Time-lapse Live Imaging and Quantification of Fast Dendritic Branch Dynamics in Developing Drosophila Neurons
Summary
Here, we describe the method we employed to image highly motile dendritic filopodia in a live preparation of the Drosophila larval brain, and the protocol we developed to quantify time-lapse 3D imaging datasets for quantitative assessments of dendrite dynamics in developing neurons.
Abstract
Highly motile dendritic filopodia are widely present in neurons at early developmental stages. These exploratory dynamic branches sample the surrounding environment and initiate contacts with potential synaptic partners. Although the connection between dendritic branch dynamics and synaptogenesis is well established, how developmental and activity-dependent processes regulate dendritic branch dynamics is not well understood. This is partly due to the technical difficulties associated with the live imaging and quantitative analyses of these fine structures using an in vivo system. We established a method to study dendrite dynamics using Drosophila larval ventral lateral neurons (LNvs), which can be individually labeled using genetic approaches and are accessible for live imaging. Taking advantage of this system, we developed protocols to capture branch dynamics of the whole dendritic arbor of a single labeled LNv through time-lapse live imaging. We then performed post-processing to improve image quality through drift correction and deconvolution, followed by analyzing branch dynamics at the single-branch level by annotating spatial positions of all branch terminals. Lastly, we developed R scripts (Supplementary File) and specific parameters to quantify branch dynamics using the coordinate information generated by the terminal tracing. Collectively, this protocol allows us to achieve a detailed quantitative description of branch dynamics of the neuronal dendritic arbor with high temporal and spatial resolution. The methods we developed are generally applicable to sparsely labeled neurons in both in vitro and in vivo conditions.
Introduction
Dendrites are specialized neuronal compartments that receive and process sensory and synaptic input. The complex and stereotyped structure of dendritic arbors has been under intense investigation since their discovery. A number of model systems, including Xenopus optic tectal neurons, chick retinal ganglion cells, and dendritic arborization (da) neurons in the Drosophila system, have been established to study the development, remodeling and plasticity of neuronal dendrites1,2,3,4. Drosophila ventral lateral neurons (LNvs) are a group of visual projection neurons initially identified for their important functions in circadian regulation of fly behaviors5. Studies also revealed the role of larval LNvs as the direct postsynaptic target of the larval photoreceptors (PRs)6,7. Importantly, culturing developing larvae in different light regimes strongly affects the size of LNvs' dendritic arbors, demonstrating the suitability of LNvs as a new model for studying dendritic plasticity7. Recent work from our group further indicates that both the size of the LNv dendrite and the dynamic behavior of the dendritic branches display experience-dependent plasticity8,9. As part of this work, we developed a new live imaging and quantification protocol to perform analysis on the dendrite dynamics of LNvs from 2nd or 3rd instar larvae.
The transparent nature of the Drosophila larval brain makes it ideal for live imaging. However, the dendritic arbors of LNvs are situated in the densely innervated larval optic neuropil (LON) in the center of the larval brain lobe6. To capture images of fine dendrite branches and filopodia in the intact brain tissue, we utilize two-photon microscopy, which increases the depth of light penetration and reduces the phototoxicity during live imaging experiments10. Using this setup, we successfully performed live imaging experiments on LNvs for over 30 min without observing obvious morphological deterioration of the neuron. In addition, genetic manipulations using the Flip-out technique enabled us to label the individual LNvs with a membrane tagged GFP, which is also critical for monitoring the movements of individual branches11,12,13.
To capture the dynamic behavior of all branches on the LNv dendritic arbor with optimal optic resolution, we performed time-lapse 3D imaging on freshly dissected larval brain explants with a high spatial resolution at 1 min per frame for 10 to 30 min. Developing LNv dendrites are highly dynamic, with a large percentage of the branches displaying observable changes within the 10 min window. This leads to one of the main technical challenges in studying dendrite dynamics, quantifying branch behavior based on the 4D image data sets. Previously established methods have various limitations, including lack of accuracy and excessive time requirement. Therefore, we developed a semi-automatic method that combines image post-processing, manual marking of the branch terminals, and automatic 4D spot tracing using an image annotation software. We calculate the movements of branch terminals at different time points based on the 3D coordinates of the spots. The data are then exported and analyzed to produce quantitative measurements of the branch dynamics. This method accurately assesses the duration and extent of extension and retraction events of existing branches, as well as the formation of new branches, allowing us to monitor dendrite dynamics in a large number of neurons.
Protocol
CAUTION: This protocol involves the use of class IV lasers and will require proper training and safety guidelines to be followed. Avoid eye or skin exposure to direct or scattered laser light.
NOTE: The protocol includes six steps. The workflow is shown in Figure 1A.
1. Labeling Individual Neurons Using the Flip-out Technique
NOTE: The single labeling of LNvs is achieved by expressing mCD8::GFP in single LNvs using flippase -mediated stochastic labeling. The genotype of the fly line is: hs-flp; Pdf-Gal4; UAS-FRT-CD2-stop-FRT-mCD8::GFP11,12,13. The frequency of obtaining a single labeled LNv is around 10%. Fly stocks are maintained in standard medium in circadian- and humidity-controlled 25 °C incubators.
- Collect 100-200 eggs within a 2 h window post fertilization on a grape juice plate with yeast supplement. Incubate the embryos at 25 °C for 24 h and collect newly hatched first instar larvae for the next step.
- Heat shock the newly hatched first instar larvae at 37.5 °C for 40 min two times, with a 40 min recovery period in between.
- Culture the larvae at 25 °C with circadian and humidity controls to the desired developmental stage(s).
2. Dissecting and Mounting Larval Brain Explants
- Dissect larval brains in the physiological external saline solution (120 mM NaCl, 4 mM MgCl2, 3 mM KCl, 10 mM NaHCO3, 10 mM Glucose, 10 mM Sucrose, 5 mM TES, 10 mM HEPES, 2 mM Ca2+, PH 7.2) under a dissection microscope (4.5x magnification power) with two pairs of #5 standard tip dissection forceps (11 cm). Use one pair of forceps to hold the larval body in place and the other to carefully dissect out the brain. Preserve the eye disks, brain lobes and the ventral nerve cord. Remove attached muscles to minimize sample movements during imaging.
- Prepare a glass slide (25 x 75 x 1.0 mm3) and use a syringe to draw a square chamber with vacuum grease.
- Add 20 µL of external saline solution to the square chamber with grease barriers.
- Transfer dissected larval brains into the chamber on the glass slide using forceps. Adjust the position of the brains under the dissection scope to ensure the dorsal side faces up.
- Cover the chamber with a glass cover slip (22 x 22 x 0.15 mm3). The larval brain is now mounted on the slide within a chamber filled with the external saline solution (Figure 1B).
NOTE: Pressing gently on the coverslip confines the brains and reduces sample drifting in the subsequent imaging session.
3. Time-lapse Live Imaging
NOTE: We perform time-lapse imaging experiments using a confocal microscope equipped with a multiphoton laser. The acquisition parameters need to be adjusted for other imaging setups.
- Identify brain explants containing individually labeled neurons using a 40X water immersion objective (NA 1.3) and an epifluorescent light source. For image collection, use a two-photon laser tuned to 920 nm and a non-descanned (NDD) detector.
- Collect images at 512 x 512 pixels per frame and 1 min per Z-stack for 10 min. Adjust the optical and digital zoom to achieve a sufficient x-y-z resolution while ensuring the coverage of the whole dendritic arbor within 1 min (Figure 2, Supplementary Video 1). The settings generate images with a typical x-y-z resolution of 0.11 x 0.11 x 0.25 µm3.
- Collect the time lapse image series within 30 min of the brain dissection. Data with excessive drift or obvious morphological deterioration should be excluded from post- processing and quantification.
4. Drift Correction and Deconvolution
NOTE: Depending on the image quality, both steps are optional but strongly recommended.
- Open an acquired image file with a drift correction software. Edit microscopic parameters to match those used for the experiment. For the software (see Table of Materials), click Edit | Edit microscopic parameters. Set microscope type as widefield for two-photon images if there is no two-photon option and follow the workflow defined by the software. For example, open the tab Deconvolution | Object Stabilizer, and choose Stabilize time frames.
- Open the drift-corrected image in the deconvolution software and follow its workflow. For the software (see Table of Materials), select the stabilized image and then click Deconvolution | Deconvolution Express. To get a better result, fine-tune the parameters using Deconvolution Wizard.
- Save the deconvolved image in a file type that is supported by the subsequent image annotation software capable of analyzing 4D data and reporting the spatial coordinates of defined spots in the image (see Table of Materials).
5. Image Annotation
- Open the deconvolved image in the image annotation software. Examine and mark branch tips at all time points in 3D. The image annotation software reports and stores the spatial and temporal coordinates of the marked branch tips. Export the coordinate information as a .csv file for subsequent calculations.
NOTE: The following two steps are specific to the annotation software we used (see Table of Materials). The workflow may be different for other software. - Within the Spots module, click "Skip automatic creation, edit manually" and check the bottom "Auto-connect to selected Spot" checkbox (Figure 3A). Go over the frames in the time series and select a branch for annotation. Hold the Shift key and click the branch terminal tip to add a spot. Click through all time points.
NOTE: The image annotation software connects the spots between frames and generates a trajectory automatically. The spatial and temporal information of the branch tips is now associated with marked spots. Repeat these steps until all branch tips are annotated (Figure 3B). - Within the Spots module, click the Statistics tab, choose Detailed and select Specific Values | Position. The recorded spatial and temporal coordinate information will be on display. Click the Save bottom to export that information as a .csv file (Figure 3C).
6. Calculating Dendritic Branch Dynamics
NOTE: Using the coordinate information, displacement of the branch tips in 3D can be easily calculated. In our study, all dendritic branch movements are categorized as Extension or Retraction. The steps below describe how to process the .csv files using custom-written R scripts (Supplementary File) and by adding the directional information through manual editing.
- Open the .csv file as a spreadsheet. Select the Track ID column, click the Sort Smallest to Largest option and accept Expand the selection. After sorting, Track ID identifies each unique dendritic branch, and spots from the same branch share the same Track ID. The Time column stores the information of different time frames.
- In the spreadsheet, add a Distance column. Calculate the distance of every two temporally adjacent spots using their coordinates and put the values in the Distance column (Figure 4A).
- Minor movements at the single voxel level. Based on the imaging settings, 0.3 µm are usually artifacts from uncorrected drifting or imperfect image annotation. Filter these movements out by resetting all Distance values smaller than 0.3 µm to 0. Process multiple .csv files manually or use our R script batch column filtering.R (Supplementary File).
- Manually generate a new column named Displacement in the spreadsheet. Copy the values from Distance column to Displacement column. Manually assign the extension and retraction events for each branch tip. If it is an extension, leave the Displacement value unchanged, which is a positive value. If it is a retraction, change the corresponding Displacement value to a negative value (e.g., 0.35 to -0.35) (Figure 4B).
- Generate a new column named Event. In this column, manually sum the Displacement values for individual extension and retraction events (Figure 4B).
- Process the modified spreadsheet and quantify the extension and retraction events based on their displacement values using R script batch column sum.R (Supplementary File). The output parameters include Number of extension events, Number of retraction events, Cumulative length extended, Cumulative length retracted, Net length changed, and Total length traveled.
Representative Results
Using the live imaging protocol described above, we capture high resolution image stacks for the subsequent analyses and quantification. Supplementary Video 1 shows the maximum intensity projected (MIP) image series collected from a representative individually labeled LNv. Figure 2B shows the corresponding montage of eight frames of the image series. In both panels, arrowheads mark retraction events and arrows mark extension events.
Next, we perform semi-automated 4D tracking of the branch terminals using an image annotation software (Table of Materials). Figure 3B shows annotations of the dynamic branch terminals from a representative individually labeled LNv. Using this software, we visualize the image stacks in 3D, manually mark all branch terminals and use the Spots module to track the terminals' movements through time. This software generates time-stamped trajectories for every annotated branch terminal, serving as visual representations of the dynamic events on the dendritic arbor.
We then use the coordinate information from annotated branch tips to calculate the direction and distance of branch movements at each time point. Screenshots in Figure 4A,B show how this is achieved. The processed spreadsheets generate output files that we use to quantify dendrite dynamics with four parameters, including percentage of dynamic branches, number of new branches, cumulative distance traveled, and number of branch movement events. Figure 4C shows comparisons of dendritic branch dynamics for individually labeled LNvs from different developmental stages. Consistent with the findings in mammalian neurons and zebrafish tectal cells, the LNv dendrite dynamics are developmentally regulated14,15. LNv dendrites in younger larvae, 48-72 h after egg laying (AEL), are significantly more dynamic compared to those in older larvae, 96-120 h AEL, as measured by all four parameters, and the developmental transition from the dynamic to stable state occur between 72 to 96 h AEL8.
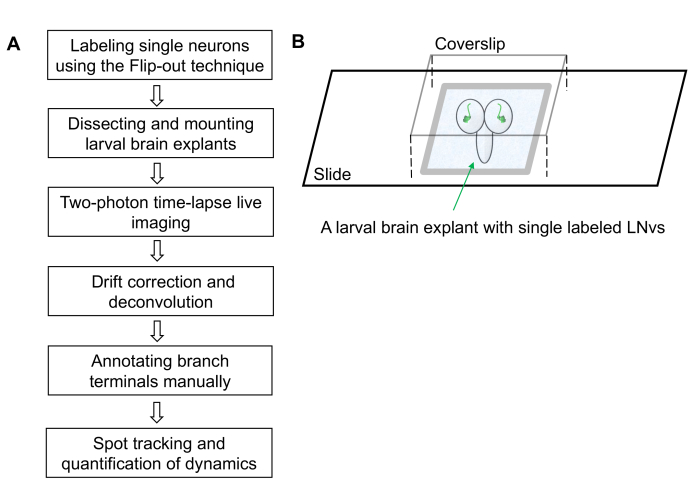
Figure 1: Workflow of the dendrite dynamics imaging and quantification protocol. (A) The protocol contains six steps covering sample preparation, image collection, image processing and semi-automated quantification of dendrite dynamics. (B) A schematic diagram illustrating an imaging chamber containing a larval brain explant mounted with the dorsal side up. The brain explant has an individually labeled LNv in each lobe and is immersed in the external saline solution. The vacuum grease barriers support the weight of the coverslip and forms a small chamber that prevents the brain from moving. Please click here to view a larger version of this figure.
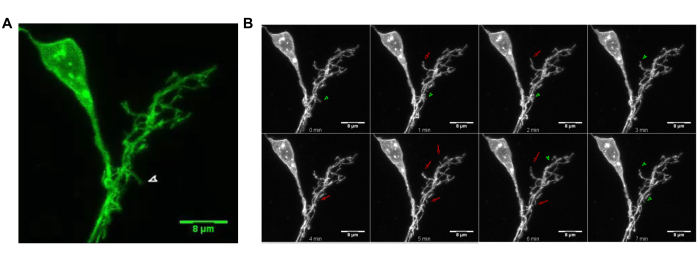
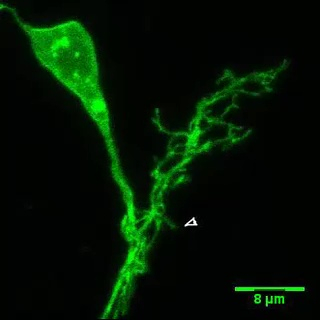
Figure 2: Time-lapse imaging of dendritic branch dynamics in 3D. (A) The dendritic arbor of an individually labeled LNv from a 3rd instar larva. (B) The corresponding montage image of (A) illustrating the representative branch movements. Arrowheads (Green) mark retraction events; arrows (Red) mark extension events. Please click here to view a larger version of this figure.
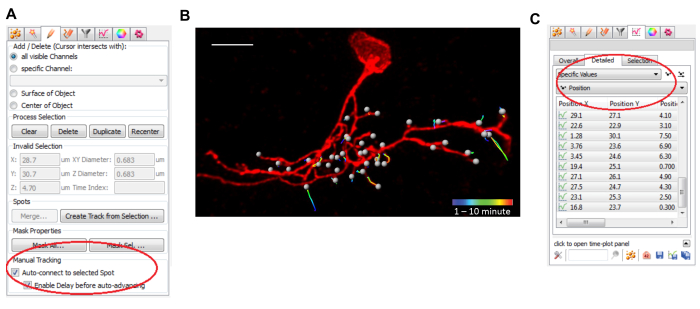
Figure 3: Manual annotation and automatic tracking of dendritic branch terminals in an image annotation software. (A) A screenshot shows the settings for branch terminal tracking using the Spots module. The red oval outlines the Manual Tracking option. (B) Representative time-stamped trajectories of annotated branch terminals (yellow spots) generated by the image annotation software in an individually labeled LNv. Scale bar = 5 µm. (C) A screenshot shows the interface for viewing and exporting the coordinate information from the Spots module. Figure 3B has been modified from Sheng, C. et al. 20188. Please click here to view a larger version of this figure.
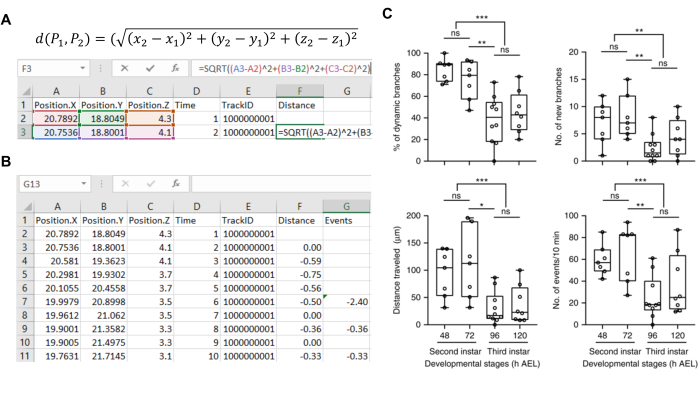
Figure 4: Quantification of dendritic branch dynamics using the coordinate information of the branch terminals. (A) A screenshot shows the spreadsheet containing the X, Y and Z coordinates, Track IDs and formula used for calculating displacement of spots in the adjacent frames. (B) A screenshot shows representative results obtained from tracking one branch. The minor movements (<0.3 µm) were filtered out. The retractions are marked by assigning their value to negative (Distance column). By summing up adjacent positive/negative values, the displacement of each biological extension/retraction is computed (Events column). (C) Representative quantification of results showing the developmental regulation of dendrite dynamics. Data are presented as a box plot (box, 25-75%; center line, median) overlaid with a dot plot (individual data points). Figure 4C has been modified from Sheng, C. et al. 20188. Please click here to view a larger version of this figure.
Supplementary Video 1: Ten maximum-intensity-projection (MIP) images played at a speed of 2 frames per second. The whole dendritic arbor was imaged at 1 min per Z-stack. Please click here to view this video (Right click to download).
Supplementary File 1: Batch Column Filtering.R Script. Please click here to view this file (Right click to download).
Supplementary File 2: Batch Column Sum.R Script. Please click here to view this file (Right click to download).
Discussion
Here, we describe a protocol we developed to record and quantify the dynamic behavior of dendritic branches in individually labeled neurons in Drosophila larval brains. Notably, our live imaging protocol contains specific parameters that enable us to capture the whole dendritic arbor of a larval LNv and provide a global view of the dynamic state of its dendrite branches. However, because our quantification methods heavily rely on the annotation of branch terminals, neurons with complex dendritic structures and condensed arborizations are not suitable subjects for this analysis.
Sample preparation, image acquisition and post-processing are the critical steps in this protocol. High quality raw images of single LNvs are essential for the accurate quantification of dendrite dynamics. Our data indicate that LNv dendrite morphology and its physiological responses are well-preserved in a carefully prepared larval brain explant within 30 min of dissection. Samples with observable brain tissue deterioration, elongated dendritic arbors and branch breakage should not be used for imaging and quantification. Although optional for datasets with low background and no observable movements, we strongly recommend including the drift correction and deconvolution steps to improve the image quality, which is important for the annotation of branch terminals and automatic spot tracking. Besides the commercial software used in our studies, ImageJ/Fiji's Correct 3D drift (Plugins | Registration | Correct 3D drift) also works well for drift correction.
One of the major differences between our protocol and existing methods is that we track the movements of individual branch tips but not the whole dendritic arbor. Reconstruction of whole dendritic arbors, although useful for measuring the total dendritic length and architecture, is not ideal for dynamic studies using 3D imaging datasets. For consecutive Z-stack slices, automatic tracing programs often generate different combinations of "branches" and reconstruct the dendritic arbor differently, making the comparison at the single branch level between different time points particularly difficult. In addition, comparing to tracing the whole dendritic arbor, terminal tracking significantly reduces the processing time and the requirements on computation power.
Several modifications can potentially improve the efficiency and analytical features of our method. To extract quantitative positional information from the time-lapse image series, accurate annotation of branch tips at all time points is critically important. This step is currently performed manually, which not only is time consuming, but also introduces variability. New developments in open source 3D visualization software could potentially address current technical limitations and make automation of this critical step possible. Several recently developed software tools provide automatic quantification function for studies on filopodia16,17,18,19,20,21 and can be potentially tested and adopted for studies on dendrite branch dynamics.
Another step that is currently performed manually is the identification of extension vs. retraction events. Automation of this step requires the 3D coordinates of branch bifurcation sites. By comparing distances to the branching site at two time points, a custom-written scrip can be developed to assign the directionality of the terminal displacements. Following the same logic, additional 3D coordinates along the branch may also be used for analyzing the extension and retraction events occurring within the branch. These add-ons will not only optimize the workflow of our protocol, but also increase the information contents regarding the structural changes of the dendritic arbors at different temporal scales.
In summary, to support our studies on experience-dependent dendrite plasticity, we developed time-lapse live imaging protocols and semi-automated quantification methods to perform assessments of fast branch dynamics in developing neurons. Based on the results generated by the methods described here, our study identified highly motile dendritic filopodia in Drosophila central neurons and revealed a developmental coordination between heightened dendrite dynamics and synaptogenesis. Future improvements at the image acquisition and automation level will greatly enhance the potential of this protocol and facilitate studies on dendrite dynamics and structural plasticity.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work is supported by the Intramural Research Program of National Institute of Neurological Disorders and Stroke, National Institutes of Health. Project number 1ZIANS003137.
Materials
| Chameleon Vision II multiphoton laser | Coherent | ||
| high vacuum grease | Dow Corning | 79751-30 | |
| LSM 780 two-photon laser scanning confocal microscope | Carl Zeiss | upright configuration | |
| Microscope Cover Glass | Fisher Scientific | 12-544-E | |
| Superfrost Plus Microscope Slides | Fisher Scientific | 12-550-15 | |
| Software | |||
| Excel | Microsoft | for processing .csv files | |
| Huygens Professional | Scientific Volume Imaging | for drift correction and deconvolution | |
| Imaris | Oxford Instruments | for 3D visualization and image annotation | |
| Reagents | |||
| Glucose | |||
| HEPES | |||
| KCl | |||
| MgCl2 | |||
| NaCl | |||
| NaHCO3 | |||
| PBS | |||
| Sucrose | |||
| TES | |||
References
- Wong, W. T., Wong, R. O. L. Changing specificity of neurotransmitter regulation of rapid dendritic remodeling during synaptogenesis. Nature Neuroscience. 4 (4), 351-352 (2001).
- Wu, G. Y., Zou, D. J., Rajan, I., Cline, H. Dendritic Dynamics In Vivo Change during Neuronal Maturation. The Journal of Neuroscience. 19 (11), 4472-4483 (1999).
- Jan, Y. N., Jan, L. Y. Branching out: mechanisms of dendritic arborization. Nature Reviews Neuroscience. 11, 316 (2010).
- Cline, H., Haas, K. The regulation of dendritic arbor development and plasticity by glutamatergic synaptic input: a review of the synaptotrophic hypothesis. The Journal of Physiology. 586 (6), 1509-1517 (2008).
- Helfrich-Forster, C., et al. Development and morphology of the clock-gene-expressing lateral neurons of Drosophila melanogaster. Journal of Comparative Neurology. 500 (1), 47-70 (2007).
- Sprecher, S. G., Cardona, A., Hartenstein, V. The Drosophila larval visual system: High-resolution analysis of a simple visual neuropil. Developmental Biology. 358 (1), 33-43 (2011).
- Yuan, Q., et al. Light-Induced Structural and Functional Plasticity in Drosophila Larval Visual System. Science. 333 (6048), 1458-1462 (2011).
- Sheng, C., et al. Experience-dependent structural plasticity targets dynamic filopodia in regulating dendrite maturation and synaptogenesis. Nature Communications. 9 (1), 3362 (2018).
- Yin, J., et al. Transcriptional Regulation of Lipophorin Receptors Supports Neuronal Adaptation to Chronic Elevations of Activity. Cell Reports. 25 (5), 1181-1192 (2018).
- Denk, W., Strickler, J., Webb, W. Two-photon laser scanning fluorescence microscopy. Science. 248 (4951), 73-76 (1990).
- Golic, K. G., Lindquist, S. The FLP recombinase of yeast catalyzes site-specific recombination in the drosophila genome. Cell. 59 (3), 499-509 (1989).
- Harrison, D. A., Perrimon, N. Simple and efficient generation of marked clones in Drosophila. Current Biology. 3 (7), 424-433 (1993).
- del Valle Rodríguez, A., Didiano, D., Desplan, C. Power tools for gene expression and clonal analysis in Drosophila. Nature Methods. 9, 47 (2011).
- Portera-Cailliau, C., Pan, D. T., Yuste, R. Activity-regulated dynamic behavior of early dendritic protrusions: evidence for different types of dendritic filopodia. Journal of Neuroscience. 23 (18), 7129-7142 (2003).
- Niell, C. M., Smith, S. J. Live optical imaging of nervous system development. Annual Review of Physiology. 66, 771-798 (2004).
- Barry, D. J., Durkin, C. H., Abella, J. V., Way, M. Open source software for quantification of cell migration, protrusions, and fluorescence intensities. The Journal of Cell Biology. 209 (1), 163-180 (2015).
- Hendricusdottir, R., Bergmann, J. H. M. F-dynamics: Automated quantification of dendrite filopodia dynamics in living neurons. Journal of Neuroscience Methods. 236, 148-156 (2014).
- Jacquemet, G., et al. FiloQuant reveals increased filopodia density during breast cancer progression. The Journal of Cell Biology. 216 (10), 3387 (2017).
- Nilufar, S., Morrow, A. A., Lee, J. M., Perkins, T. J. FiloDetect: automatic detection of filopodia from fluorescence microscopy images. BMC Systems Biology. 7 (1), 66 (2013).
- Tsygankov, D., et al. CellGeo: A computational platform for the analysis of shape changes in cells with complex geometries. The Journal of Cell Biology. 204 (3), 443-460 (2014).
- Urbančič, V., et al. Filopodyan: An open-source pipeline for the analysis of filopodia. The Journal of Cell Biology. 216 (10), 3405-3422 (2017).

