Field Postmortem Rabies Rapid Immunochromatographic Diagnostic Test for Resource-Limited Settings with Further Molecular Applications
Summary
We present a complete protocol for postmortem diagnosis of animal rabies under field conditions using a rapid immunochromatographic diagnostic test (RIDT), from brain biopsy sampling to final interpretation. We also describe further applications using the device for molecular analysis and viral genotyping.
Abstract
Functional rabies surveillance systems are crucial to provide reliable data and increase the political commitment necessary for disease control. To date, animals suspected as rabies-positive must be submitted to a postmortem confirmation using classical or molecular laboratory methods. However, most endemic areas are in low- and middle-income countries where animal rabies diagnosis is restricted to central veterinary laboratories. Poor availability of surveillance infrastructure leads to serious disease underreporting from remote areas. Several diagnostic protocols requiring low technical expertise have been recently developed, providing opportunity to establish rabies diagnosis in decentralized laboratories. We present here a complete protocol for field postmortem diagnosis of animal rabies using a rapid immunochromatographic diagnostic test (RIDT), from brain biopsy sampling to the final interpretation. We complete the protocol by describing a further use of the device for molecular analysis and viral genotyping. RIDT easily detects rabies virus and other lyssaviruses in brain samples. The principle of such tests is simple: brain material is applied on a test strip where gold conjugated antibodies bind specifically to rabies antigens. The antigen-antibody complexes bind further to fixed antibodies on the test line, resulting in a clearly visible purple line. The virus is inactivated in the test strip, but viral RNA can be subsequently extracted. This allows the test strip, rather than the infectious brain sample, to be safely and easily sent to an equipped laboratory for confirmation and molecular typing. Based on a modification of the manufacturer's protocol, we found increased test sensitivity, reaching 98% compared to the gold standard reference method, the direct immunofluorescence antibody test. The advantages of the test are numerous: rapid, easy-to-use, low cost and no requirement for laboratory infrastructure, such as microscopy or cold-chain compliance. RIDTs represent a useful alternative for areas where reference diagnostic methods are not available.
Introduction
Canine rabies is the main cause of human rabies, globally responsible for approximately 59,000 human deaths per year, nearly all occurring in low- and middle-income countries (LMICs) in Asia and Africa1. The main etiological agent is a neurotropic canine-associated classical rabies virus (RABV, family Rhabdoviridae, genus Lyssavirus, species Rabies lyssavirus). However, other rabies-related lyssaviruses, mostly circulating in bat species, also cause disease2,3. In affected regions, disease surveillance and control are often hampered by low level political commitment likely due to lack of reliable data4,5,6. One reason for disease underreporting is the absence of laboratory diagnosis, due in part to limited access to equipped laboratories and trained staff as well as the difficulties of shipment of the specimens. Laboratory diagnosis is necessary to confirm rabies cases and additionally allows for genetic characterization of the involved strains, providing insight on virus transmission at the regional level4,5,7.
The current gold standards for postmortem rabies diagnosis, approved by both the World Health Organization (WHO) and the World Organization for Animal Health (OIE), are the direct fluorescent antibody test (DFAT), the direct rapid immunohistochemistry test (DRIT) and molecular methods (e.g., reverse transcription polymerase chain reaction (RT-PCR))4,8. However, proper application in LMICs remains limited due to inadequate laboratory facilities with inconsistent power supply, uncooled sample transportation, and lack of a quality management system. Because animal rabies diagnosis is typically only conducted at central veterinary laboratories in LMICs, existing surveillance data mainly reflects the rabies situation in urban areas.
Recently developed low technology diagnostic alternatives offer opportunities to establish rabies diagnosis in remote areas and decentralized rabies laboratories4,8,9. The rapid immunochromatographic diagnostic test (RIDT) is a lateral flow test based on immunochromatography using gold conjugated detector antibodies and is a very promising rabies diagnostic tool10,11,12,13. The principle is simple: after dilution, brain material is mixed in the provided buffer, and a few drops are applied on the test strip where gold conjugated monoclonal antibodies bind specifically to rabies antigens, mainly the nucleoproteins (Figure 1). The antigen-antibody complexes then undergo lateral flow migration, binding at the test line (T-line) to fixed antibodies against rabies antigens, resulting in a clearly visible purple line. The remaining gold conjugated antibodies not bound to rabies antigens continue migrating and fix to the membrane through additional targeting antibodies, resulting in a clearly visible purple control line (C-line).
The one-step, low cost method is rapid, extremely easy and does not require expensive equipment or special storage conditions. With modification of the manufacturer protocol to eliminate the dilution step, nearly all equipment and reagents required to perform the test are included in the kit14. The result is read after 5-10 minutes without a microscope. This is a major advantage over the DFAT test, which requires a fluorescence microscope and immunofluorescence conjugate, along with refrigerated transportation and sample storage. Even the DRIT test, which can be performed using a light microscope, requires a continuous cold chain to store the anti-rabies antibodies, which are also not yet commercially available. In comparison to the DRIT, the RIDT requires no toxic chemicals, a particular advantage in countries where waste disposal is poorly regulated. The rapid test is less time-consuming with much easier interpretation compared to the gold standard tests DFAT and DRIT. This allows for on-site testing by personnel with limited technical expertise.
Based on these test properties, prompt diagnosis of suspected animals in remote areas becomes feasible, facilitating implementation of post exposure prophylaxis (PEP) for exposed people as soon as possible. In addition, distance transport of rabies samples is not necessary, resulting in better sample quality at the time of testing. However, the results obtained with the RIDT tests should currently be confirmed using a reference diagnostic test such as DFAT or DRIT.
RIDT techniques for detection of RABV and other lyssaviruses have been evaluated. One of the first studies was conducted by Korean researchers in 200710. Compared to the DFAT method, in 51 animal samples and 4 RABV isolates, the RIDT showed a sensitivity and specificity of 91.7% and 100%, respectively. These results were later confirmed with 110 animal brain samples from Korea, with sensitivity and specificity, compared to DFAT, of 95% and 98.9%, respectively15. More recently, other studies assessed the performance of this RIDT using virus isolates and/or infected brain samples from various animals with different geographical origins. A panel of 21 samples, including African RABV and other African lyssaviruses (Duvenhage virus (DUVV), Lagos bat virus (LBV) and Mokola virus (MOKV)), were successfully detected, with sensitivity of 100% compared to the DFAT16. Similar high sensitivity (96.5%) and specificity (100%) values were obtained from a panel of 115 brain samples from Ethiopia17. Another study evaluated European RABV isolates, two other European lyssaviruses (European bat lyssavirus type 1 (EBLV-1) and type 2 (EBLV-2)), and the Australian bat lyssavirus (ABLV)18. Based on analysis of 172 animal brain samples, the RIDT kit had 88.3% sensitivity and 100% specificity compared to DFAT, and the three rabies-related lyssaviruses were successfully detected. In this study, some of the false negative results came from brain samples stored in glycerol buffer, suggesting that improper glycerol removal influenced capillary flow or antibody binding. A recent analysis of 43 clinical samples from Australian bats confirmed previous test results, with complete concordance to DFAT19. Two studies were conducted in India using the RIDT on a limited number of clinical samples (11 and 34 samples). Compared to DFAT, sensitivity was between 85.7% and 91.7% and specificity was 100%20,21. Another evaluation of this kit using 80 animal brain samples from Africa, Europe and the Middle East obtained complete concordance with DFAT for specificity (100%) but a higher sensitivity (96.9%) compared to the previous studies22. In a recent inter-laboratory comparison of this RIDT performed in 22 different laboratories using a panel of 10 samples, overall concordance was 99.5%23.
Only one recent multicentric study showed unsatisfactory overall RIDT performance24. Samples from three different datasets were tested and provided variable sensitivity and specificity values compared to DFAT. For example, sensitivity and specificity obtained with the first panel (n=51) and the second panel (n=31) of samples from experimental infected animals, all tested in laboratory A, gave a sensitivity of 16% and 43%, respectively, whereas the specificity was 100% for both. Conversely, the results of the third panel (n=30) of field clinical samples analyzed by laboratory B provided a complete concordance with the results of DFAT, which was further nearly completely confirmed by laboratory A (85% sensitivity and 100% specificity). Batch-to-batch variation was suggested as a possible explanation for the fluctuating relatively low sensitivity with RIDT24.
At the same time, another study performed a similar validation process of the above described RIDT, with a modification of the manufacturer recommended protocol14. The pre-dilution step (1:10) in PBS was omitted during preparation of the brain material. Based on this simpler modified protocol, the authors obtained sensitivity and specificity of 95.3% and 93.3%, respectively, compared to DFAT by testing, under laboratory conditions, a dataset of 73 animal brain samples, naturally or experimentally infected with various RABV strains. The study presented the first evaluation of this RIDT in a field setting (Chad, Africa). In 48 clinical brain samples, sensitivity and specificity were 94.4% and 100%, respectively. The discrepancies between DFAT and RIDT were due to false positive results with DFAT, determined after confirmation with RT-PCR. When these results were deleted, there was complete concordance, and it demonstrated that the RIDT was more reliable that DFAT under these field conditions14. No batch-to-batch variation was observed using the modified protocol. When the modified protocol was applied to a small number of the DFAT/ RIDT divergent samples (n=8) in the study of Eggerbauer et al.24, all were found concordant (100% sensitivity).
Another major advantage of the RIDT is secondary use for detecting viral RNA fixed on the strip using molecular techniques (such as RT-PCR) and subsequent genotyping14,24. Following an extraction step, Léchenne et al.14 demonstrated viral RNA fixed on the Anigen device membrane using RT-PCR with 86.3% sensitivity in a panel of 51 samples (including 18 samples tested and shipped from Chad at ambient temperature). Subsequent genotyping was possible in 93% of the 14 samples tested. Sanger sequencing of PCR amplicons of at least 500 nucleotides in length were used. In addition to RABV isolates, the test detected four other lyssavirus species, DUVV, EBLV-1, EBLV-2 and Bokeloh bat lyssavirus (BBLV), during a fully concordant international inter laboratory test14. The sensitivity of viral RNA detection was even higher (100%) in the study of Eggerbauer et al., based on laboratory samples examination24. The latter study also demonstrated that the buffer used in the RIDT kit inactivated virus. Thereby, the devices can be shipped easily, at ambient temperature without specific biosafety precautions to reference laboratories, for molecular confirmation and genotyping.
Based on the previous evaluations, RIDT tools offer numerous advantages for use in field settings, especially when the reference diagnostic techniques are not available. However, this test also has some limitations, in particular, low sensitivity of antigen detection14,24. The test is applicable for samples containing high quantities of viral antigens, such as brain samples. However, it is not appropriate for other samples such as saliva or other body fluids. Another drawback is cost of the device (around 5-10 Euros in Europe), which is less expensive compared to the cost of performing DFAT, RT-PCR or DRIT, but which still remains high for LMICs. However, future development and validation of similar RIDTs from other companies could lead to a price decrease. One study reported batch-to-batch variations. Although not reported by others, strict quality controls should nevertheless be performed when testing a new batch, as for any reagent used in a quality management environment. The use of the modified protocol was not altered when using different batches14. All except one study demonstrated that the sensitivity of RDIT was high compared to DFAT (around 90%-95%). Because rabies is always fatal, it is still strongly recommended to confirm any negative results with RDIT using a reference diagnostic test such as DFAT, DRIT or RT-PCR14.
In this manuscript, we present a complete protocol for field postmortem diagnosis of animal rabies based on an example of a commercialized RIDT, from brain sample collection to application of a modified protocol compared to the manufacturer recommendations (which were previously validated14) and subsequent molecular analysis. This protocol was applied and validated many times under field conditions in West- and Central Africa, where the RIDT was used routinely for rabies diagnosis alongside the DFAT test. We additionally demonstrate a second application for the device, in laboratory settings, for extraction and detection using RT-PCR of viral RNA fixed on the device.
Protocol
1. Sample collection via the foramen magnum (occipital route)25
NOTE: This technique can be implemented under laboratory conditions or in field settings. Samples should be processed as soon as possible after death of the suspected animal or kept at cool temperature (refrigerated or frozen, if possible) to avoid decomposition which could affect the results. Similar to other reference techniques based on lyssavirus antigens detection such as DFAT and DRIT, decomposed samples should not be tested because it can affect the result (risk of false negative result).
CAUTION: All samples should be considered as potentially infectious. Safety regulations and procedures should be strictly followed, even in field settings4. In particular, wear appropriate personal protective equipment including mask, glasses, gloves and a lab coat. Use appropriate disinfectant for material and sample decontaminations (e.g., sodium hypochlorite with recommended manufacturer dilutions, 70% alcohol – ethanol or isopropanol, 1% soap solution). All personnel handling samples should be vaccinated against rabies.
- Remove the animal head with a knife before the first cervical vertebra (atlas vertebra) to access the foramen magnum.
NOTE: To minimize infective aerosol, avoid using a manual saw or similar tool. - Collect brainstem (medulla oblongata) sample using a disposable plastic pipette (Figure 2A), a drinking straw (Figure 2B), a clamp (Figure 2C) or a dropper (supplied with the RIDT) (Figure 2D).
NOTE: Special attention must be paid when collecting the sample, because it is an utmost important step for the reliability of the results. In addition to the associated video which shows in a simple way how to collect the part of the brainstem of interest, a training step is highly recommended to make sure to collect the correct anatomical section. - Optionally and in addition of brain stem (medulla oblongata), collect other parts of the brainstem or the brain (cerebellum, hippocampus, thalamus and cortex) by the same occipital route by pushing and rotating the plastic pipette or straw towards the eye socket (Figure 3).
- If using a straw or pipette, gently squeeze it to deposit the brain sample (0.5-2 g) in a tube for subsequent analysis and/or biobanking.
NOTE: Sample storage in glycerol is not recommended, as it seems to affect capillary flow or the antibody binding step of the RIDT18.
2. Execution of the modified RIDT protocol14
NOTE: This modification omits a dilution step (1:10) into PBS, as specified in manufacturer protocol (all versions), and can be implemented under laboratory or field settings.
- Use the swab/dropper to collect the equivalent of half a peanut or pea (0.1-0.5 g) of brain material and place it in the buffer sample tube.
NOTE: For the modified protocol, all reagents/consumables are included in the kit (no PBS or additional tube is needed) (Figure 4). Document the batch number of the kit and check validity of the expiration date. - Carefully crush the brain material directly in the tube with the swab or the dropper for about 30 s until a homogeneous suspension is obtained.
NOTE: The buffer reaction inactivates the infectivity of the virus in the conditions of the manufacturer's protocol24. - Using the dropper, deposit four drops (approximately 100 µL) of the suspension in the sample inlet on the test device.
- Wait for complete sample migration (1-5 min) before reading the test device. The migration should start rapidly after deposit of the sample (1-5 min).
- In case of delay (due to high viscosity suspension) or to accelerate the start of the migration, gently scratch the bottom of the deposit site of the device with the dropper (1-5 times) and eventually add 1-2 more drops. Migration should start immediately thereafter.
- Read the test result in the detection window after 5-10 min, and no more than 20 min, after the end of the migration.
- Interpret the result based on presence or absence of the control line (C-line) and test line (T-line) (purple lines) in the detection window, according to Figure 5. Consider the sample positive when two lines are visible (Figure 5A), negative if only the C-line is present (Figure 5B) and invalid if only the T-line is present or if no lines are visible (Figure 5C).
NOTE: Invalid results should be repeated at least once. Other techniques should be performed if results remain invalid. Negative results obtained with RIDT need to be subsequently confirmed using a gold standard reference method, like DFAT, DRIT and molecular methods (polymerase chain reaction or PCR). Even though the sensitivity of this test is high (see representative results), it is not 100%. - Store used devices at room temperature, or refrigerate/freeze when possible, for subsequent molecular analysis (see section 4). Freeze the remaining sample suspension at -20 °C/-80 °C in the buffer tube to repeat the test if necessary or for subsequent molecular analysis.
3. RNA extraction and detection by RT-qPCR from the RIDT device
NOTE: This step can only be implemented under laboratory conditions with adapted environment and suitable equipment for molecular diagnosis. It can be done soon after the RIDT test or retrospectively on archived RIDT devices, stored at room temperature (15-30 °C), refrigerated or frozen.
- RNA extraction
NOTE: To monitor the extraction step, it is recommended to use an internal control that can be an endogenous mRNA (such as ß-actin) or an exogenous control (such as eGFP synthetic RNA) directly spiked into the sample during the first steps of the extraction26,27.- Carefully open the device and remove the filter paper.
- Cut the deposit area of the sample and place it into a tube containing 1 mL of Tri-Reagent LS. Incubate at RT for 1 hour with gentle regular manual agitation.
- Perform the extraction in accordance with manufacturer recommendations, as previously described27. At this step, the exogenous internal control can be added.
- During the process, add 2 µL of glycogen for facilitating precipitation of RNA, according to the manufacturer recommendations.
- Adjust the final volume for RNA resuspension in nuclease-free water, with a volume of 50 µL generally used.
NOTE: At the end of the centrifugation step for aqueous and organic phase separation (after addition of 200 µL of chloroform into the Tri-Reagent LS), the piece of membrane from the device will be at the bottom of the tube and not interfere with collection of the upper aqueous phase. Alternatively, other easy and rapid protocols can be used, for instance, using phenol-based reagents and silica membranes28.
- Detection by RT-qPCR26
NOTE: Detection of potential viral RNA present in extracted samples can be done using different molecular techniques, such as reverse-transcription PCR, conventional (endpoint) or real time PCR (qPCR). Several methods are available, such as conventional RT-PCR27,29 or RT-qPCR26,30 targeting the viral nucleoprotein or polymerase gene. One example will be presented below based on a dual combined pan-lyssavirus RT-qPCR targeting a conserved region among the viral polymerase. This RT-qPCR technique associates two different RT-qPCR: one based on the TaqMan probe technology (pan-RABV RT-qPCR) and the other using the SyBR Green detection (pan-lyssa RT-qPCR). In addition, the detection of an exogenous internal control (eGFP RNA) directly spiked during the extraction process is done by a specific TaqMan probe-based RT-qPCR (eGFP RT-qPCR). Careful on-site validation of the molecular techniques selected for detection of viral RNA is important, in particular, to verify that primers, and probes for real-time RT-PCR, are adapted for detection of the strains circulating in the region of interest4.- Dilute RNA sample to 1:10 in nuclease free water. Test each RNA sample in duplicate, using a 96-well reaction plate or other formats. Use positive and negative controls for each assay and test at least in duplicate.
- Prepare the master mix reaction solution for the three different RT-qPCR assays according to Table 1, and with the primers/probes indicated in Table 2.
- Add 5 µL of diluted RNA samples and 15 µL of master mix to each of the three different assays. The pan-RABV RT-qPCR assay and the eGFP RT-qPCR assay can cycle in the same plate.
- Run the different assays following the thermal cycling conditions indicated in Table 3. If only one PCR thermal cycler is available, start with the pan-RABV RT-qPCR and keep the plate for the pan-lyssa RT-qPCR at 4 °C until the end of the pan-RABV RT-qPCR.
- Analyze the results obtained with the three assays according to Table 4.
4. Genotyping after RNA extraction from the RIDT device
- Reverse transcription RT27,29
- Prepare a master mix with 6 µL of RNA, 2 µL of pd(N)6 random primers (200 µg/µL) and 2 µL of nuclease-free water for a final volume of 10 µL.
- Incubate at 65 °C for 10 min in a heat-block and then store on ice.
- Prepare a master mix with 6 µL of 5x First-Strand Buffer, 2 µL of 0.1 M dithiothreitol (DTT), 1 µL (200 U) of Superscript II reverse transcriptase, 2 µL (80 U) of RNasin, 2 µL of dNTP mix (10 µM) and complete with nuclease-free water to obtain a final volume of 20 µL for each sample.
- Add the master mix (20 µL) to the sample (10 µL) (final volume of 30 µL) and incubate at 42 °C for 90 min in a heat-block.
- Proceed to the next step with PCR amplification or store the cDNA at -20 °C.
- Conventional PCR27,29,31
NOTE: Different techniques of conventional PCR are available for genotyping. Two are presented, both hemi-nested PCR, targeting a part of the nucleoprotein or a part of the viral protein of the lyssavirus. The protocol is the same for each of these assays, except for the primers and cycling conditions. Positive (positive RNA) and negative (negative cDNA and/or nuclease-free water) controls should be included in each series and each round of PCR.- Prepare for each sample in a 0.2 mL microtube a master mix reaction solution for the first PCR step. This mix contains 5 µL of 10x NH4 Reaction Buffer, 2.5 µL of MgCl2 solution (50 mM), 1 µL of dNTP Mix (10 µM), 1 µL of each primer (10 µM), 0.2 µL (1 U) of Biotaq DNA polymerase and 37.3 µL of nuclease-free water (final volume of 48 µL). The primers are indicated in Table 5.
- Add 2 µL of cDNA in every tube and cycle on a separate conventional PCR thermal cycler for each assay, according to Table 6.
- Prepare a second master mix reaction solution identical to the previous one with using the appropriate primers (Table 5) for the hemi-nested PCR reaction.
- Add 2 µL of the first round PCR product and cycle on a conventional PCR thermal cycler using the cycling parameters indicated in Table 6.
- Visualize the different PCR products (first and second round PCR) after loading them on a 1% agarose gel (100 mL of Tris-acetate EDTA buffer 1x – TAE 1x) with ethidium bromide (final concentration around 0.01%) and run the gel during 30 min at 120 V. A positive PCR result is observed in the form of a bright band of the expected size (Table 5).
- Sanger sequencing
- Perform a Sanger sequencing of the amplicons obtained with the pan-lyssavirus hemi-nested PCR and complete the genotyping analysis.
Representative Results
As with any diagnostic method, sample collection is of paramount importance for reliability of the results, especially when performed in field settings. The collection process needs to be as simple as possible to guarantee collection of high-quality samples. The collection of a brain biopsy (brainstem with medulla oblongata) via the foramen magnum route for postmortem diagnosis of animal rabies fulfills this requirement, as indicated in Figure 2A-D25.
After collection, the brain sample is submitted to the modified protocol of the RIDT, summarized in Figure 6. As indicated in the Protocol section, the major adaptation from the manufacturer provided protocol is omission of the dilution step in PBS, which simplifies the procedure and necessary consumables/reagents, thus all included in the kit (Figure 4).
This modified protocol was implemented and evaluated in five different laboratories, including one WHO collaborative center on rabies (Lab 1, France), one FAO reference center for rabies (Lab 5, Italy) and three reference laboratories located in enzootic African countries, Chad (Lab 2), Ivory Coast (Lab 3) and Mali (Lab 4). In Chad, an evaluation of the RIDT was done in both laboratory and field settings.
Compared to the reference technique DFAT, sensitivity and specificity of the RDIT were high for all laboratories, with 96% to 100% and 93.7% to 100%, respectively (Table 7). The lowest sensitivity and specificity of the RDIT was obtained for Lab 1 (France) during the laboratory validation step. Based on the cumulative number of tested samples (n=162) (Supplementary Table 1), the overall sensitivity and specificity compared to DFAT were 98.2% and 95.8%, respectively (Table 7). However, these preliminary but promising results were obtained on a limited sample dataset and need to be further confirmed on a large number of positive and negative samples, especially for those tested in enzootic areas, to avoid any potential underestimating or bias due to the current heterogenous datasets.
The RIDT test is suitable to detect lyssavirus in brain biopsies from infected animals, where the level of lyssavirus antigens is important. However, the test limit of detection remains high when testing titrated virus suspension (Table 8; Figure 7).
Table 9 (from Léchenne 201614) shows an example of results obtained after RNA detection by the dual combined pan-lyssavirus RT-qPCR targeting the viral polymerase of lyssavirus. A panel of 51 positive RIDT tests performed in laboratory conditions (Lab 1, n=32) or in Chad (Lab 2, n=19) and then shipped at ambient temperature to Lab 1, was tested. Positive detection was obtained for 18 (94.7%), 26 (81.2%) and 44 (86.3%) samples from Lab 1, Lab 2 and the two combined, respectively. In addition, genotyping was performed for 14 of these samples (10 from Lab 1 and 4 from Lab 2) using the hemi-nested PCR targeting the partial nucleoprotein gene and was successful for 13 of them (93%) (from Léchenne et al. 201614).
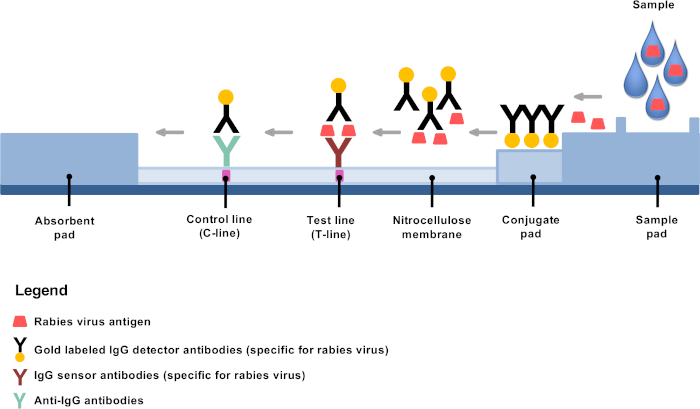
Figure 1: Schematic representation of the structure of an RIDT for rabies diagnosis. Please click here to view a larger version of this figure.
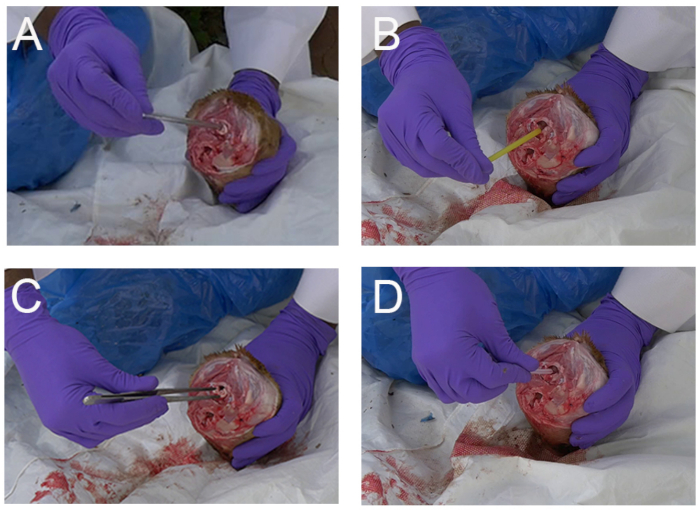
Figure 2: Examples of rapid simple techniques for collection of brain samples (brainstem with medulla oblongata) in animals (dog shown here) via the occipital foramen in field settings (Mali). (A) Collection with a disposable plastic pipette (B) Collection with a plastic drinking straw (C) Collection with a clamp (D) Collection with the disposal dropper provided in the RIDT kit. Please click here to view a larger version of this figure.
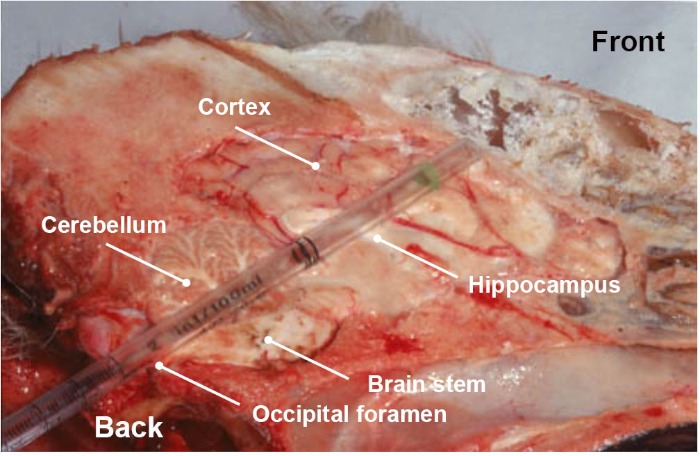
Figure 3: Longitudinal anatomical section of dog head, showing the different parts of the brain (brainstem, cerebellum, hippocampus, thalamus and cortex) collected when pushing, in a rotational movement, a disposable plastic pipette through the occipital foramen route. Please click here to view a larger version of this figure.

Figure 4: Description of the contents of RIDT kit, including the device, a disposable plastic dropper, a disposable swab, and the assay diluent. The tube where the sample will be collected and stored is not provided. Please click here to view a larger version of this figure.
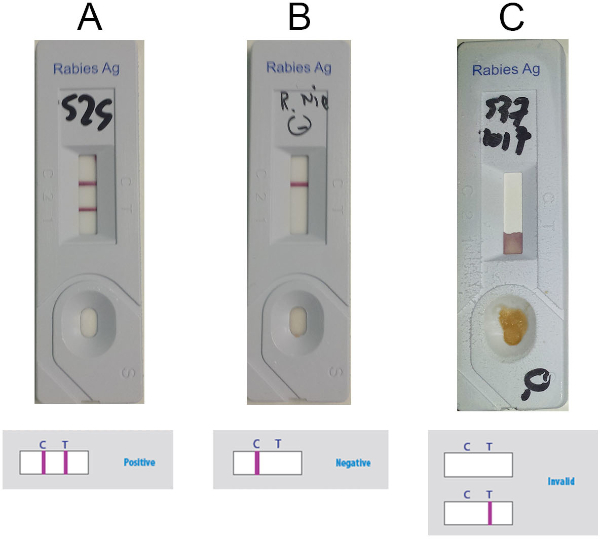
Figure 5: Representative results for interpretation of the Anigen RIDT. (A) Positive results (visible presence of two lines, C-line and T-line) (B) Negative results (visible presence of C-line only) (C) Invalid results (absence of visible C-line). Please click here to view a larger version of this figure.
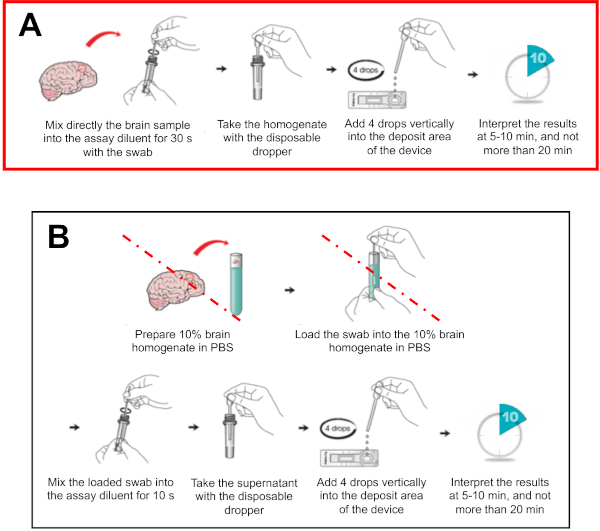
Figure 6: Schematic representation of RIDT protocol, adapted from manufacturer instructions. (A) Modified version of the protocol, with deletion of the dilution step recommended by the manufacturer (B) Initial protocol recommended by manufacturer, with a pre 1:10 dilution step in PBS of the brain samples. The steps deleted in the modified version of the protocol (presented in Figure 6A) are indicated with a red line. Please click here to view a larger version of this figure.
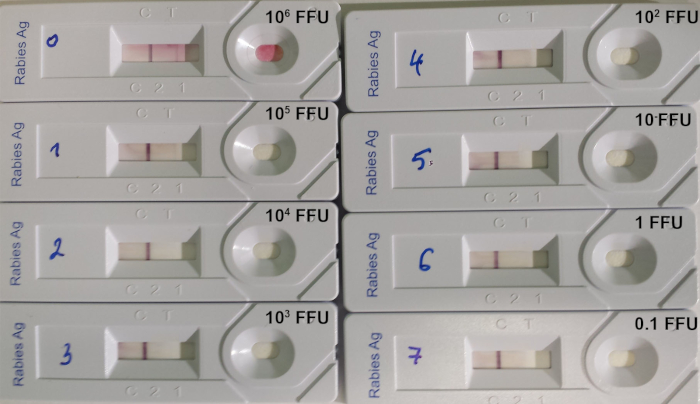
Figure 7: Example of determination of the limit of detection of RIDT14. A serial 10:1 dilution of a titrated rabies virus of the strain 9704ARG was used. The quantity of virus deposited on each device is indicated in FFU (fluorescent focus-forming units). Please click here to view a larger version of this figure.
| Pan-RABV RT-qPCR assay | |
| Reagent | μL/Reaction |
| 2X Reaction Mix (a buffer containing 0.4 mM of each dNTP and 6 mM MgSO4) | 10 |
| Nuclease free water | 1.5 |
| Taq3long (Forward) [10 μM] | 1 |
| Taq17revlong (Reverse) [10 μM] | 1 |
| RABV4 [10 μM] | 0.3 |
| RABV5 [10 μM] | 0.3 |
| MgSO4 [50-mM] (provided in the kit) | 0.25 |
| ROX Reference Dye (25 μM) (provided in the kit) | 0.05 |
| RNasin (40U/µL) (Promega) | 0.2 |
| SuperScript III RT/Platinum Taq Mix | 0.4 |
| Total per reaction | 15 |
| eGFP RT-qPCR assay | |
| Reagent | μL/Reaction |
| 2X Reaction Mix (a buffer containing 0.4 mM of each dNTP and 6 mM MgSO4) | 10 |
| Nuclease free water | 2.8 |
| EGFP1F (Forward) [10 μM] | 0.5 |
| EGFP2R (Reverse) [10 μM] | 0.5 |
| eGFP probe [10 μM] | 0.3 |
| MgSO4 [50-mM] (provided in the kit) | 0.25 |
| ROX Reference Dye (25 μM) (provided in the kit) | 0.05 |
| RNasin (40U/µL) (Promega) | 0.2 |
| SuperScript III RT/Platinum Taq Mix | 0.4 |
| Total per reaction | 15 |
| Pan-lyssa RT-qPCR assay | |
| Reagent | μL/Reaction |
| 2x SYBR Green Reaction Mix | 10 |
| Nuclease free water | 2.1 |
| Taq5long (Forward) [10 μM] | 1 |
| Taq16revlong (Reverse) [10 μM] | 1 |
| MgSO4 [50-mM] (provided in the kit) | 0.25 |
| ROX Reference Dye (25 μM) | 0.05 |
| RNasin (40U/µL) (Promega) | 0.2 |
| SuperScript III RT/Platinum Taq Mix | 0.4 |
| Total per reaction | 15 |
Table 1: Description of the master mix reaction solution for the three different RT-qPCR assays (pan-RABV RT-qPCR, pan-lyssa RT-qPCR and eGFP RT-qPCR).
| RT-qPCR assay | Name | Type | Length | Sequence (5’-3’) | Sense | Position |
| pan-RABV RT-qPCR assay | Taq3long | Primer | 22 | ATG AGA AGT GGA AYA AYC ATC A | S | 7273-7294a |
| Taq17revlong | Primer | 25 | GAT CTG TCT GAA TAA TAG AYC CAR G | AS | 7390-7414a | |
| RABV4 | Probe (FAM/TAMRA) | 29 | AAC ACY TGA TCB AGK ACA GAR AAY ACA TC | AS | 7314-7342a | |
| RABV5 | Probe (FAM/TAMRA) | 32 | AGR GTG TTT TCY AGR ACW CAY GAG TTT TTY CA | S | 7353-7384a | |
| Pan-lyssa RT-qPCR assay | Taq5long | Primer | 23 | TAT GAG AAA TGG AAC AAY CAY CA | S | 7272-7294a |
| Taq16revlong | Primer | 25 | GAT TTT TGA AAG AAC TCA TGK GTY C | AS | 7366-7390a | |
| eGFP RT-qPCR assay | EGFP1F | Primer | 20 | GAC CAC TAC CAG CAG AAC AC | S | 637-656b |
| EGFP2R | Primer | 19 | GAA CTC CAG CAG GAC CAT G | AS | 768-750b | |
| EGFP | Probe (FAM/TAMRA) | 22 | AGC ACC CAG TCC GCC CTG AGC A | S | 703-724b |
Table 2: Description of the primers/probes for the three different RT-qPCR assays (pan-RABV RT-qPCR, pan-lyssa RT-qPCR and eGFP RT-qPCR). aAccording to the Pasteur virus (PV) RABV genome sequence (GenBank accession number M13215). bAccording to the cloning vector pEGFP-1 sequence (GenBank accession number U55761).
| Pan-RABV RT-qPCR and eGFP RT-qPCR assays | ||||
| Step | Cycle | Temp | Time | Data Collection |
| Reverse Transcription | 1 | 45 °C | 15 min | |
| RT inactivation/initial denaturation | 1 | 95 °C | 3 min | |
| Amplification | 40 | 95 °C | 15 s | |
| 61 °C | 1 min | End point | ||
| Pan-lyssa RT-qPCR assay | ||||
| Step | Cycle | Temp | Time | Data Collection |
| Reverse Transcription | 1 | 45 °C | 15 min | |
| RT inactivation/initial denaturation | 1 | 95 °C | 3 min | |
| Amplification | 40 | 95 °C | 15 s | |
| 55 °C | 1 min | End point | ||
| Dissociation curve | 1 | 95 °C | 15 s | Increase 0.1 °C/s, 55–95 °C |
| 55 °C | 1 min | |||
| 95 °C | 15 s | |||
| 55 °C | 15 s | |||
Table 3: Description of the thermal cycling conditions for the three different RT-qPCR assays (pan-RABV RT-qPCR, pan-lyssa RT-qPCR and eGFP RT-qPCR).
| Assay | Analysis | Results | Interpretation |
| eGFP RT-qPCR | Cq in the interval of acceptance | Extraction validated | Analysis of other assays can be done |
| Cq out of the interval of acceptance | Extraction not validated | Retest the sample (repeat the run or/and the extraction), request another sample if necessary | |
| pan-RABV RT-qPCR | Cq <38 | Positive | Positive detection of viral RNA |
| Cq ≥38 | Negative | Analysis the pan-lyssa RT-qPCR assay | |
| pan-lyssa RT-qPCR | Melting curve considered as positive | Positive | Positive detection of viral RNA |
| Melting curve considered as negative | Negative | Absence of detection of viral RNA |
Table 4: Overall interpretation of the dual combined pan-lyssavirus RT-qPCR assay.
| Hemi-nested conventional PCR assay | PCR round | Name | Length | Sequence (5’-3’) | Sense | Positiona | Amplicon size (bp) |
| Hemi-nested PCR targeting the polymerase gene | 1st round | PVO5m | 20 | ATG ACA GAC AAY YTG AAC AA | S | 7170-7189 | 320 |
| PVO9 | 19 | TGA CCA TTC CAR CAR GTN G | AS | 7471-7489 | |||
| 2nd round | PVO5m | 20 | ATGA CAG ACA AYY TGA ACA A | S | 7170-7189 | 250 | |
| PVO8 | 22 | GGT CTG ATC TRT CWG ARY AAT A | AS | 7398-7419 | |||
| Hemi-nested PCR targeting the nucleoprotein gene | 1st round | N127 | 20 | ATG TAA CAC CTC TAC AAT GG | S | 55-74 | 1532 |
| N8m | 19 | CAG TCT CYT CNG CCA TCT C | AS | 1568-1586 | |||
| 2nd round | N127 | 20 | ATG TAA CAC CTC TAC AAT GG | S | 55-74 | 845 | |
| N829 | 19 | GCC CTG GTT CGA ACA TTC T | AS | 881-899 |
Table 5: Description of the primers used for the conventional hemi-nested PCR.
| Hemi-nested PCR targeting the polymerase gene | ||||
| Step | Cycle | Temperature | Time | |
| First and second rounds | Initial denaturation | 1 | 94 °C | 3 min |
| Denaturation | 35 | 94 °C | 30 s | |
| Hybridation | 56 °C | 45 s | ||
| Elongation | 72 °C | 40 s | ||
| Final elongation | 1 | 72 °C | 3 min | |
| Hemi-nested PCR targeting the nucleoprotein gene | ||||
| Step | Cycle | Temperature | Time | |
| First round | Initial denaturation | 1 | 94 °C | 3 min |
| Denaturation | 35 | 94 °C | 30 s | |
| Hybridation | 56 °C | 30 s | ||
| Elongation | 72 °C | 45 s | ||
| Final elongation | 1 | 72 °C | 3 min | |
| Second round | Initial denaturation | 1 | 94 °C | 3 min |
| Denaturation | 35 | 94 °C | 30 s | |
| Hybridation | 58 °C | 30 s | ||
| Elongation | 72 °C | 30 s | ||
| Final elongation | 1 | 72 °C | 3 min | |
Table 6: Description of the thermal cycling conditions for the conventional hemi-nested PCR.
| Lab | Country | Period of evaluation | Nb of samples | DFAT results | RIDT results | Sensitivity | Specificity | ||
| Pos | Neg | Pos | Neg | ||||||
| Lab 1 | France | 2015 | 82 | 50 | 32 | 50 | 32 | 96% | 93.7% |
| Lab 2 | Chad | 2012-2015 | 44 | 33 | 11 | 33 | 11 | 100% | 100% |
| Lab 3 | Ivory Coast | 2017 | 10 | 8 | 2 | 8 | 2 | 100% | 100% |
| Lab 4 | Mali | 2017 | 18 | 15 | 3 | 15 | 3 | 100% | 100% |
| Lab 6 | Italy | 2016 | 8 | 8 | 0 | 8 | 0 | 100% | – |
| All | 2015-2017 | 162 | 114 | 48 | 114 | 48 | 98.2% | 95.8% | |
Table 7: Determination of the intrinsic parameters (sensitivity, specificity) of the RIDT test compared to the reference DFAT method, based on the analysis of a total of 162 samples and with the participation of 5 different laboratories.
| Virus straina | Original host | Location | Initial concentration (FFU/mL)b | Limit of detection (FFU/mL)c |
| 9147FRA | Red fox | France | 3.1 x 107 | 106 |
| CVS | Lab isolate | – | 1.6 x 107 | 106 |
| 8743THA | Human | Thailand | 8.1 x 107 | > 8.1 x 106 |
| 9508CZK (SAD) | Lab isolate | – | 5.4 x 108 | 107 |
| PV | Lab isolate | – | 4.3 x 107 | 106 |
| 9001FRA | Dog | French Guiana | 2.4 x 106 | > 2.4 x 105 |
| 9704ARG | Bat | Argentina | 9.5 x 107 | 105 |
| 04030PHI | Human | Philippines | 2.5 x 107 | 105 |
Table 8: Limit of detection of the RIDT using 8 different titrated rabies virus suspensions (from Léchenne et al. 201614). aCVS: Challenge virus strain, SAD: Street Alabama Dufferin, PV: Pasteur virus. bNumber of fluorescent focus-forming units (FFU) per mL. cNumber of fluorescent focus-forming units (FFU) deposited on the strip.
| RIDT performed in | ||||||||||
| Lab 1 | Lab 2 | Combined | ||||||||
| Positive | Negative | Total | Positive | Negative | Total | Positive | Negative | Total | ||
| Viral RNA detection | Positive | 18 | 1 | 19 | 26 | 0 | 32 | 44 | 7 | 51 |
| Negative | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 3 | 3 | |
| Total | 18 | 1 | 19 | 26 | 0 | 35 | 44 | 10 | 54 | |
Table 9: Detection of viral RNA with RT-qPCR on Anigen test strip used in laboratory conditions (Lab 1), in field conditions and shipped at ambient temperature (Lab 2) or combined (from Léchenne et al. 201614).
Supplementary Table 1: Description of the 162 samples tested with the RIDT test for determination of its intrinsic parameters presented in Table 7. Please click here to view this table (Right click to download).
Discussion
The RIDT is a simple, rapid and low-cost method for postmortem rabies diagnosis and a promising field alternative to laboratory testing. The application of such a test, especially for decentralized areas of low- and middle-income countries, would improve understanding of rabies virus prevalence and transmission on a local and potentially national scale. When combined with the rapid brain sample collection method (without full necropsy), a great advantage is that the test can be entirely performed in the field setting, away from laboratory facilities. Brain samples collected via the foramen magnum can be used for testing, thus it is not required to completely open the animal skull. The test is simple to perform and interpret and is particularly suitable for field surveillance activities14. Other advantages of the RIDT over the DFAT or DRIT are no need for positive and negative controls and kit storage at room temperature. In addition, the modified protocol, where the dilution step (1:10) into PBS is omitted, does not require extra reagents to perform the test and further simplifies the procedure under field conditions.
A key point is the quality of the brain samples. Samples should be collected and tested as soon as possible after death of the suspected animal, or kept at cool temperature before testing, to avoid degradation. Decomposed samples should not be tested because it can affect the result (risk of false negative result). Although no data are yet available regarding the loss of sensitivity of RIDT over time for brain samples, we hypothesize that it is similar compared to the DFAT test32. However, time between the death of the animal and performing the test can be reduced, as the test can be done quickly and directly in the field. Thus, there is in general a lower risk of decomposed samples.
Another critical step within the protocol is the sample suspension migration. The migration should start directly after deposit of the sample (1-5 min). High viscosity of the suspension could therefore negatively influence the migration. Gently scratching the bottom of the device deposit site with the dropper and adding 1-2 more drops often solves this problem, and the migration begins immediately after.
Most of the RIDT tests performed in African laboratories (Chad, Ivory Coast and Mali) were performed at ambient temperature which can exceed 30 °C, whereas the range of temperature for storage and use recommended by the manufacturer is 15 °C – 30 °C. Although we did not identify any impact of high temperature on RIDT test performance, it is necessary to evaluate it more carefully. Similarly, the impact of high temperature during storage and transportation of the device after use for viral RNA detection and genotyping needs additional evaluation. The sensitivity of the viral RNA detection by RT-qPCR from the RIDT strip can be affected by the quality of the brain sample initially used in the test, but also by the condition of storage of the RIDT tests after use. For example, the sensitivity of the RNA detection was higher when used RIDT tests were stored under controlled laboratory conditions (94.7%) compared to under field conditions (e.g., Chad) (81.2%)14. These conditions might also affect the integrity (especially the length) of RNA fixed on the strip, possibly explaining the moderate sensitivity for genotyping based on longer PCR amplicons (e.g., >500 nucleotides)14. The sensitivity of RT-qPCR performed on the test strip was lower than that obtained using FTA Whatman cards (80.6%)14. Similar to other molecular techniques, the viral load can also impact the success of genotyping based on RDIT strips, with potential negative results for samples with low viral load14.
The test is not currently recommended by WHO and OIE for routine diagnosis and disease surveillance, and a result cannot be used on its own to guide PEP decision making. Further test validation is still needed. However, accurate quick rabies diagnosis is a crucial element of well-functioning continuous rabies surveillance systems and is instrumental to increase political commitment, which is eminently important for successful sustainable rabies control33. RIDT tests offer new rabies diagnostic opportunities in this context and are a useful tool to expand animal rabies surveillance in the field in low- or middle-income enzootic areas.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported through the Global Alliance for Vaccines and Immunisation (GAVI), the Wolfermann Nägeli Foundation, the Swiss African Research Cooperation (SARECO), the SWF Stiftung für wissenschaftliche Forschung, the Freiwillige Akademische Gesellschaft (FAG) Basel, the Bilateral Science and Technology Cooperation Programme of Switzerland with Asia and the Novartis Foundation for biomedical research.
We thank especially the dog owners, the veterinary personnel and the laboratory staff for their great commitment. We also want to acknowledge Lisa Crump for the language editing.
Materials
| Anti-Rabies Nucleocapsid Conjugate (lyophilizied, adsorbed) | Bio-Rad, France | 3572112 | Fluorescein-5-isothiocyanate (FITC) conjugated polyclonal antibody against the nucleocapsid of rabies virus. Use for the DFAT reference tecnnique. |
| Applied Biosystems 7500 Real-Time PCR System | Applied Biosystems, France | 4351104 | Amplification of the viral RNA by RT-qPCR after extraction from the RIDT device. |
| Disposable plastic pipette, drinking straw, clamp, dropper | – | – | Equipment used for the collection of the brain stem (medulla oblongata) via the foramen magnus (occipital route). |
| Evans Blue Solution 1% | Bio-Rad, France | 3574911 | Counter-coloration used for the DFAT to facilite the reading under UV microscope. |
| Primer eGFPF1 | Eurofins Genomics, Germany | – | Forward primer for the amplification of the internal control eGFP by RT-qPCR after RNA extraction from the RIDT device, sequence: 5'-GACCACTACCAGCAGAACAC-3'. |
| Primer eGFPR2 | Eurofins Genomics, Germany | – | Reverse primer for the amplification of the internal control eGFP by RT-qPCR after extraction from the RIDT device, sequence: 5'-GAACTCCAGCAGGACCATG-3'. |
| Primer Taq17 revlong | Eurofins Genomics, Germany | – | Reverse primer for the amplification of the viral RNA by RT-qPCR after RNA extraction from the RIDT device, sequence: 5'-ATGAGAAGTGG AAYAAYCATCA-3'. |
| Primer Taq3 long | Eurofins Genomics, Germany | – | Forward primer for the amplification of the viral RNA by RT-qPCR after RNA extraction from the RIDT device, sequence: 5'-GATCTGTCTGAA TAATAGAYCCARG-3'. |
| Probe eGFP FAM/TAMRA | Eurofins Genomics, Germany | – | Forward primer for the amplification of the internal control by RT-qPCR after extraction from the RIDT device, sequence: 5'-AGCACCCAGT CCGCCCTGAGCA-3'. |
| Probe RABV4 FAM/TAMRA | Eurofins Genomics, Germany | – | Probe for the amplification of the viral RNA by RT-qPCR after RNA extraction from the RIDT device, sequence: 5'-AACACYTGATCBA GKACAGARAAYACATC-3'. |
| Probe RABV5 FAM/TAMRA | Eurofins Genomics, Germany | – | Probe for the amplification of the viral RNA by RT-qPCR after RNA extraction from the RIDT device, sequence: 5'-AGRGTGTTTTCYAG RACWCAYGAGTTTTTYCA-3'. |
| Rapid Rabies Ag Test Kit | BioNote Inc., Republic of Korea | RG18-01DD | Rapid immunochromatographic diagnostic test (RIDT, also named lateral flow device or LFD) for the post-mortem diagnosis of rabies. |
| Recombinant RNasin Ribonuclease Inhibitor | Promega, USA | N2515 | Enzyme used with the kit for the amplification of the viral RNA by RT-qPCR after extraction from the RIDT device. |
| SuperScript III Platinum One-Step qRT-PCR Kit | Invitrogen, France | 11732-020 | Kit for the amplification of the viral RNA by RT-qPCR after extraction from the RIDT device. |
| TRIzol Reagent | Invitrogen, France | 15596026 | Phenol/chloroforme based total RNA extraction using the cellulose membrane of the RIDT. |
References
- Hampson, K., et al. Correction: Estimating the Global Burden of Endemic Canine Rabies. PLoS Neglected Tropical Diseases. 9 (5), e0003786 (2015).
- Rupprecht, C., Kuzmin, I., Meslin, F. Lyssaviruses and rabies: current conundrums, concerns, contradictions and controversies. F1000Research. 6, 184 (2017).
- Walker, P. J., et al. ICTV Virus Taxonomy Profile: Rhabdoviridae. The Journal of General Virology. 99 (4), 447-448 (2018).
- World Health Organization (WHO). WHO Expert Consultation on Rabies, third report. HO Technical Report Series, No. 1012. , (2018).
- Dacheux, L., et al. More Accurate Insight into the Incidence of Human Rabies in Developing Countries through Validated Laboratory Techniques. PLoS Neglected Tropical Diseases. 4 (11), e765 (2010).
- Welburn, S. C., Beange, I., Ducrotoy, M. J., Okello, A. L. The Neglected Zoonoses – The Case for Integrated Control and Advocacy. Clinical Microbiology and Infection: The Official Publication of the European Society of Clinical Microbiology and Infectious Diseases. , (2015).
- Dacheux, L., Bourhy, H. Diagnostic tests for human rabies. Revue Scientifique Et Technique (International Office of Epizootics). 37 (2), 581-593 (2018).
- Kasempimolporn, S., Saengseesom, W., Lumlertdacha, B., Sitprija, V. Detection of rabies virus antigen in dog saliva using a latex agglutination test. Journal of Clinical Microbiology. 38 (8), 3098-3099 (2000).
- Kang, B., et al. Evaluation of a rapid immunodiagnostic test kit for rabies virus. Journal of Virological Methods. 145 (1), 30-36 (2007).
- Nishizono, A., et al. A simple and rapid immunochromatographic test kit for rabies diagnosis. Microbiology and Immunology. 52 (4), 243-249 (2008).
- Kasempimolporn, S., Saengseesom, W., Huadsakul, S., Boonchang, S., Sitprija, V. Evaluation of a rapid immunochromatographic test strip for detection of Rabies virus in dog saliva samples. Journal of Veterinary Diagnostic Investigation: Official Publication of the American Association of Veterinary Laboratory Diagnosticians, Inc. 23 (6), 1197-1201 (2011).
- Ahmed, K., et al. Evaluation of a monoclonal antibody-based rapid immunochromatographic test for direct detection of rabies virus in the brain of humans and animals. The American Journal of Tropical Medicine and Hygiene. 86 (4), 736-740 (2012).
- Léchenne, M., et al. Validation of a Rapid Rabies Diagnostic Tool for Field Surveillance in Developing Countries. PLoS Neglected Tropical Diseases. 10 (10), e0005010 (2016).
- Yang, D. K., et al. Comparison of four diagnostic methods for detecting rabies viruses circulating in Korea. Journal of Veterinary Science. 13 (1), 43-48 (2012).
- Markotter, W., et al. Evaluation of a rapid immunodiagnostic test kit for detection of African lyssaviruses from brain material. The Onderstepoort Journal of Veterinary Research. 76 (2), 257-262 (2009).
- Reta, T., et al. Evaluation of Rapid Immunodiagnostic Test for Rabies Diagnosis Using Clinical Brain Samples in Ethiopia. Journal of Veterinary Science & Medical Diagnosis. 2 (3), 1-3 (2013).
- Servat, A., Picard-Meyer, E., Robardet, E., Muzniece, Z., Must, K., Cliquet, F. Evaluation of a Rapid Immunochromatographic Diagnostic Test for the detection of rabies from brain material of European mammals. Biologicals: Journal of the International Association of Biological Standardization. 40 (1), 61-66 (2012).
- Certoma, A., et al. Assessment of a Rabies Virus Rapid Diagnostic Test for the Detection of Australian Bat Lyssavirus. Tropical Medicine and Infectious Disease. 3 (4), (2018).
- Ahmad, A., Singh, C. K. Comparison of rapid immunodiagnosis assay kit with molecular and immunopathological approaches for diagnosis of rabies in cattle. Veterinary World. 9 (1), 107-112 (2016).
- Sharma, P., Singh, C. K., Narang, D. Comparison of immunochromatographic diagnostic test with Hheminested Reverse transcriptase polymerase chain reaction for detection of rabies virus from brain samples of various species. Veterinary World. 8 (2), 135-138 (2015).
- Voehl, K. M., Saturday, G. A. Evaluation of a rapid immunodiagnostic rabies field surveillance test on samples collected from military operations in Africa, Europe, and the Middle East. U.S. Army Medical Department Journal. , 27-32 (2014).
- Servat, A., Robardet, E., Cliquet, F. An inter-laboratory comparison to evaluate the technical performance of rabies diagnosis lateral flow assays. Journal of Virological Methods. 272, 113702 (2019).
- Eggerbauer, E., et al. Evaluation of Six Commercially Available Rapid Immunochromatographic Tests for the Diagnosis of Rabies in Brain Material. PLoS Neglected Tropical Diseases. 10 (6), e0004776 (2016).
- Barrat, J., Meslin, F. X., Kaplan, M. M., Koprowski, H. Simple technique for the collection and shipment of brain specimens for rabies diagnosis. Laboratory techniques in rabies. , 425-432 (1996).
- Dacheux, L., et al. Dual Combined Real-Time Reverse Transcription Polymerase Chain Reaction Assay for the Diagnosis of Lyssavirus Infection. PLoS Neglected Tropical Diseases. 10 (7), e0004812 (2016).
- Dacheux, L., et al. A reliable diagnosis of human rabies based on analysis of skin biopsy specimens. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America. 47 (11), 1410-1417 (2008).
- Rupprecht, C., Fooks, A. R., Abela-Ridder, B. Application of next generation sequencing to rabies virus and other lyssaviruses. Laboratory techniques in rabies. 2, 49-61 (2019).
- Rupprecht, C., Fooks, A. R., Abela-Ridder, B. Conventional pan-lyssavirus reverse transcriptase polymerase chain reaction. Laboratory techniques in rabies. 2, 1-16 (2019).
- Rupprecht, C., Fooks, A. R., Abela-Ridder, B. Rabies real-time reverse transcriptase polymerase chain reaction. Laboratory techniques in rabies. 2, 17-34 (2019).
- Talbi, C., et al. Evolutionary history and dynamics of dog rabies virus in western and central Africa. The Journal of General Virology. 90 (Pt 4), 783-791 (2009).
- McElhinney, L. M., Marston, D. A., Brookes, S. M., Fooks, A. R. Effects of carcase decomposition on rabies virus infectivity and detection. Journal of Virological Methods. 207, 110-113 (2014).
- Vigilato, M. A. N., et al. Progress towards eliminating canine rabies: policies and perspectives from Latin America and the Caribbean. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 368 (1623), 20120143 (2013).

