siRNA Electroporation to Modulate Autophagy in Herpes Simplex Virus Type 1-Infected Monocyte-Derived Dendritic Cells
Summary
In this study, we present inhibitor- and siRNA-based strategies to interfere with autophagic flux in Herpes simplex virus type-1 (HSV-1)-infected monocyte-derived dendritic cells.
Abstract
Herpes simplex virus type-1 (HSV-1) induces autophagy in both, immature dendritic cells (iDCs) as well as mature dendritic cells (mDCs), whereas autophagic flux is only observed in iDCs. To gain mechanistic insights, we developed efficient strategies to interfere with HSV-1-induced autophagic turnover. An inhibitor-based strategy, to modulate HSV-1-induced autophagy, constitutes the first choice, since it is an easy and fast method. To circumvent potential unspecific off-target effects of such compounds, we developed an alternative siRNA-based strategy, to modulate autophagic turnover in iDCs upon HSV-1 infection. Indeed, electroporation of iDCs with FIP200-specific siRNA prior to HSV-1 infection is a very specific and successful method to ablate FIP200 protein expression and thereby to inhibit autophagic flux. Both presented methods result in the efficient inhibition of HSV-1-induced autophagic turnover in iDCs, whereby the siRNA-based technique is more target specific. An additional siRNA-based approach was developed to selectively silence the protein expression of KIF1B and KIF2A, facilitating autophagic turnover upon HSV-1 infection in mDCs. In conclusion, the technique of siRNA electroporation represents a promising strategy, to selectively ablate the expression of distinct proteins and to analyze their influence upon an HSV-1 infection.
Introduction
The generation of human monocyte-derived dendritic cells (DCs) constitutes an appropriate in vitro model to study the functions and biology of this important immune cell type. Isolation as well as differentiation of monocytes into DCs has been well established in recent years1,2. The infection of DCs with the α-herpesvirus herpes simplex virus type-1 (HSV-1) serves as a model system to study HSV-1-mediated modulations of DC biology2,3,4,5,6. This is particularly important to elucidate how herpesviruses dampen or inhibit potent antiviral immune responses, to establish latency in immune-privileged niches inside the host7,8. In this respect, herpesviruses are very successful pathogens that are wide-spread throughout the population reaching a sero-prevalence of up to 90 % according to the geographic region9. To understand and possibly prevent this, more insights into the HSV-1-mediated modulations of the host’s immune system, and especially of immune cells such as DCs, are required.
A completely new observation regarding the interplay of DCs with HSV-1 was recently published by Turan et al.10. The authors demonstrated that the accomplishment of HSV-1 replication is strictly dependent on the maturation status of DCs. In iDCs, complete replication of HSV-1 is facilitated by autophagy-dependent mechanisms. While HSV1 induces autophagy in both, iDCs and mDCs, autophagic flux is observed only in iDCs. This in turn facilitates nuclear egress of viral capsids via autophagic degradation of nuclear lamins in iDCs. To gain mechanistic insights into this HSV-1-induced degradation pathway in iDCs versus mDCs, new and efficient strategies are critical to investigate autophagic flux.
Macroautophagy (autophagy) is a well-conserved multistep process targeting intracellular proteins or whole organelles for lysosomal digestion11. Simplistically, autophagy can be divided into the (i) initiation, (ii) membrane nucleation, (iii) vesicle expansion, and (iv) autophagosome-lysosome fusion phase12. During initiation (i), components such as the activated ULK1/2 kinase complex, containing the focal adhesion kinase family interacting protein of 200 kD (FIP200), are critical to activate the beclin-1-Vps34-AMBRA1 complex. Subsequently, membrane nucleation (ii) initiates phagophore formation13, which engulfs cytoplasmic cargos that are marked by molecules such as p6214. During vesicle expansion and autophagophore maturation (iii) microtubule-associated protein light chain 3 (LC3)-I is converted into its lipidated form LC3-II that is inserted into the autophagosomal membrane. Thus, LC3-I to -II conversion rates are an indicator for autophagy induction by mirroring the formation of mature autophagosomes15,16. Upon autophagosome-lysosome fusion (iv), not only the autophagic cargo but also associated p62 and LC3-II proteins undergo degradation (e.g., by hydrolysis). Thus, loss of p62 and LC3-II serve as markers for autophagic flux17. The fusion of autophagosomes with lysosomes, and thus following autophagic turnover, is highly dependent on the intracellular lysosomal localization. This is, among others, regulated by the kinesin family members KIF1B and KIF2A, which were shown to negatively affect autophagosome-lysosome fusion18. Interestingly, protein expression of KIF1B and KIF2A is induced upon DC maturation and is thereby responsible for the inefficient autophagic flux in HSV-1-infected mDCs, which hampers complete HSV-1 replication10.
Experimental attempts to modulate autophagy include the usage of compounds known to induce or inhibit this particular pathway19,20,21. In this study, we describe two inhibitor-based strategies to block autophagic turnover in HSV-1-infected iDCs. The first compound used in our experiments is specific and potent autophagy inhibitor-1 (spautin-1), which was described to promote beclin-1-Vps34-AMBRA1 complex degradation during the initiation phase of autophagy22. The second compound used in the present study is bafilomycin-A1 (BA1), a V-ATPase inhibitor that blocks the late autophagic events (i.e., autophagosome-lysosome fusion as well as autolysosome acidification)23,24. The usage of either of these two inhibitors prior to iDC infection with HSV-1 potently inhibits autophagy, but does not disturb efficient viral gene expression. Thus, this inhibitor-based strategy prior to HSV-1 infection offers a powerful tool to inhibit HSV-1-induced autophagic flux that can easily be expanded for a plethora of different cell types and viruses, which also potentially induce autophagy.
To overcome a major downside of an inhibitor-based approach (i.e., unspecific off-target effects), we developed an siRNA-based method to block autophagic flux in (HSV-1-infected) iDCs. The technique of siRNA electroporation represents a powerful alternative strategy, via selective ablation of the expression of distinct proteins (i.e., autophagic components). In our experiments iDCs were electroporated with FIP200-specific siRNA using the electroporation apparatus I (see Table of Materials) and a modified protocol described by Gerer et al. (2017) and Prechtel et al. (2007), to inhibit autophagy during the initiation phase25,26. This technique allowed us to specifically knockdown FIP200 expression in iDCs, without interfering with cell viability and their immature phenotype two days post electroporation. Noteworthy, HSV-1 infection was established in these electroporated iDCs mirrored by efficient viral protein expression. This siRNA-based technique offers a unique benefit (i.e., that a variety of different autophagic components, even in combination), can be specifically targeted for the ablation of their expression.
In this study, we further describe an siRNA-based method to induce autophagic flux also in HSV-1-infected mDCs. In this case, iDCs were electroporated with siRNA targeted against KIF1B and KIF2A prior to DC maturation using the electroporation apparatus II (see Table of Materials). Since both proteins are upregulated during DC maturation and known to negatively regulate fusion of autophagosomes with lysosomes10,18, their knockdown strongly induced autophagic flux in mDCs upon HSV-1 infection. Thus, the siRNA-based technique enabled us to specifically induce autophagic turnover via interfering with KIF protein expression in mDCs, and could thereby mimic their expression levels in iDCs.
In summary, we present two distinct methods to inhibit autophagic flux in HSV-1-infected iDCs. While the first inhibitor-based approach constitutes an easy, cheap and fast way to interfere with autophagic degradation, the second siRNA-based technique is more specific and a very suitable method to support and verify the results of inhibitor-based experiments. In addition, we describe a method to induce autophagic flux also in HSV-1-infected mDCs, via the siRNA-mediated knockdown of two KIF proteins.
Protocol
Monocyte-derived DCs were generated from leukapheresis products of healthy donors. For this, a positive vote from the local ethics committee has been obtained (reference number 4556). The experiments of the present study were performed in accordance with the recommendations of the ethics committee of the "Friedrich-Alexander-Universität Erlangen-Nürnberg" (reference number 4556). All donors approved a written informed consent, including the accordance with the Declaration of Helsinki.
1. Generation and handling of immature dendritic cells (iDCs) and mature dendritic cells (mDCs)
- Isolate human peripheral blood mononuclear cells (PBMCs) from leukoreduction system chambers (LRSCs) as previously described27. Avoid cryopreservation of PBMCs and use them directly upon isolation to obtain higher DC yields.
- Generate human DCs from PBMCs of different healthy donors in T175 cell culture flasks as previously described10,27. Briefly, use 350-400 millions of PBMCs in 30 mL of DC medium (RPMI 1640 without L-glutamine, 1% (v/v) AB-serum, 100 U/mL penicillin, 100 mg/mL streptomycin, 0.4 mM L-glutamine, 10 mM HEPES) per cell culture flask for isolation of monocytes by adherence. After 1 h, wash off nonadherent fraction using RPMI 1640. Add fresh DC medium supplemented with 800 U/mL GM-CSF and 250 U/mL IL-4, and incubate for 3 days.
- On day 3 post adherence, add 5 mL of fresh DC medium containing GM-CSF and IL-4 with a final concentration of 400 U/mL and 250 U/mL per cell culture flask, respectively, for DC differentiation.
- To harvest iDCs, gently rinse loosely-adherent iDCs from the bottom of the cell culture flask, on day 4 post adherence. Repeat this step 2 times. For generation of mDCs, add maturation cocktail composed as follows: GM-CSF (final concentration: 40 U/mL), IL-4 (final concentration: 250 U/mL), IL-6 (final concentration: 1000 U/mL), IL-1β (final concentration: 200 U/mL), TNF-α (final concentration: 10 ng/mL), prostaglandin E2 (PGE2; final concentration: 1 μg/mL).
- Six days post adherence (two days post induction of maturation using a cytokine cocktail), rinse mDCs from the bottom of the cell culture flask. Repeat this step two times.
NOTE: Immature and mature DCs can be sequentially generated from identical donors in 1 cell culture flask. To do so, (i) separate the appropriate number of iDCs and (ii) induce maturation of the remaining cells in the flasks using the cytokine cocktail listed in step 1.2.2.
- Transfer iDCs or mDCs in the respective cell culture medium into 50 mL tubes. Harvest the cells via centrifugation at 300 x g for 5 min.
- Gently resuspend (=wash) DCs in 5-10 mL of RPMI 1640 per cell culture flask. Combine respective DC suspensions in one tube.
- Define the cell number using a counting chamber or an alternative method. Avoid temperature alterations when handling iDCs, to reduce the risk of phenotypic changes.
2. Flow cytometric analyses to monitor the phenotypic maturation status
- Transfer iDCs or mDCs (0.5 x 106) from step 1.3.1 into a 1.5 mL tube. Harvest the cells via centrifugation at 3390 x g for 1.5 min. Wash the cells once with FACS buffer (PBS supplemented with 2% fetal calf serum (FCS)).
- Resuspend the cells in 100 µL of antibody staining solution (FACS buffer) containing specific fluorochrome-labeled antibodies against defined surface molecules.
- Use the following antibodies to verify purity (CD3-FITC, CD14-PE) as well as maturation status of DCs (CD80-PacBlue/-PE-Cy5, CD11c-PE-Cy5, CCR7-PE-Cy7, CD83-APC, CD86-PE, MHCII-APC-Cy7).
- Prepare one unstained sample in 100 µL of FACS buffer as a control.
- Stain the cells on ice in the dark for 30 min.
- Subsequently wash the cells two times in 1 mL of FACS buffer and centrifuge at 3390 x g for 1.5 min.
- Finally, resuspend the cells in 200 µL of FACS buffer supplemented with 2% PFA and analyze the cells by flow cytometry. Fixed cells can be stored at 4° C in the dark up to 2 days.
3. Infection procedure of DCs with Herpes simplex virus type-1 (HSV-1) and interference of HSV-1-induced autophagic flux via spautin-1 or bafilomycin-A1
NOTE: The strain HSV-1/17+/CMV-EGFP/UL43 (HSV-1 EGFP) used in this study was obtained from the laboratory strain HSV-1 strain 17+. The HSV-1 EGFP strain expresses the enhanced green fluorescent protein (EGFP) which has been inserted into the UL43 gene locus under control of the CMV promoter. EGFP serves as a marker for HSV-1 infection. Moreover, the strain HSV1-RFPVP26 was used for DC infection studies (previously described in Turan et al., 2019). This virus expresses the capsid surface protein VP26 fused to monomer red fluorescent protein (mRFP).
- Transfer iDCs or mDCs (2 x 106) from step 1.3 into a 2 mL tube. Subsequently, centrifuge the cells at 3390 x g for 1.5 min and discard the supernatant.
- Gently resuspend the cells in infection medium (RPMI 1640 supplemented with 20 mM HEPES).
- To inhibit the autophagosomal-lysosomal degradation pathway, pre-treat DCs with spautin-1 or bafilomycin-A1 1 h prior to infection. Add either 10 µM spautin-1 or 1 µM BA1, or DMSO as untreated control, to the infection medium. Incubate the cells in a heating block at 300 rpm shaking at 37 °C for 1 h.
- For infection studies, inoculate the cells with HSV-1 virions at a multiplicity of infection (MOI) of 2. Add the respective volume of MNT buffer (30 mM 2-(N-morpholino)ethanesulfonic acid (MES), 100 mM NaCl, 20 mM Tris) as mock control. Incubate the cells in a heating block at 300 rpm shaking at 37 °C for 1 h.
- 1 h post infection (hpi), collect the cells at 3390 x g for 1.5 min. Aspirate inoculum and gently resuspend the cells in DC medium containing 40 U/mL of GM-CSF, 250 U/mL of IL-4 and either 10 µM spautin-1, 1 µM BA1, or DMSO as control. Seed mock-treated and HSV-1-infected cells at a final concentration of 1 x 106/mL into a 6-well plate.
- At 16-24 hpi, harvest the cells by rinsing (mDCs) or using a cell scraper (iDCs). Transfer the cells into a 1.5 mL safelock tube.
- Collect the cells via centrifugation at 3390 x g for 1.5 min and wash the pellet once by adding 1 mL of PBS.
- Vigorously resuspend the cells in lysis mix containing 29 µL of 2x Roti-Load, 1 µL 100 mM MgCl2 and 12.5 U/mL benzonase.
- For cell lysis and DNA digestion using benzonase, incubate the samples at 37 °C for 10 min. Subsequently, denature the proteins at 95 °C for 10 min.
- Perform SDS-PAGE and Western blot analyses to verify protein levels of LC3BI/II, p62, ICP0/5, and GAPDH.
4. Interference of HSV-1-induced autophagic flux via electroporation of iDCs using FIP200-siRNA
NOTE: The present protocol for siRNA electroporation was modified from Prechtel et al. (2007) and Gerer et al. (2017).
- Transfer iDCs (12 x 106) at day 3.5 post adherence into a 50 mL tube. Subsequently, centrifuge the cells at 300 x g for 5 min and discard the supernatant. In parallel, perform flow cytometric analysis to monitor the maturation status as described in step 2 (Use Life/Dead violet instead of CD80-PacBlue).
- Gently wash iDCs in 5 mL of OptiMEM without phenol red and centrifuge the cells at 300 x g for 5 min. Discard the supernatant and gently resuspend iDCs in 200 µL of OptiMEM without phenol red, adjusting a cell concentration of 6 x 106/100 µL. Do not place the cells on ice and avoid temperature alterations. Move on quickly and avoid long incubation periods of iDCs in OptiMEM without phenol red.
- Transfer either 75 pmol of FIP200-specific siRNA or 75 pmol of scrambled siRNA, as a control, into 4 mm electro cuvettes and add 100 µL (6×106 cells) of the cell suspension. Directly pulse iDCs using the electroporation apparatus I, applying the following settings: 500 V for 1 ms.
- Prior to the experimental procedure, prepare siRNA suspensions according to the manufacturer’s instruction, aliquot and store them at -20 °C. Thaw and keep them on ice when using these siRNAs for electroporation. Before electroporating the samples, perform a test pulse.
- After electroporation, directly transfer iDC into 6-well plates with fresh pre-warmed DC medium (supplemented with 40 U/mL of GM-CSF and 250 U/mL of IL-4). Seed the cells at a final concentration of 1 x 106/mL and place them into an incubator. Do not rinse the cells out of the electro cuvette.
- After 48 h, first examine the morphology of electroporated iDCs microscopically. Then, harvest the cells using a cell scrapper and transfer them into 15 mL tubes. Rinse the wells with 1 mL of PBS supplemented with 0.01% EDTA and transfer the solution in the respective tubes.
- Subsequently, split 6 x 106 iDCs per siRNA condition as described in the next steps.
- Use 0.5 x 106 cells to assess the maturation status and cell viability as described in step 2. Use the following antibodies: CD11c-PE-Cy5, CCR7-PE-Cy7, CD83-APC, MHCII-APC-Cy7 and Life/Dead violet.
- Use 1 x 106 cells for Western blot analyses to verify FIP200-specific knockdown efficiency. Transfer and harvest the cells into a 1.5 mL safelock tube by centrifugation at 3390 x g for 1.5 min. Prepare cell lysates as described in step 3.4 and perform Western blot analyses.
- Use the remaining cells (4.5 x 106) for HSV-1 infection experiments. For each experimental condition, transfer 2.25 x 106 iDCs into 2 mL tubes and either infect them with HSV1 at an MOI of 2 or add MNT buffer as a mock control. Perform the infection as described in step 3.3.
- At 20 hpi, harvest the cells using a cell scraper and prepare cell lysates for Western blot analyses as described in step 3.4.
5. Modulation of the autophagosome-lysosomal pathway in HSV-1-infected mDCs using KIF1B/2A-siRNA electroporation
- Transfer iDCs (24 x 106) at day 4 post adherence into a 50 mL tube. Subsequently, centrifuge the cells at 300 x g for 5 min and discard the supernatant.
- Gently resuspend (= wash) iDCs in 8 mL of PBS, to adjust a final cell concentration of 3 x 106/mL. Transfer 3 x 106 cells into 1.5 mL tubes and harvest the cells at 3390 x g for 1.5 min.
- Resuspend iDCs in 100 µL of buffer P3 containing the supplement mix (according to the manufacturer’s instructions; electroporation kit apparatus II) and either (i) 75 pmol of KIF1B-specific siRNA, (ii) 75 pmol of KIF2A-specific siRNA, or (iii) both. Use (iv) the respective amount of scrambled siRNA as a control. Prepare two tubes for each siRNA condition (6 x 106 cells) and transfer the suspensions into separate electro cuvettes. Directly pulse iDCs applying the pulse "EH-100" using the electroporation apparatus II.
- Prior to the experimental procedure, prepare siRNA suspensions according to the manufacturer’s instruction, aliquot and store them at -20 °C. Thaw and keep them on ice when using these siRNAs for electroporation. Do not place iDCs on ice and avoid temperature alterations. Move on quickly and avoid long incubation of iDCs in PBS or buffer P3.
- Directly after electroporation, add 500 µL of pre-warmed RPMI 1640 to electro cuvettes. Incubate the cells in an incubator for 5-10 min. Transfer iDCs into 6-well plates with fresh pre-warmed DC medium (supplemented with 40 U/mL of GM-CSF and 250 U/mL of IL-4). Combine respective conditions into one well, seed the cells at a final concentration of 1·106/mL and place them into an incubator.
- 4 h after incubation, add the maturation cocktail containing the cytokines listed in step 1.2.2.
NOTE: Prepare one sample from non-electroporated DCs (1 x 106) as control for flow cytometric analyses 2 days post electroporation. Treat control cells analogously to electroporated samples. - Two days post electroporation, harvest cells by resuspension and transfer into 15 mL tubes. Rinse the wells with 1 mL of PBS and transfer the suspensions in the respective tubes. Split 6·106 DCs per siRNA condition as described in the following steps:
- Use 0.25 x 106 DCs (electroporated and non-electroporated) to check for maturation status and cell viability as described in step 2. Use the following antibodies: CD80-PE-Cy5, CD83-APC, CD86-PE, MHCII-APC-Cy7, and Life/Dead violet.
- Use 0.75 x 106 cells for Western blot analyses to assess KIF1B/2A-specific knockdown efficiency. Collect the cells into a 1.5 mL safelock tube by centrifugation at 3390 x g for 1.5 min. Prepare cell lysates as described in step 3.4 and perform Western blot analyses.
- Use the remaining cells (5 x 106) from each siRNA condition and perform HSV-1 infection experiments. For each experimental condition, transfer 2.5 x 106 iDCs into 2 mL tubes and either infect them with HSV-1 at an MOI of 2 or add MNT buffer as a mock control. Perform the infection as described in step 3.3.
- At 20 h post infection, harvest the cells by resuspension and prepare cell lysates for Western blot analyses to verify the induction of HSV-1-induced autophagic turnover, as described above in step 3.4.
Representative Results
In this manuscript, we describe methods to interfere with HSV-1-induced autophagy in dendritic cells. This includes the generation of human monocyte-derived iDCs and mDCs, which were phenotypically analyzed by flow cytometry (Figure 1). On day 4 post adherence, DCs show an immature phenotype characterized by weak expression of CD80, CCR7, and CD83 as well as high CD11c and intermediate MHCII expression. Since CD3 and CD14 signals are missing, T cell and monocyte contaminations can be excluded. On day 6 post adherence (i.e., day 2 post induction of maturation), DCs show a mature phenotype reflected by a significant increase in CD80, CCR7, CD83, and MHC-II surface expression. Infection with an eGFP-expressing HSV-1 strain (Figure 2) results in an almost complete infection of either iDCs (Figure 2A upper panel) or mDCs (Figure 2A lower panel, Figure 2B), based on strong GFP signals analyzed by fluorescence microscopy as well as flow cytometry.
As demonstrated in our recent report, HSV-1 induces autophagy both in iDCs and mDCs, however, autophagic turnover occurs in iDCs only10. In a first approach, we treated iDCs and mDCs with spautin-1 (Figure 3A) – to block autophagy initiation – or bafilomycin-A1 (BA1; Figure 3B) – to inhibit final autophagosome-lysosome fusion. Upon HSV-1 infection of iDCs in the absence of spautin-1 and BA1, autophagic flux is mirrored by the decline of p62 and LC3B expression, respectively. In contrast, HSV-1 infection of mDCs in the absence of spautin-1 does not affect p62 expression, while spautin-1 and BA1 treatment induce an accumulation of LC3B-II. This reflects the induction of autophagy but a failure of autophagic turnover in mDCs. In iDCs, spautin-1 pre-treatment strongly restores autophagic degradation of p62 upon HSV-1 infection, due to the inhibition of autophagy during the initiation phase. Upon pre-treatment with BA1, mock- and HSV-1-infected iDCs show a strong accumulation of LC3B-II protein levels, indicating successful inhibition of autophagic turnover via blocking late autophagosome-lysosome fusion. Consistent with this, spautin-1 and BA1 pre-treatment of mDCs also results in stable p62 and increased LC3B-II protein levels, respectively.
In a second method to impair autophagic flux, siRNA electroporation targeting FIP200 is examined regarding its capacity to block autophagic flux in HSV-1-infected iDCs. As shown in Figure 4A, strongly reduced FIP200 protein levels were detected in iDCs 48 h post electroporation, compared to control siRNA. At this time point, iDCs do not show any signs of cell death (Figure 4B) and maintain their immature phenotype (Figure 4C). Infection of FIP200-silenced iDCs with HSV-1 reveals a strong decrease in autophagic flux compared to their control siRNA-treated counterparts (Figure 4D). This is accompanied by increased protein levels of LC3B as well as p62 when FIP200 is silenced in HSV-1-infected iDCs.
In a reverse attempt, we studied whether siRNA-mediated ablation of KIF1B and KIF2A protein expression enables autophagosomal-lysosomal turnover also in HSV-1-infected mDCs. Thus, iDCs were electroporated using specific siRNAs targeting either one or both of these proteins, and cells were subsequently matured (Figure 5). 2 days post electroporation, mDCs show a strong reduction in KIF1B and/or KIF2A protein expression, when specific siRNAs were used (Figure 5A). This method also did neither lead to prominent cell death (Figure 5B) nor to changes in their phenotypic maturation status (Figure 5C). Supporting the importance of KIF1B and KIF2A during autophagosomal-lysosomal degradation, their depletion prior to HSV-1 infection facilitates an increased autophagic flux in mDCs. This is reflected by decreased residual p62 protein levels, in contrast to the respective control condition (Figure 5D).
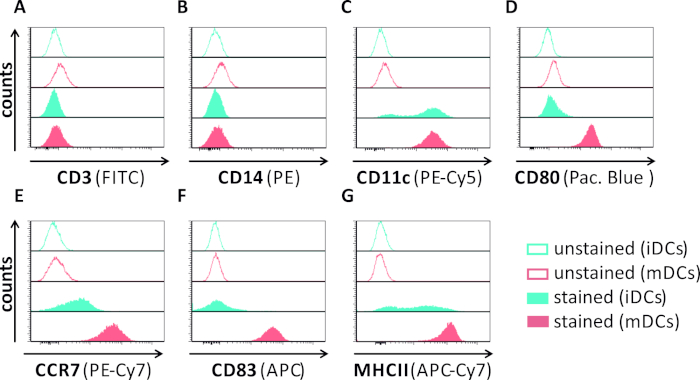
Figure 1: Phenotypic characterization of human monocyte-derived iDCs and mDCs using flow cytometry. DCs were generated and stained with specific antibodies to verify their purity: (A) CD3 to exclude T cell contaminations, (B) CD14 to exclude contamination with monocytes, and (C) CD11c as a marker for DCs. To assess their phenotypic maturation status the following antibodies were used: (D) CD80, (E) CCR7, (F) CD83, and (G) MHCII. These molecules are highly expressed on mDCs and thus allow the discrimination between the immature and mature DC phenotype. Data were analyzed using FCS express 5.0. Please click here to view a larger version of this figure.
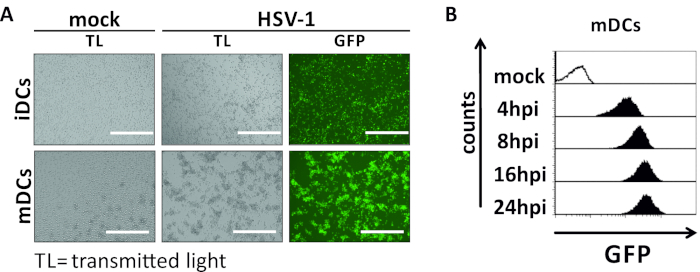
Figure 2: Microscopic as well as flow cytometric analyses of HSV-1-infected iDCs and mDCs. iDCs and mDCs were infected with an HSV-1 strain expressing EGFP (HSV-1 EGFP), to allow quantification of the infection rate based on the GFP signal. (A) Microscopic analyses of GFP-positive HSV-1-infected iDCs and mDCs infected at an MOI of 2, compared to their uninfected counterparts, at 24 hpi. To visualize infected cells, GFP fluorescence was monitored. Scale bar represents 400 µm. (B) Flow cytometric measurement of mock- or HSV-1-infected mDCs during infection kinetics. Upper panels (black lined histograms) show mock condition, lower panels (black filled histograms) show HSV-1-infected cells after the indicated time points post infection. Data were analyzed using FCS express 5.0. Please click here to view a larger version of this figure.
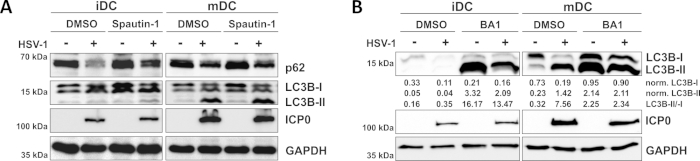
Figure 3: Spautin-1 and b afilomycin-A1 modulate the autophagic flux in HSV-1-infected iDCs. iDCs and mDCs were treated with (A) spautin-1 or (B) bafilomycin-A1 (BA1) for 1 h prior to infection. Cells were subsequently mock- or HSV-1-infected (HSV1-RFPVP26) using an MOI of 2. After 16-18 h, DCs were harvested and protein lysates were subjected to Western blotting to determine expression of p62 or LC3B-I/-II as autophagic markers, ICP0 as infection control, and GAPDH as loading control. LC3B-I and LC3B-II protein levels were quantified and normalized to the reference protein GAPDH using Bio1D (optical density). The ratio of normalized LC3B-II to normalized LC3B-I signals is shown. This figure has been modified and adapted from ©2019 Turan et al. originally published in JCB. https://doi.org/10.1083/jcb.20180115110. Please click here to view a larger version of this figure.
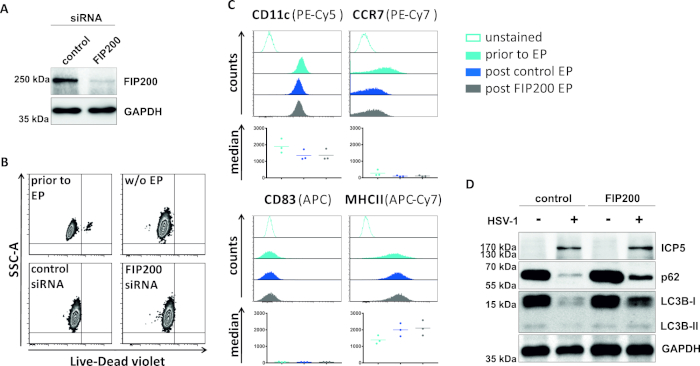
Figure 4: Analysis of autophagic flux in HSV-1-infected iDCs upon FIP200-siRNA electroporation. iDCs were electroporated with control siRNA or FIP200-specific siRNA using the electroporation apparatus I. (A) DCs were analyzed regarding the efficiency of FIP200 knockdown 48 h post electroporation, by performing Western blot analyses. (B) Cell viability as well as (C) maturation status was analyzed prior to electroporation (light blue histograms) and 48 h post (dark blue and grey histograms) electroporation by flow cytometry. Median values for three different donors are shown. After confirming efficient knockdown of FIP200 and the immature phenotype, cells were HSV-1-infected (HSV-1 EGFP) using an MOI of 2. Data were analyzed using FCS express 5.0. (D) At 20 h post infection, cells were subjected to Western blot analyses to determine expression of LC3B-I/-II and p62 as autophagic markers. ICP5 was detected as infection control, and GAPDH as loading control. Panels A and D have been modified and adapted from ©2019 Turan et al. originally published in JCB. https://doi.org/10.1083/jcb.20180115110. Please click here to view a larger version of this figure.
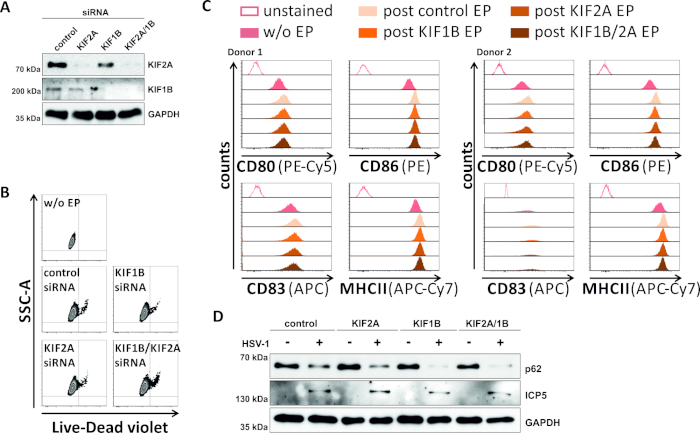
Figure 5: siRNA-mediated ablation of KIF1B and/or KIF2A modulates autophagic turnover in HSV-1-infected mDCs. iDCs were electroporated with KIF1B-specific and/or KIF2A-specific siRNA, as well as control siRNA, using the electroporation apparatus II. At 4 h post electroporation, maturation was induced via addition of a maturation cocktail. At 48 h post electroporation, DCs were analyzed regarding (A) the efficiency of KIF knockdown via Western blotting, (B) cell viability as well as their (C) phenotypic maturation status using flow cytometric analyses (two different donors are shown). "w/o EP" means without electroporation, but post induction of maturation; "post control EP" means post electroporation using control siRNA; "post KIF1B, KIF2A, KIF1B/2A EP" means post electroporation using KIF1B- and/or KIF2A-specific siRNA. After confirming efficient knockdown of KIF1B and/or KIF2A and the mature phenotype, cells were HSV-1-infected (HSV-1 EGFP) using an MOI of 2. Data were analyzed using FCS express 5.0. (D) Cells were subjected to Western blot analyses 20 h post infection, in order to assess expression of p62 as an autophagic marker. ICP5 was used as an infection control, and GAPDH as loading control. Figures A and D have been modified and adapted from ©2019 Turan et al. originally published in JCB. https://doi.org/10.1083/jcb.20180115110. Please click here to view a larger version of this figure.
Discussion
The scope of the present protocol includes (i) the handling of human monocyte-derived iDCs as well as mDCs, (ii) their infection with HSV-1, (iii) their treatment with compounds known to inhibit autophagy, and (iv) their electroporation with siRNA using two different technical setups. Using the present protocol, autophagic flux can either be blocked in HSV-1-infected iDCs or induced in HSV-1-infected mDCs.
Since DCs, and especially iDCs, are very vulnerable cells, working with these cells involves rather delicate steps. For DC generation, we recommend to use freshly isolated PBMCs, and to avoid their cryopreservation, in order to obtain higher cell yields. Furthermore, when handling iDCs during experiments, including their subsequent cultivation, prevent harsh or prolonged temperature alterations. Otherwise, iDCs might undergo phenotypic changes and thus it is necessary to verify their immature phenotype by flow cytometry. Note, in contrast to their mature counterparts, iDCs lack distinct markers, such as CD80, CD83, and CD8628,29. The infection of iDCs and mDCs with HSV-1 is a well-established method2,3,4,5,6,10. We and others showed that DCs are highly susceptible for HSV-1 infection, when an MOI of 1 or 2 has been used (Figure 2). In our hands, keeping the volume of the infection medium at low levels (1-3 x 106 cells in 250-350 µL) will lead to better infection efficiencies.
A classical approach to interfere with a given distinct cellular pathway is the usage of specific compounds. A variety of different modulators of autophagy, i.e. activators as well as inhibitors, are currently available30. Regarding HSV-1-induced autophagy in DCs, Turan et al., (2019) recently showed the inhibitory effects of spautin-1 and bafilomycin-A1 (BA1) on autophagic turnover in iDCs10. This technique for autophagy inhibition is suitable for the combination with a subsequent HSV-1 infection, since neither the infection rate nor the maturation status of DCs (especially iDCs) is impaired. In future applications, this inhibitor-based approach could be applied also in combination with other infectious agents, stress conditions, such as starvation, as well as for different cell types. However, when using inhibitors, limitations arise in determining the suitable concentration for efficient autophagy inhibition, without severely affecting cell viability. The major limitation when using inhibitors is, however, the occurrence of potential off-target or adverse effects, which could lead to misleading results31,32.
The second approach to interfere with autophagy, covered in the present protocol, is the specific knockdown using siRNA33,34,35. On the one hand, we used the electroporation apparatus I to specifically ablate the expression of FIP200, thereby inhibiting HSV-1-induced autophagic turnover in iDCs. On the other hand, we silenced two different KIF proteins (i.e., KIF1B and KIF2A), using the electroporation apparatus II, to facilitate autophagic flux in HSV-1-infected mDCs. Both electroporation protocols resulted in an almost complete ablation of FIP200 in iDCs, and KIF1B/KIF2A in mDCs, which was verified via Western blot analyses (Figure 4A, Figure 5A). In contrast to the electroporation apparatus I, which does not affect the viability of DCs, electroporation of mDCs using the electroporation apparatus II results in slightly higher rates of dead cells (Figure 4B, Figure 5B). Hence, in future applications, the electroporation apparatus I should be preferentially used for both, iDCs and mDCs. Remarkably, both siRNA-based techniques, to modulate autophagic flux, are compatible with subsequent HSV-1 infection of either iDCs or mDCs. Furthermore, neither the immature phenotype of iDCs nor the mature phenotype of mDCs is altered post electroporation.
Electroporation of iDCs using FIP200-specific siRNA is an efficient and highly specific method for gene knockdown as well as inhibition of autophagic flux upon HSV-1 infection. In addition to the specific silencing of FIP200, this protocol can be adapted to silence other autophagic components, participating at different steps during the autophagic cascade. However, identifying the appropriate target for efficient siRNA-mediated inhibition of autophagy includes several aspects of concern. Firstly, knockdown efficiency of autophagy-related genes (ATG) does not necessarily positively correlate with efficient inhibition of autophagy and is highly dependent on the specific ATG protein that is silenced36. Secondly, distinct ATG proteins are additionally involved in pathways distinct from autophagy, thus their ablation could also lead to adverse side effects37,38,39. Thirdly, different ATGs may have redundant functions, thus knockdown of one component may not be sufficient to inhibit autophagy (e.g., beclin-1 and beclin-2)40.
In addition, the electroporation apparatus I -based electroporation protocol of DCs is also suitable for mRNAs, and could be used for a variety of additional primary cell types, such as PBMCs25. This system thus provides a general strategy to deliver distinct RNA species into different primary cell types. In conclusion, we present two protocols to inhibit autophagic flux, by either using an inhibitor- or siRNA-based approach combined with subsequent HSV-1 infection of iDCs. Furthermore, we describe an siRNA electroporation approach to induce autophagic flux in mDCs upon HSV-1 infection.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by the German Research Council (DFG) via the project STE 432/11-1 awarded to AS and by the ELAN Program from the Faculty of Medicine (Friedrich-Alexander-Universität Erlangen-Nürnberg) via the project 18-12-21-1, granted to LG.
Materials
| 4D-Nucleofector Core Unit (electroporation apparatus II) | Lonza (Basel, Switzerland) | AAF-1002B | |
| AB-Serum | Sigma Aldrich Chemie GmbH (Steinheim, Germany) | H4522 | Dendritic cell cultivation |
| ACD-A | Sigma-Aldrich Chemie GmbH (Steinheim, Germany) | 9007281 | |
| Amaxa P3 Primary Cell 4D-Nucleofector X Kit L (electroporation kit apparatus II) | Lonza (Basel, Switzerland) | V4XP-3024 | |
| Amersham ECL Prime Western Blotting Detection Reagent | GE Healthcare (Solingen, Germany) | RPN2232 | Western Blot Detection |
| Ammonium persulfate (APS) | Sigma Aldrich Chemie GmbH (Steinheim, Germany) | A3678 | |
| anti-mouse-IgG (mouse, polyclonal, HRP) | Cell Signaling (Leiden, Netherlands) | 7076 | Western Blot detection |
| anti-rabbit-IgG (goat, polyclonal, HRP) | Cell Signaling (Leiden, Netherlands) | 7074 | Western Blot detection |
| Bafilomycin A1 | Sigma-Aldrich Chemie GmbH (Steinheim, Germany) | tlrl-baf1 | inhibition of autophagy and lysosomal degradation |
| BD FACS Canto II Flow Cytometer | BD Biosciences (Heidelberg, Germany) | 338962 | |
| Benzonase | Sigma-Aldrich Chemie GmbH (Steinheim, Germany) | E1014 | |
| Blotting Chamber Fastblot B44 | Biometra (Göttingen, Germany) | 846-015-100 | |
| CCR7 (mouse, Pe-Cy7) | BioLegend (Fell, Germany) | 557648 | Flow cytometry Dilution: 1:100 Clone: G043H7 |
| CD11c (mouse, Pe-Cy5) | BD Biosciences (Heidelberg, Germany) | 561692 | Flow cytometry Dilution: 1:100 Clone: B-ly6 |
| CD14 (mouse, PE) | BD Biosciences (Heidelberg, Germany) | 555398 | Flow cytometry Dilution: 1:100 Clone: M5E2 |
| CD3 (mouse, FITC) | BD Biosciences (Heidelberg, Germany) | 555332 | Flow cytometry Dilution: 1:100 Clone: UCHT1 |
| CD80 (mouse, V450) | BD Biosciences (Heidelberg, Germany) | 560442 | Flow cytometry Dilution: 1:100 Clone: L307.4 |
| CD83 (mouse, APC) | eBioscience Thermo Fisher Scientific (Langenselbold, Germany) | 17-0839-41 | Flow cytometry Dilution: 1:200 Clone: HB15e |
| CD86 (mouse, PE) | BD Biosciences (Heidelberg, Germany) | 553692 | Flow cytometry Dilution: 1:100 |
| EVOS FL Cell Imaging | System AMG/Life Technologies (Carlsbad, USA) | AMF4300 | |
| FIP200 (rabbit) | Cell Signaling (Leiden, Netherlands) | 12436 | Western Blot detection Dilution: 1:1000 Clone: D10D11 |
| GAPDH (mouse) | Merck Millipore (Massachusetts, USA) | AB2302 | Western Blot detection Dilution: 1:5000 Clone: MAB374 |
| Gene Pulser II apparatus (electroporation apparatus I) | BioRad Laboratories GmbH (München, Germany) | 165-2112 | |
| GM-CSF (4×104 U/mL) | Miltenyi Biotec (Bergisch Gladbach, Germany) | 130-093-868 | |
| HLA‐DR (mouse, APC-Cy7) | BioLegend (Fell, Germany) | 307618 | Flow cytometry Dilution: 1:200 Clone: L243 |
| HSV-1/17+/CMV-EGFP/UL43 | BioVex | DC infection |
|
| ICP0 (mouse) | Santa Cruz Biotechnology (St. Cruz; Dallas, Texas, USA) | sc-53070 | Western Blot detection Dilution: 1:1000 Clone: 11060 |
| ICP5 (mouse) | Santa Cruz Biotechnology (St. Cruz; Dallas, Texas, USA) | sc-56989 | Western Blot detection Dilution: 1:1000 Clone: 3B6 |
| IL-1β (0.1×106 U/mL) | Cell Genix GmbH (Freiburg, Germany) | 1411-050 | |
| IL-4 (1×106 U/mL) | Miltenyi Biotec (Bergisch Gladbach, Germany) | 130-093-924 | |
| IL-6 (1×106 U/mL) | Cell Genix GmbH (Freiburg, Germany) | 1404-050 | |
| ImageQuant LAS 4000 | GE Healthcare (Solingen, Germany) | 28955810 | |
| KIF1B (mouse) | Santa Cruz Biotechnology (St. Cruz; Dallas, Texas, USA) | sc-376246 | Western Blot detection Dilution: 1:1000 Clone: E-12 |
| KIF2A (mouse) | Santa Cruz Biotechnology (St. Cruz; Dallas, Texas, USA) | sc-271471 | Western Blot detection Dilution: 1:1000 Clone: D-7 |
| LC3B (rabbit) | Cell Signaling (Leiden, Netherlands) | 3868 | Western Blot detection Dilution: 1:1000 Clone: D11 |
| L-glutamine | Lonza (Basel, Switzerland) | 17-605E | |
| LIVE/DEAD Fixable Violet dead cell stain kit | Life Technologies (Carlsbad, CA, USA) | L34964 | L/D staining in Flow cytometry |
| Lymphoprep | Alere Technologies AS (Oslo, Norway) | 04-03-9391/01 | |
| Magnesiumchloride | Carl Roth GmbH (Karlsruhe, Germany) | A537.1 | |
| Megafuge 2.0 RS | Heraeus (Hanau, Germany) | 75015505 | |
| N, N, N', N'-Tetramethylethylendiamine (TEMED) | Sigma-Aldrich Chemie GmbH (Steinheim, Germany) | T9281 | |
| Neubauer counting chamber | Brand (Wertheim, Germany) | 717805 | |
| Nunc Cell culture flasks (175.0 cm2) | Thermo Scientific (Rockford, USA) | 159910 | |
| p62 (rabbit) | Cell Signaling (Leiden, Netherlands) | 88588 | Western Blot detection Dilution: 1:1000 Clone: D5L7G |
| PageRuler prestained protein ladder | Thermo Fisher Scientific (Langenselbold, Germany) | 26616 | |
| Paraformaldehyde, 16 % | Alfa Aesar, Haverhill, USA | 43368.9M | |
| PerfectSpin 24 Plus | Peqlab (Erlangen, Germany) | C2500-R-PL | |
| PGE2 (1 mg/mL) | Pfizer (Berlin, Germany) | BE130681 | |
| Phosphate buffered saline (PBS) | Lonza (Basel, Switzerland) | 17-512F | |
| Protein gel system MiniProtean II | Bio-Rad Laboratories GmbH (München, Germany) | 1652960 | |
| RestoreTM Western Blot Stripping Buffer | Thermo Scientific, Rockford, USA | 21059 | |
| Rocking Platform wt 15 | Biometra (Göttingen, Germany) | 042-590 | |
| RotiBlock | Carl Roth GmbH (Karlsruhe, Germany) | A151.4 | |
| Roti-Load 1 (4x) | Carl Roth GmbH (Karlsruhe, Germany) | K929.3 | |
| Rotiphorese Gel 30 (37.5:1) | Carl Roth GmbH (Karlsruhe, Germany) | 3029.1 | |
| RPMI 1640 | Lonza (Basel, Switzerland) | 12-167F | |
| Sodium dodecyl Sulfate (SDS) | Carl Roth GmbH (Karlsruhe, Germany) | 2326.2 | |
| Thermomixer comfort | Eppendorf (Hamburg, Germany) | 5355 000.011 | |
| TNF-α (10 μg/mL) | Peprotech (Hamburg, Germany) | 300-01A | |
| Tris | Carl Roth GmbH (Karlsruhe, Germany) | 4855.3 | |
| Trypan blue solution (0.4 %) | Sigma-Aldrich Chemie GmbH (Steinheim, Germany) | T8154 | |
| Tween 20 | Carl Roth GmbH (Karlsruhe, Germany) | 9127.1 | |
| Whatman 0.2 μm nitrocellulose membrane | GE Healthcare (Solingen, Germany) | 10600001 | |
| WhatmanTM Chromatography Paper 3 mm Chr | Fisher Scientific GmbH (Schwerte, Germany) | 3030917 |
References
- Chapuis, F., Rosenzwajg, M., Yagello, M., Ekman, M., Biberfeld, P., Gluckman, J. C. Differentiation of human dendritic cells from monocytes in vitro. European Journal of Immunology. 27 (2), 431-441 (1997).
- Kummer, M., et al. Herpes simplex virus type 1 induces CD83 degradation in mature dendritic cells with immediate-early kinetics via the cellular proteasome. Journal of Virology. 81 (12), 6326-6338 (2007).
- Kruse, M., et al. Mature dendritic cells infected with herpes simplex virus type 1 exhibit inhibited T-cell stimulatory capacity. Journal of Virology. 74 (15), 7127-7136 (2000).
- Salio, M., Cella, M., Suter, M., Lanzavecchia, A. Inhibition of dendritic cell maturation by herpes simplex virus. European Journal of Immunology. 29 (10), 3245-3253 (1999).
- Prechtel, A. T., et al. Infection of mature dendritic cells with herpes simplex virus type 1 dramatically reduces lymphoid chemokine-mediated migration. Journal of General Virology. 86, 1645-1657 (2005).
- Theodoridis, A. A., Eich, C., Figdor, C. G., Steinkasserer, A. Infection of dendritic cells with herpes simplex virus type 1 induces rapid degradation of CYTIP, thereby modulating adhesion and migration. Blood. 118 (1), 107-115 (2011).
- Cohrs, R. J., Gilden, D. H. Human herpesvirus latency. Brain Pathology. 11 (4), 465-474 (2001).
- Grinde, B. Herpesviruses: latency and reactivation – viral strategies and host response. Journal of Oral Microbiology. 5, (2013).
- Whitley, R. J., Roizman, B. Herpes simplex virus infections. The Lancet. 357 (9267), 1513-1518 (2001).
- Turan, A., et al. Autophagic degradation of lamins facilitates the nuclear egress of herpes simplex virus type 1. The Journal of Cell Biology. 218 (2), 508-523 (2019).
- Takeshige, K., Baba, M., Tsuboi, S., Noda, T., Ohsumi, Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. The Journal of Cell Biology. 119 (2), 301-311 (1992).
- Yin, Z., Pascual, C., Klionsky, D. J. Autophagy: machinery and regulation. Microbial Cell. 3 (12), 588-596 (2016).
- Bodemann, B. O., et al. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell. 144 (2), 253-267 (2011).
- Bjorkoy, G., et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. The Journal of Cell Biology. 171 (4), 603-614 (2005).
- Kabeya, Y., et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. The EMBO Journal. 19 (21), 5720-5728 (2000).
- Kabeya, Y., Mizushima, N., Yamamoto, A., Oshitani-Okamoto, S., Ohsumi, Y., Yoshimori, T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. Journal of Cell Science. 117, 2805-2812 (2004).
- Pankiv, S., et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. The Journal of Biological Chemistry. 282 (33), 24131-24145 (2007).
- Korolchuk, V. I., Rubinsztein, D. C. Regulation of autophagy by lysosomal positioning. Autophagy. 7 (8), 927-928 (2011).
- Li, Y., et al. A cell-based quantitative high-throughput image screening identified novel autophagy modulators. Pharmacological Research. 110, 35-49 (2016).
- Pampaloni, F., et al. A Novel Cellular Spheroid-Based Autophagy Screen Applying Live Fluorescence Microscopy Identifies Nonactin as a Strong Inducer of Autophagosomal Turnover. SLAS Discovery. 22 (5), 558-570 (2017).
- Deng, Y., Zhu, L., Cai, H., Wang, G., Liu, B. Autophagic compound database: A resource connecting autophagy-modulating compounds, their potential targets and relevant diseases. Cell Proliferation. 51 (3), 12403 (2018).
- Liu, J., et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 147 (1), 223-234 (2011).
- Mauvezin, C., Neufeld, T. P. Bafilomycin A1 disrupts autophagic flux by inhibiting both V-ATPase-dependent acidification and Ca-P60A/SERCA-dependent autophagosome-lysosome fusion. Autophagy. 11 (8), 1437-1438 (2015).
- Yoshimori, T., Yamamoto, A., Moriyama, Y., Futai, M., Tashiro, Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. Journal of Biological Chemistry. 266 (26), 17707-17712 (1991).
- Gerer, K. F., Hoyer, S., Dorrie, J., Schaft, N. Electroporation of mRNA as Universal Technology Platform to Transfect a Variety of Primary Cells with Antigens and Functional Proteins. Methods in Molecular Biology. 1499, 165-178 (2017).
- Prechtel, A. T., Turza, N. M., Theodoridis, A. A., Steinkasserer, A. CD83 knockdown in monocyte-derived dendritic cells by small interfering RNA leads to a diminished T cell stimulation. The Journal of Immunology. 178 (9), 5454-5464 (2007).
- Pfeiffer, I. A., et al. Leukoreduction system chambers are an efficient, valid, and economic source of functional monocyte-derived dendritic cells and lymphocytes. Immunobiology. 218 (11), 1392-1401 (2013).
- Villadangos, J. A., Heath, W. R. Life cycle, migration and antigen presenting functions of spleen and lymph node dendritic cells: limitations of the Langerhans cells paradigm. Seminars in Immunology. 17 (4), 262-272 (2005).
- Lechmann, M., Berchtold, S., Hauber, J., Steinkasserer, A. CD83 on dendritic cells: more than just a marker for maturation. Trends in Immunology. 23 (6), 273-275 (2002).
- Yang, Y. P., et al. Application and interpretation of current autophagy inhibitors and activators. Acta Pharmacologica Sinica. 34 (5), 625-635 (2013).
- Redmann, M., et al. Inhibition of autophagy with bafilomycin and chloroquine decreases mitochondrial quality and bioenergetic function in primary neurons. Redox Biology. 11, 73-81 (2017).
- Yan, Y., et al. Bafilomycin A1 induces caspase-independent cell death in hepatocellular carcinoma cells via targeting of autophagy and MAPK pathways. Scientific Reports. 6, 37052 (2016).
- Hale, C. M., et al. Identification of modulators of autophagic flux in an image-based high content siRNA screen. Autophagy. 12 (4), 713-726 (2016).
- Lipinski, M. M., et al. A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Developmental Cell. 18 (6), 1041-1052 (2010).
- Orvedahl, A., et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 480 (7375), 113-117 (2011).
- Staskiewicz, L., Thorburn, J., Morgan, M. J., Thorburn, A. Inhibiting autophagy by shRNA knockdown: cautions and recommendations. Autophagy. 9 (10), 1449-1450 (2013).
- Lee, I. H., et al. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science. 336 (6078), 225-228 (2012).
- Fremont, S., Gerard, A., Galloux, M., Janvier, K., Karess, R. E., Berlioz-Torrent, C. Beclin-1 is required for chromosome congression and proper outer kinetochore assembly. EMBO Reports. 14 (4), 364-372 (2013).
- Bestebroer, J., V’kovski, P., Mauthe, M., Reggiori, F. Hidden behind autophagy: the unconventional roles of ATG proteins. Traffic. 14 (10), 1029-1041 (2013).
- Galluzzi, L., Kroemer, G. Common and divergent functions of Beclin 1 and Beclin 2. Cell Research. 23 (12), 1341-1342 (2013).

