C with shaking (180 rpm) for several hours.
- Growing S. cerevisiae to exponential and stationary phase.
- In the afternoon, inoculate 10 mL of YPAD medium with a small amount of fresh budding yeast biomass and incubate overnight at 30 °C with shaking (180 rpm).
- The morning of imaging day, dilute the culture to OD 0.1 in 10 mL of YPAD medium and grow to the required OD (e.g., OD 1 for exponential phase). Perform any culture dilutions as described in step 2.1.2. Proceed to staining (Section 3).
3. Lipid Droplet Staining
- Prepare a microscope cover slip for each sample to be imaged. Spread 1 µL of slide coating solution onto a clean cover slip using the long side of a horizontally positioned pipette tip. Allow the coating solution to dry completely and store the cover slips in a dust-free environment.
NOTE: Glass slides and coverslips can be cleaned prior to use if required. The cleaning procedure consists of washing with dishwashing detergent, rinsing with water, overnight soaking in 3% hydrochloric acid, and washing with distilled water. Cleaned slides and coverslips are stored in pure ethanol until use. - Measure the OD of cell culture or number of cells/mL, as required. Try to reach similar values among all tested strains to ensure comparable experimental conditions.
- Pipette 1 mL of each cell culture to a 1.5 mL microcentrifuge tube. For S. cerevisiae only, add 5 µL of the slide coating solution, vortex briefly, and incubate at 30 °C with shaking for 5 min.
- Add 1 µL of the lipid staining solution to each culture aliquot and vortex briefly. Then add 10 µL of the cell boundary visualization solution and vortex briefly.
NOTE: Do not prepare pre-mixed solutions of both stains as this leads to fluorescence quenching of BODIPY 493/503. - Collect the cells by centrifugation (1,000 x g, 3 min, RT) and remove almost all supernatant (~950 µL). Resuspend the cells in the remaining supernatant.
- Pipette 2 µL of the dense cell suspension on a lectin-coated cover slip and place onto a clean microscope slide. The cells should form a monolayer. Proceed to microscopy (Section 4) as quickly as possible to minimize artefacts in imaging; process maximum of two samples at a time.
4. Setting up the Microscope and Imaging
- Optimize imaging conditions.
NOTE: Setting up the microscope requires long exposures to strong light sources that could cause damage to the sample and skew results. Therefore, set up the imaging conditions using a dedicated sample slide that will not be further used for LD quantification.- Focus on the cells using phase contrast or differential interference contrast (DIC).
NOTE: Phase contrast or DIC images may be taken for reference, but they are not used during the automated image analysis step. - Set z-stack settings to span the whole cell volume. The total vertical distance depends on the cell size; the number of optical slices depends on the numerical aperture of the objective (point spread function in z-axis). Set the focus to move relative to the central focal plane.
NOTE: The optimal number of slices is often set by the microscope control software and does not need to be calculated manually. The typical cell widths are 3-5 µm for S. pombe, 4-7 µm for S. japonicus, and 3-7 µm for S. cerevisiae. - To image LDs, set light intensity and exposure time in the green channel (excitation and emission maxima of BODIPY are 493 and 503 nm, respectively).
NOTE: BODIPY 493/503 is a very bright fluorochrome; however, it may get bleached rapidly with overly strong light intensity. Moreover, LDs are mobile in live cells, thus minimize exposure time and capture the full green-channel z-stack first (before switching to the blue channel) to prevent blurring artifacts. Also, take into account the linear range of the camera for signal intensity to avoid saturated pixels. - To image cell boundaries, set light intensity and exposure time in the blue channel (excitation and emission maxima of Cascade Blue dextran are 400 and 420 nm, respectively).
NOTE: Signal intensity in the blue channel is required for image segmentation, but it is not used for LD quantification itself. Therefore, optimal settings in this channel are not crucial for analysis. - If possible, create an automated experimental workflow in the microscope control software to facilitate imaging of multiple samples under standardized conditions.
- Focus on the cells using phase contrast or differential interference contrast (DIC).
- Once imaging conditions have been optimized, image samples to be used for quantification. Focus on the cells and image them in green and blue channels as described in Step 4.1.
NOTE: All images must be acquired using the same settings to allow comparison between samples. Image multiple fields of view per sample to obtain robust, representative data. - Save the blue and green channel z-stack images as 16-bit multi-layer TIFF files (i.e., two files per field of view). Include words “green” or “blue” in the corresponding file names. Proceed with image analysis (Section 5).
5. Image Analysis
- Visually check the quality of acquired images.
- Open microscopic images in ImageJ29,30 or other suitable image analysis software.
- Remove any image stacks containing a considerable number of cells that moved during acquisition (and thus created blurring artifacts).
- Remove any image stacks containing highly fluorescent non-cell particles in the blue channel (e.g., dirt on microscope slide or cover slip, impurities in cultivation medium).
NOTE: Very bright non-cell objects in the blue channel may create cell detection artifacts or may interfere with detection of cells in their vicinity. - Remove any image stacks containing a large proportion of dead cells (i.e., cells with increased blue fluorescence compared to live cells).
NOTE: While the presence of a small proportion of dead cells in the sample is typically not a problem and these cells are automatically discarded during analysis, some dead or dying cells may occasionally be recognized as live cells by the segmentation algorithm and thus skew the reported results.
- Analyze images in the MATLAB software.
- Create a main folder and copy all MATLAB scripts to this location.
- Create a sub-folder (“pombe”, “cerevisiae” or “japonicus”) and copy input TIFF image files to this location.
- Start MATLAB, open script MAIN.m and run it. In the menu select the yeast species to be analyzed and start image processing.
NOTE: Some of the parameters required for cell and LD detection are pre-set for the particular species, others are determined automatically during image processing. The pre-set values were determined empirically and depend on several factors such as objective magnification, camera type and sensitivity, and imaging settings. If required, users may edit the script files to change the organism-specific presets to better reflect their experimental setup. Namely, during cell recognition acceptable object sizes are given by the “minArea” and “maxArea” parameters, and the minimum fraction of filled volume within the object boundaries is given by the “Solidity” parameter. For LD recognition, the brightness threshold is given by the “th” parameter (its value is affected mostly by image bit depth and fluorescence signal intensity), and maximum acceptable LD size is given by the “MaxArea” parameter. - Inspect and process the output files as required using a spreadsheet editor or statistical package; the workflow produces semicolon-separated CSV files, and segmented TIFF files with detected cell objects and LDs.
NOTE: The workflow segments images into background and cell objects, where each cell object may be composed of multiple adjacent cells. Therefore, the output in “xxxx_cells.csv” files does not represent single-cell data and should only be used to calculate per-unit-of-cell-volume metrics.
Representative Results
The whole procedure is summarized in Figure 1 for the fission yeasts (the budding yeast workflow is analogous), and below we provide examples of how the workflow can be used to study LD content in three different yeast species under various conditions known to affect cellular LD content. Each example represents a single biological experiment.
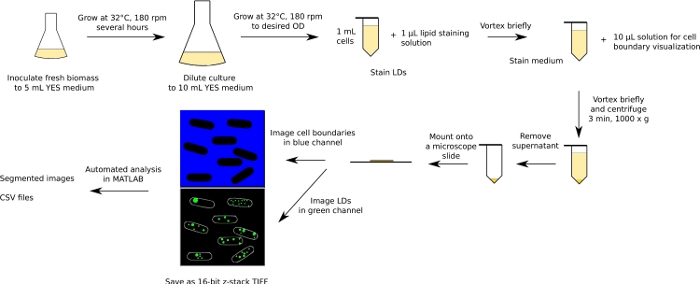
Figure 1: Schematic diagram of the experimental and analytical workflow. The workflow for fission yeasts is shown as an example. Please click here to view a larger version of this figure.
First, we analyzed S. pombe cells (Figure 2). Wild-type (WT; h+s) cells were grown to exponential phase in either the complex YES medium or defined EMM medium. Compared to YES, fewer LDs and higher LD staining intensity per unit of cell volume were detected in EMM (Figure 2A-C). Moreover, individual LDs formed in EMM medium were larger and displayed increased total staining intensity (Figure 2D, E). This is in agreement with previous findings of increased storage lipid content in cells grown in EMM24. The ppc1 gene encodes a phosphopantothenate-cysteine ligase required for coenzyme A synthesis. The temperature-sensitive ppc1-88 mutant shows a marked decrease in LD content when grown at the restrictive temperature31, providing an example of cells with low BODIPY 493/503 signal (Figure 2A). Accordingly, compared to wild type (grown at 32°C), smaller LDs with lower total staining intensity were detected in ppc1-88 cells grown in YES following a shift to 36°C (Figure 2D, E), without any apparent change in LD number per unit of cell volume (Figure 2B).
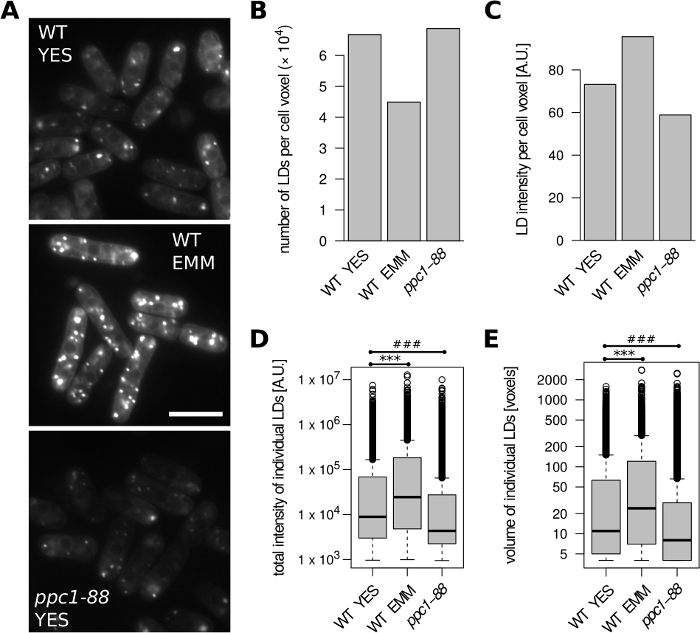
Figure 2: Impact of growth media and lipid metabolism mutation on LD content in S. pombe. Wild type (WT) and ppc1-88 cells were grown to exponential phase in the complex YES or defined EMM medium, as indicated. WT cells were grown at 32 °C. The temperature-sensitive ppc1-88 cells were grown at 25°C and shifted to 36°C for 2 hours prior to analysis. (A) Representative unprocessed microscopic images of LDs stained with BODIPY 493/503. A single optical slice is shown for each condition; 10% overlay with inverted blue channel was added to better visualize cell boundaries. Scale bar = 10 µm. (B) Number of identified LDs per unit of cell volume. (C) Fluorescence intensity of identified LDs per unit of cell volume. (D) Distributions of total fluorescence intensities of all identified LDs. ***, ### unpaired Wilcoxon test p = 1.7 x 10-107, p = 3.7 x 10-132, respectively. (E) Distributions of volumes of all identified LDs. ***, ### unpaired Wilcoxon test p = 6.8 x 10-71, p = 1 x 10-64, respectively. Data in panels B-E were derived from 242, 124 and 191 cell objects for the WT YES, WT EMM and ppc1-88 samples, respectively. Please click here to view a larger version of this figure.
Next, we quantified LD content in S. japonicus cells (h+ matsj-2017)32 from exponential and early-stationary cultures grown in YES (Figure 3A). Cells entering stationary phase showed markedly decreased number of LDs per unit of cell volume compared to exponentially growing cells (Figure 3B), while volume-normalized LD fluorescence intensity decreased slightly between the two conditions (Figure 3C). The early stationary-phase LDs were typically moderately larger in size and had moderately higher total fluorescence intensity compared to LDs from exponentially growing cells (Figure 3D, E).
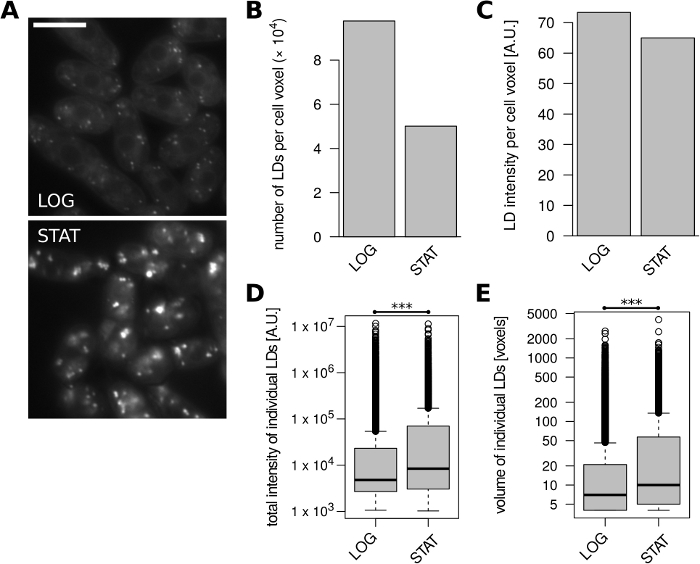
Figure 3: LD content in S. japonicus cells changes with growth phase. Exponentially growing (LOG) and early stationary phase (STAT) cells were analyzed. (A) Representative unprocessed microscopic images of LDs stained with BODIPY 493/503. A single optical slice is shown for each condition; 10% overlay with inverted blue channel was added to better visualize cell boundaries. Scale bar represents 10 µm. (B) Number of identified LDs per unit of cell volume. (C) Fluorescence intensity of identified LDs per unit of cell volume. (D) Distributions of total fluorescence intensities of all identified LDs. *** unpaired Wilcoxon test p = 1.3 x 10-114. (E) Distributions of volumes of all identified LDs. *** unpaired Wilcoxon test p = 2.4 x 10-85. Data in panels B-E were derived from 274 and 187 cell objects for the LOG and STAT samples, respectively. Please click here to view a larger version of this figure.
Finally, we analyzed S. cerevisiae cells of the widely used BY4741 laboratory strain (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) grown to exponential and stationary phase, respectively, in the complex YPAD medium. Budding yeast cells typically accumulate storage lipids upon entry into stationary phase1, and we were able to recapitulate these findings (Figure 4). Stationary cells contained somewhat fewer LDs per unit of volume compared to exponentially growing cells (Figure 4B), but their volume-normalized LD fluorescence intensity almost doubled (Figure 4C). This sharp increase in overall LD content was due to the much higher fluorescence intensity and volume of individual LDs in stationary phase (Figure 4D, E).
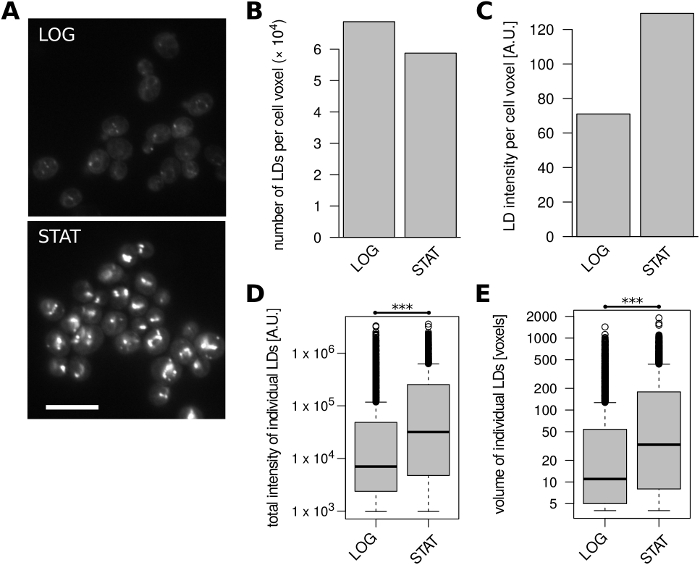
Figure 4: LD content in S. cerevisiae cells changes with growth phase. Exponentially growing (LOG) and stationary phase (STAT) cells were analyzed. (A) Representative unprocessed microscopic images of LDs stained with BODIPY 493/503. A single optical slice is shown for each condition; 10% overlay with inverted blue channel was added to better visualize cell boundaries. Scale bar represents 10 µm. (B) Number of identified LDs per unit of cell volume. (C) Fluorescence intensity of identified LDs per unit of cell volume. (D) Distributions of total fluorescence intensities of all identified LDs. *** unpaired Wilcoxon test p = 4.6 x 10-78. (E) Distributions of volumes of all identified LDs. *** unpaired Wilcoxon test p = 3.7 x 10-63. Data in panels B-E were derived from 430 and 441 cell objects for the LOG and STAT samples, respectively. Please click here to view a larger version of this figure.
Thus, our analysis workflow can detect changes in LD number, size and lipid content in three different and morphologically distinct yeast species under various conditions that positively or negatively affect cellular LD content.
Discussion
The understanding of lipid metabolism and its regulation is important for both basic biology, and clinical and biotechnological applications. LD content represents a convenient readout of lipid metabolism state of the cell, with fluorescence microscopy being one of the major methods used for LD content determination. The presented protocol allows automated detection and quantitative description of individual LDs in three different and morphologically distinct yeast species. To our knowledge, no similar tools exist for the fission yeasts. The MATLAB scripts required for image processing are included as Supplementary files, and are also available from the Figshare repository (DOI 10.6084/m9.figshare.7745738) together with all raw and processed image and tabular data from this manuscript, detailed descriptions of the CSV output files, and R scripts for downstream data analysis and visualization. Also, the latest version of the MATLAB scripts is available from GitHub (https://github.com/MartinSchatzCZ/LipidDots-analysis).
Successful LD analysis is largely dependent on the quality of the raw fluorescence images obtained. For optimal performance of the segmentation algorithms, clean glass slides devoid of dust particles should be used for microscopy, the cells should form a monolayer (the actual number of cells per field of view is not a critical parameter), and should not contain a large proportion of dead cells. Also, the z-stack imaging should start slightly below and end slightly above the cells. Depending on the particular microscopic setup, users may need to adjust some of the parameters in the image processing scripts (such as “th” for image background intensity threshold). While the current method is able to detect and describe individual LDs in the segmented cell objects, the workflow does not produce truly single-cell data due to difficulties with automated separation of all individual cells. Instead, LD content per unit of cell volume generalized for the whole sample is reported. This limitation may hamper data interpretation in analyses of heterogeneous cell populations. Also, care should be taken when working with cells with altered transport of small molecules (e.g., efflux pump mutants), as this might affect the intracellular BODIPY 493/503 concentration and LD staining, as observed for the Nile Red lipophilic dye33,34.
Staining the medium with the cell-impermeable Cascade Blue fluorescent dextran is a convenient way of distinguishing cells from the background35, which can be applied to many (if not all) yeast species. It also helps with automated removal of dead cells from the analysis as these will turn blue upon staining. Any dying or sick (and thus partially permeable for dextran) cells detected as alive can be removed during data analysis steps based on the “IntensityMedianBlue” value of the detected cell objects. In principle, the whole workflow can be used to detect various other cellular structures, such as DNA repair foci, provided the structures can be labelled with suitable fluorophores. The workflow should also be applicable to cells of other (yeast) species, further broadening its utility.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by Charles University grants PRIMUS/MED/26, GAUK 1308217 and SVV 260310. We thank Ondřej Šebesta for help with microscopy and development of the image analysis pipeline. We thank the ReGenEx lab for S. cerevisiae strains, and JapoNet and Hironori Niki’s lab for S. japonicus strains. The ppc1-88 strain was provided by The Yeast Genetic Resource Center Japan. Microscopy was performed in the Laboratory of Confocal and Fluorescence Microscopy co-financed by the European Regional Development Fund and the state budget of the Czech Republic (Project no. CZ.1.05/4.1.00/16.0347 and CZ.2.16/3.1.00/21515).
Materials
| 12-bit monochromatic CCD camera Hamamatsu ORCA C4742-80-12AG | Hamamatsu | or equivalent | |
| Adenine hemisulfate salt, ≥99% | Merck | A9126-25G | |
| BODIPY 493/503 (4,4-Difluoro-1,3,5,7,8-Pentamethyl-4-Bora-3a,4a-Diaza-s-Indacene) | Thermo Fisher Scientific | D3922 | for neutral lipid staining |
| D-(+) – Glucose, ≥99.5% | Merck | G7021 | |
| Dextran, Cascade Blue, 10,000 MW, Anionic, Lysine Fixable | Thermo Fisher Scientific | D1976 | for negative staining of cells |
| Dimethyl sulfoxide, ≥99.5% | Merck | D4540 | or higher purity, keep anhydrous on molecular sieves |
| EMM broth without dextrose | Formedium | PMD0405 | medium may also be prepared from individual components |
| Fiji/ImageJ software | NIH | or equivalent; for visual inspection of microscopic data | |
| High precision cover glasses, 22×22 mm, No 1.5 | VWR | 630-2186 | use any # 1.5 cover glass |
| Image Processing Toolbox for MATLAB, version 10.0 | Mathworks | ||
| Lectin from Glycine max (soybean) | Merck | L1395 | for cell immobilization on slides |
| MATLAB software, version 9.2 | Mathworks | ||
| Microscope slide, 26 x 76 mm, 1 mm thickness | Knittel Glass | L762601.2 | use any microscope slide fitting your microscope stage, clean thoroughly before loading cells |
| Olympus CellR microscope with automatic z-axis objective movement | Olympus | or equivalent | |
| pentaband filter set | Semrock | F66-985 | brightfield, green and blue channels are sufficient |
| Signal Processing Toolbox for MATLAB, version 7.4 | Mathworks | ||
| SP supplements | Formedium | PSU0101 | |
| standard office computer capable of running MATLAB | |||
| Statistics and Machine Learning Toolbox for MATLAB, version 11.1 | Mathworks | ||
| Universal peptone M66 for microbiology | Merck | 1070431000 | |
| UPLSAPO 60XO objective | Olympus | or equivalent | |
| Yeast extract | Formedium | YEA03 | |
| Yeast nitrogen base without amino acids | Formedium | CYN0405 |
References
- Koch, B., Schmidt, C., Daum, G. Storage lipids of yeasts: a survey of nonpolar lipid metabolism in Saccharomyces cerevisiae, Pichia pastoris, and Yarrowia lipolytica. FEMS Microbiology Reviews. 38 (5), 892-915 (2014).
- Krahmer, N., Farese, R. V., Walther, T. C. Balancing the fat: lipid droplets and human disease. EMBO Molecular Medicine. 5 (7), 973-983 (2013).
- Lazar, Z., Liu, N., Stephanopoulos, G. Holistic Approaches in Lipid Production by Yarrowia lipolytica. Trends in Biotechnology. 36 (11), 1157-1170 (2018).
- Kim, D. U., et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nature Biotechnology. 28 (6), 1628-1629 (2010).
- Giaever, G., Nislow, C. The yeast deletion collection: a decade of functional genomics. Genetics. 197 (2), 451-465 (2014).
- Meyers, A., et al. The protein and neutral lipid composition of lipid droplets isolated from the fission yeast, Schizosaccharomyces pombe. Journal of Microbiology (Seoul, Korea). 55 (2), 112-122 (2017).
- Meyers, A., et al. Lipid Droplets Form from Distinct Regions of the Cell in the Fission Yeast Schizosaccharomyces pombe. Traffic (Copenhagen, Denmark). 17 (6), 657-659 (2016).
- Long, A. P., et al. Lipid droplet de novo formation and fission are linked to the cell cycle in fission yeast. Traffic (Copenhagen, Denmark). 13 (5), 705-714 (2012).
- Yang, H. J., Osakada, H., Kojidani, T., Haraguchi, T., Hiraoka, Y. Lipid droplet dynamics during Schizosaccharomyces pombe sporulation and their role in spore survival. Biology Open. , 8 (2016).
- Aoki, K., Shiwa, Y., Takada, H., Yoshikawa, H., Niki, H. Regulation of nuclear envelope dynamics via APC/C is necessary for the progression of semi-open mitosis in Schizosaccharomyces japonicus. Genes To Cells: Devoted To Molecular & Cellular Mechanisms. 18 (9), 733-752 (2013).
- Karolin, J., Johansson, L. B. A., Strandberg, L., Ny, T. Fluorescence and Absorption Spectroscopic Properties of Dipyrrometheneboron Difluoride (BODIPY) Derivatives in Liquids, Lipid Membranes, and Proteins. Journal of the American Chemical Society. 116 (17), 7801-7806 (1994).
- Bozaquel-Morais, B. L., Madeira, J. B., Maya-Monteiro, C. M., Masuda, C. A., Montero-Lomeli, M. A new fluorescence-based method identifies protein phosphatases regulating lipid droplet metabolism. PloS One. 5 (10), e13692 (2010).
- Sitepu, I. R., et al. An improved high-throughput Nile red fluorescence assay for estimating intracellular lipids in a variety of yeast species. Journal of Microbiological Methods. 91 (2), 321-328 (2012).
- Rostron, K. A., Lawrence, C. L. Nile Red Staining of Neutral Lipids in Yeast. Methods in Molecular Biology (Clifton, N.J.). 1560, 219-229 (2017).
- Romero-Aguilar, L., Montero-Lomeli, M., Pardo, J. P., Guerra-Sánchez, G. Lipid Index Determination by Liquid Fluorescence Recovery in the Fungal Pathogen Ustilago Maydis. Journal of Visualized Experiments. (134), 1-6 (2018).
- Gupta, A., Dorlhiac, G. F., Streets, A. M. Quantitative imaging of lipid droplets in single cells. The Analyst. , (2018).
- Wolinski, H., Bredies, K., Kohlwein, S. D. Quantitative imaging of lipid metabolism in yeast: from 4D analysis to high content screens of mutant libraries. Methods in Cell Biology. , 108-365 (2012).
- Campos, V., Rappaz, B., Kuttler, F., Turcatti, G., Naveiras, O. High-throughput, nonperturbing quantification of lipid droplets with digital holographic microscopy. Journal of Lipid Research. 59 (7), 1301-1310 (2018).
- Ranall, M. V., Gabrielli, B. G., Gonda, T. J. High-content imaging of neutral lipid droplets with 1,6-diphenylhexatriene. BioTechniques. 51 (1), 35-42 (2011).
- Schnitzler, J. G., et al. Nile Red Quantifier: a novel and quantitative tool to study lipid accumulation in patient-derived circulating monocytes using confocal microscopy. Journal of Lipid Research. 58 (11), 2210-2219 (2017).
- Bombrun, M., Gao, H., Ranefall, P., Mejhert, N., Arner, P., Wählby, C. Quantitative high-content/high-throughput microscopy analysis of lipid droplets in subject-specific adipogenesis models. Cytometry. Part A the journal of the International Society for Analytical Cytology. 91 (11), 1068-1077 (2017).
- Capus, A., Monnerat, M., Ribeiro, L. C., de Souza, W., Martins, J. L., Sant’Anna, C. Application of high-content image analysis for quantitatively estimating lipid accumulation in oleaginous yeasts with potential for use in biodiesel production. Bioresource Technology. 203, 309-317 (2016).
- Lv, X., et al. Identification of gene products that control lipid droplet size in yeast using a high-throughput quantitative image analysis. Biochimica et biophysica acta. Molecular and Cell Biology Of Lipids. 1864 (2), 113-127 (2018).
- Zach, R., Tvarůžková, J., Schätz, M., Ťupa, O., Grallert, B., Převorovský, M. Mitotic defects in fission yeast lipid metabolism “cut” mutants are suppressed by ammonium chloride. FEMS Yeast Research. 18 (6), 1-7 (2018).
- Petersen, J., Russell, P. Growth and the Environment of Schizosaccharomyces pombe. Cold Spring Harbor Protocols. 2016 (3), (2016).
- Aoki, K., Furuya, K., Niki, H. Schizosaccharomyces japonicus: A Distinct Dimorphic Yeast among the Fission Yeasts. Cold Spring Harbor Protocols. (12), (2017).
- Curran, B. P. G., Bugeja, V. Basic investigations in Saccharomyces cerevisiae. Methods in Molecular Biology (Clifton, N.J.). , 1-14 (2014).
- Sabatinos, S. A., Forsburg, S. L. Molecular genetics of Schizosaccharomyces pombe. Methods in Enzymology. 470 (10), 759-795 (2010).
- Schindelin, J., Rueden, C. T., Hiner, M. C., Eliceiri, K. W. The ImageJ ecosystem: An open platform for biomedical image analysis. Molecular Reproduction and Development. 82 (7-8), 518-529 (2015).
- Schindelin, J., et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Nakamura, T., Pluskal, T., Nakaseko, Y., Yanagida, M. Impaired coenzyme A synthesis in fission yeast causes defective mitosis, quiescence-exit failure, histone hypoacetylation and fragile DNA. Open Biology. 2 (9), 120117 (2012).
- Furuya, K., Niki, H. Isolation of heterothallic haploid and auxotrophic mutants of Schizosaccharomyces japonicus. Yeast. 26 (4), 221-233 (2009).
- Ivnitski-Steele, I., et al. Identification of Nile red as a fluorescent substrate of the Candida albicans ATP-binding cassette transporters Cdr1p and Cdr2p and the major facilitator superfamily transporter Mdr1p. Analytical Biochemistry. 394 (1), 87-91 (2009).
- Wolinski, H., Kohlwein, S. D. Microscopic analysis of lipid droplet metabolism and dynamics in yeast. Methods in Molecular Biology (Clifton, N.J.). 457 (1), 151-163 (2008).
- Graml, V., et al. A genomic Multiprocess survey of machineries that control and link cell shape, microtubule organization, and cell-cycle progression. Developmental Cell. 31 (2), 227-239 (2014).

