A Co-Culture Method to Study Neurite Outgrowth in Response to Dental Pulp Paracrine Signals
Summary
We describe the isolation, dispersion and plating of dental pulp (DP) primary cells with trigeminal (TG) neurons cultured atop overlying transwell filters. Cellular responses of DP cells can be analyzed with immunofluorescence or RNA/protein analysis. Immunofluorescence of neuronal markers with confocal microscopy permits the analysis of neurite outgrowth responses.
Abstract
Tooth innervation allows teeth to sense pressure, temperature and inflammation, all of which are crucial to the use and maintenance of the tooth organ. Without sensory innervation, daily oral activities would cause irreparable damage. Despite its importance, the roles of innervation in tooth development and maintenance have been largely overlooked. Several studies have demonstrated that DP cells secrete extracellular matrix proteins and paracrine signals to attract and guide TG axons into and throughout the tooth. However, few studies have provided detailed insight into the crosstalk between the DP mesenchyme and neuronal afferents. To address this gap in knowledge, researchers have begun to utilize co-cultures and a variety of techniques to investigate these interactions. Here, we demonstrate the multiple steps involved in co-culturing primary DP cells with TG neurons dispersed on an overlying transwell filter with large diameter pores to allow axonal growth through the pores. Primary DP cells with the gene of interest flanked by loxP sites were utilized to facilitate gene deletion using an Adenovirus-Cre-GFP recombinase system. Using TG neurons from the Thy1-YFP mouse allowed for precise afferent imaging, with expression well above background levels by confocal microscopy. The DP responses can be investigated via protein or RNA collection and analysis, or alternatively, through immunofluorescent staining of DP cells plated on removable glass coverslips. Media can be analyzed using techniques such as proteomic analyses, although this will require albumin depletion due to the presence of fetal bovine serum in the media. This protocol provides a simple method that can be manipulated to study the morphological, genetic, and cytoskeletal responses of TG neurons and DP cells in response to the controlled environment of a co-culture assay.
Introduction
Tooth innervation allows teeth to sense pressure, temperature and inflammation, all of which are crucial to the use and maintenance of the tooth organ. Failure to sense tooth pain associated with dental caries and trauma leads to disease progression. Thus, proper innervation is a requirement for normal tooth growth, function and care.
While most organs are fully functional and innervated by the time of birth, tooth development extends into adult life, with tooth innervation and mineralization occurring in concert during postnatal stages1,2. Interestingly, the dental pulp (DP) mesenchyme initially secretes repellant signals during embryogenesis to prevent axon entry into the developing tooth organ, which later shifts to the secretion of attractant factors as the tooth nears eruption3,4. During postnatal stages, afferent axons from the trigeminal (TG) nerve penetrate into and throughout the tooth around the time dentin deposition begins (reviewed in Pagella, P. et al.5). Several in vivo studies have demonstrated that neuronal-mesenchymal interactions guide tooth innervation in mice (reviewed in Luukko, K. et al.6), but few details of the molecular mechanisms are available.
Cell co-cultures provide controlled environments in which investigators can manipulate interactions between neuronal and mesenchymal populations. Co-culture experiments make it possible to delve deeper into the signaling pathways guiding tooth innervation and development. However, several of the conventional methods used to study cells in co-culture present technical challenges. For instance, crystal violet staining of neurite outgrowth can non-specifically stain Schwann cells included in TG bundle dispersions, and there may be peaks in color intensity with relatively small responses7. Microfluidic chambers offer an attractive option, but are considerably more expensive than transwell filters8,9 and only permit the investigation of neuronal responses to DP secretions. To address these issues, we have developed a protocol that allows for: a) precise staining and imaging of TG neurite outgrowth in response to DP secretions, b) genetic modification of DP cells and/or TG neurons to investigate specific signaling pathways, and c) investigation of DP cell responses to factors secreted by TG neurons. This protocol provides the ability to precisely investigate several features of tooth innervation in the controlled environment of an in vitro co-culture assay.
Protocol
All experiments with mice were approved by the UAB Institutional Animal Care and Use Committee (IACUC).
1. Plate Preparation
NOTE: Coverslips can be used to image DP cells at the end of the assay. Be sure the plate lid is on during all incubation and rinsing steps outside of the sterile tissue culture hood to prevent contamination during sample processing.
- Coverslip preparation
- Autoclave circular coverslips.
- Filter ultrapure water under the hood.
- Transfer the coverslips to a 24-well plate and cover each one with 400 μL of 0.1 mg/mL poly-D-lysine.
- Allow the coverslips to soak, immersed for 5 min, on a rocker at 12 rpm or orbital shaker at 40-50 rpm. Be sure lid is on to prevent contamination.
- Rinse the coverslips with filtered ultrapure water for about 5 min on a rocker at 12 rpm or shaker at 50 rpm. Repeat.
- Allow the coverslips to dry for at least 2 h under the hood with the plate lid off to facilitate evaporation. These coverslips should be used within 48 h.
- Seed DP cells atop coverslips and process at the end of the assay for immunofluorescence (section 3.3).
- Coating filters
- Dilute laminin to 10 μg/mL. Filter the diluted laminin under the hood.
- Pipette 450-500 μL of 10 μg/mL laminin into each well of a 24-well plate.
- Place transwell filter, 3 μm porosity, in a well so that it contacts the laminin solution and leave it in a 37 °C incubator for either 2 h or overnight. Filter pores should allow some of the solution to diffuse and coat both the top and bottom of the filter. These filters should be used within 48 h or be refrigerated at 4 °C for up to 1 week.
2. Cell Plating with Optional Genetic Manipulation
- Mice: Genetic alteration of DP cells can be performed (but is not required) to study mesenchymal-neuronal interactions. See sections 2.3 and 2.4 for suggested genetic variations.
- DP dissection, dispersion and plating (Figure 1)
NOTE: For DP dissection, use ultra-fine, straight-edge forceps. The ultra-fine edges will allow the user to wedge the forcep edge between the mineralized structure and DP tissue.- Harvest P5-P8 mice. At this stage, the teeth should be mineralizing, and the root is open.
- Anesthetize the neonates via hypothermia by placing them in a dish in the 4 °C refrigerator until they no longer move or respond to touch. Euthanize neonates by decapitation and in accordance with IACUC procedures at the designated facility.
- Prepare a 3-5 mL aliquot of 0.25% trypsin-EDTA in a 50 mL conical tube to collect the DP from each mouse. This will facilitate the digestion of dental pulp from postnatal mice. Use more than 3 mL if digesting tissue from more than 10 postnatal mice.
- Place the head on a disposable underpad so that the mouth is toward the ceiling and the base of the neck is flat on the work surface. Use a razor blade in a sawing motion to separate the mandible from the maxilla.
- Optionally remove the tongue either with scissors or with forceps to allow easier access to the molars.
- Place the opened head in a dish atop a sterile gauze pad and place the specimen under a dissecting microscope (Figure 1D).
- Remove the alveolar bone tissue surrounding first molars. Submandibular teeth are not fully erupted at this point. Insert forceps into alveolar opening and tease the tissue away from the tooth toward the buccal (cheek) or lingual (tongue) side of the mouth. Maxillary teeth will require full removal of the cleft around the tooth for exposure and removal.
- Gently transfer the submandibular and maxillary first molars (M1s) to a separate cell culture dish with 1x phosphate-buffered saline (PBS).
- Repeat 2.2.4-2.2.8 until all M1s are collected. Keep the dish containing the M1s on ice during the harvesting.
- Remove the Enamel Outer Organ (EOE) surrounding the outside of each M1. This can alternatively be done after step 2.2.11.
- With a set of forceps, rotate the M1 so the cusps are down and the open root is exposed. There will be an oval opening on the bottom of the tooth, and opaque DP tissue encapsulated by a thin layer of dentin and enamel.
- Using the tip of the forceps, gently loosen the DP by running one arm of the forceps around the internal circumference of the mineralized tissue. Remove DP tissue out of the mineralized structure and transfer it to a third dish containing 1x PBS. Remove the EOE if it was not already separated (Figure 1E).
- Transfer all DP tissue to 0.25% trypsin-EDTA in a 50 mL conical tube. Vortex the mixture and place in a 37 °C warm water bath for 10 min. This can be done with the same forceps or with long, vial forceps. The tissue will be difficult to disperse and will require vortexing every 3-4 min. Do NOT exceed 10 min trypsinization since the trypsin can damage cell membranes.
- Under a sterile hood, add warmed co-culture media (Table 1) to a final ratio of at least 1:1 media to trypsin to inactivate the enzyme. Larger ratios are acceptable if more tissue dispersion is desired.
- Pipette the media up and down multiple times with a 10 mL pipette to further disperse the DP in the media. Be careful to avoid large bubbles. Complete dispersion is nearly impossible due to the sticky nature of the tissue. However, it is also not necessary since cells will migrate outward from the tissue once plated.
- Transfer 1 mL of the dispersed DP to each well of a 24-well tissue culture plate (Figure 1F).
- Place the plate in an incubator at 37 °C and allow cells to attach and migrate out from the undispersed tissue for 48 h before changing media. Primary cells need to be plated at relatively high concentrations in order to reach 85-90% confluence within 1 week. If this is not be achieved after 1 week, discard the plate.
- Optional Genetic Manipulation of DP Cells
- To alter cell signaling pathways, harvest DP cells from genetic knockout mice or from mice in which a gene of interest is flanked by loxP sites. In the latter case, the gene can be deleted using Adenovirus-Cre-GFP (Ad-Cre-GFP) recombinase to remove the flanked gene, as described below. Use Adenovirus-eGFP (Ad-eGFP) as a control virus to ensure that the viral infection does not cause a cellular response.
NOTE: Ad-eGFP is controlled by the CMV promoter enhancer, which is very strong. Ad-Cre-GFP is regulated by an IRES, an internal regulatory region between the Cre and GFP, which is not very strong. This results in brighter fluorescence in Ad-eGFP cells than Ad-Cre-GFP cells. Confirm equivalent infection levels based on the total numbers of cells fluorescing, not on the levels of cellular fluorescence. - Prepare 500 μL of media containing virus and 10 μg/mL of polybrene per well. Polybrene assists with viral infection in cells near confluence10. This protocol is based on a multiplicity of infection (MOI) of 100 for Ad-eGFP and 200 Ad-Cre-GFP for efficient gene infection with little or no effect on cell viability. The estimated cell number is 4 x 104 cells/well of a 24-well plate:
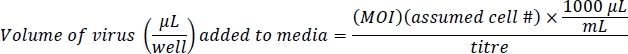
- Mix media with brief vortexing or pipetting and add 500 μL to each well.
- After 24 h, add an additional 500 μL of co-culture media that does not contain additional polybrene or virus.
- After a total of 48 h, aspirate virus-containing media and replace with fresh co-culture media. At this point, TG neurons can be added atop transwell filters.
- To alter cell signaling pathways, harvest DP cells from genetic knockout mice or from mice in which a gene of interest is flanked by loxP sites. In the latter case, the gene can be deleted using Adenovirus-Cre-GFP (Ad-Cre-GFP) recombinase to remove the flanked gene, as described below. Use Adenovirus-eGFP (Ad-eGFP) as a control virus to ensure that the viral infection does not cause a cellular response.
- Trigeminal neuron dissection, dispersion and plating
NOTE: In this protocol, the imaging of neurite outgrowth in co-culture with DP cells was optimized using adolescent (6-week-old) B6.Cg-Tg(Thy1–YFP)16Jrs/J mice. Central and peripheral nervous systems of Thy1-YFP mice have a yellow fluorescent protein (YFP) tag whose expression begins around P6-P10 in neurons and increases exponentially throughout the nervous system during postnatal and adult life11,12. YFP and GFP have conserved sequences that allow these nerves to be stained with anti-GFP antibodies, resulting in a pan-neuronal stain. Ultimately, these mice allow for better visualization and quantification of the neurons used and grown in cell culture.- Euthanize adolescent mice with carbon dioxide followed by cervical dislocation.
- Decapitate the mice and remove the skin from the skull. Be sure to include equivalent numbers of males and females.
- Insert the tip of a pair of micro-dissecting scissors into the base of the skull. Cut along the sagittal suture of the skull (Figure 1A).
- Make four small horizontal cuts: two along the coronal sutures by the ears, and two along the lambdoid sutures at the base of the skull. This should create two flaps of bone.
- Use the forceps to peel back the two flaps of bone. This should reveal the brain.
- Remove the brain. Transfer the head to a tissue culture dish with 1x PBS and put under the microscope.
- Locate the TG ganglia, which is easily visible in rodents13, housed in the dura matter between the brain and bone of the maxillary process (Figure 1B).
- Cut the three branches that travel to the eyes, maxillae and mandible and transfer the ganglia to cold 1x PBS using straight-edge fine forceps. Keep the dish containing the TG ganglia on ice during harvesting.
- Once all TG bundles are harvested, transfer ganglia to a 50 mL conical tube containing 5 mg/mL sterile-filtered collagenase type II using vial forceps.
- Vortex the collagenase with the TG bundle and place the tube in a 37 °C water bath for 25-30 min. During this time, take the conical tube out of the water bath, vortex, and return to the bath every 5-10 min.
- Centrifuge the collagenase-TG neuron solution for 2 min at 643 x g.
- Under a tissue culture hood, gently aspirate the collagenase with a micropipette.
- Add 5 mL of 1% sterile-filtered trypsin type II and vortex. Place the conical tube in a 37 °C water bath for 5 min.
- Centrifuge the trypsin-TG mix for 5 min at 643 x g. Remove the top portion of trypsin with a micropipette, so that the TG neurons are not removed. There will still be liquid in the tube.
- Add enough media to deactivate the remaining trypsin (at a 1:1 or lower ratio of trypsin to media).
- Count the number of cells and dilute the solution to 200,000 cells/mL (250 μL of cell-containing 50,000 cells).
- Place coated transwell filters from section 1.2 into wells with DP.
- Dilute the cell-containing solution so that there are 200,000 cells/mL. Pipette 250 μL onto the transwell filter, and culture the cells at 37 °C overnight (Figure 1F).
- The next day, replace the media with 1 mL of Co-culture media with 1 μM uridine and 15 μM 5'-fluor-2'deoxyuridine to stop the over-proliferation of mesenchymal cells that may prevent neurite outgrowth. Optional: Add growth factors or inhibitors in this media if attempting further manipulation.
- Culture the cells for additional desired time points. This protocol was optimized for 5 total days of culture with the media changed only on day 2 to add mitotic inhibitors. Longer time periods will require additional media changes.
3. Sample Collection and Processing
- Trigeminal staining
- Pipette 1 mL aliquots of sterile 1x PBS into a 24-well plate under a tissue culture hood for each transwell filter to be processed.
- Remove the liquid atop the filter be removed with a 200-1,000 μL pipette, being careful to leave cell layers intact. Attached cells should remain attached and unattached cells will come loose during this gentle pipetting. A vacuum filter can damage the cell layer and is not recommended for aspiration.
- Transfer the filter to the 1x PBS plate, being sure to remove the top layer of media as mentioned in the previous step.
- Place the plate with all transwell filters and lids on a rocker at 12 rpm or 40-50 rpm on an orbital shaker for 10 min.
- Aspirate the PBS, including the top layer as described above, add 500 μL of 1xPBS and place on the rocker or orbital shaker for an additional 5-10 min rinse.
- Aspirate the 1x PBS once more and replace with 4% paraformaldehyde (PFA). Use at least 500 μL so that the entire filter surface is submerged. Place the plate on a rocker at 12 rpm or orbital shaker at 40-50 rpm at room temperature for 1 h.
- Remove the PFA and rinse the plates twice for 5-10 min each on a rocker with 500 μL of 1x PBS.
- Block with 10% bovine serum albumin (BSA) + 5% goat/donkey serum, depending on the secondary antibody host animal, in 0.05% Tween-1xPBS (PBST). Use 450-500 μL of solution so that the filter is submerged in the liquid.
NOTE: At this stage, these plates can be stored at 4 °C for several months to process at a later time. Use parafilm to seal the plate to ensure evaporation does not occur and check regularly for evaporation so that that filter surfaces remain submerged. - Remove the block and add 450-500 μL of primary antibody in 1% BSA-PBST without an additional rinse. Incubate at 4 °C overnight, gently rocking.
- Remove primary antibody solution and rinse with 500 μL of 1xPBST on a rocker, 3 times, at room temperature
- Remove the PBST, add secondary antibody and incubate on a rocker overnight at 4 °C wrapped in aluminum foil to protect fluorophores from light degradation. Rinse again, 3 times, with 1x PBST on a rocker at room temperature. Replace with 1x PBS.
NOTE: At this point, the plate can be stored for several months at 4 °C if wrapped in aluminum foil to prevent fluorophore degradation. Use parafilm to seal the plate to ensure evaporation does not occur and check regularly for evaporation so that that filter surfaces remain submerged. It is best to image within one month. - For optimal imaging, utilize Thy1-YFP mouse neurons with an anti-GFP antibody to specifically stain the neuronal afferents well above background levels. Use Anti-Neurofilament 200 to precisely stain axonal structures if Thy1-YFP mice are not available.
- Image as desired. Filters do not need to be plated and can instead be placed atop a coverslip or slide for imaging with an inverted microscope.
NOTE: Areas of the filter will not contain afferent structures. Take several images with a z-stack depth that captures all afferent structures. Use stitching software to visualize the neurite outgrowth over large areas, or (preferably) entire filter regions.
- RNA, protein, and media collection
- While the transwell filters are processing, collect the media in RNAse/DNAse-free tubes and freeze for later assays (ELISA, proteomics, etc.).
- Immediately after media collection, aliquot lysis buffer or Radio Immuno Precipitation Assay (RIPA) buffer with proteinase and phosphatase inhibitors into the wells. This protocol was optimized for 100 μL/well in a 24-well plate.
- Allow buffers to lyse cells for 5 min, then scrape each filter with a new pipette tip, and collect the cell samples in RNAse/DNAse-free tubes. Freeze the lysis for future assays (semi-quantitative and/or quantitative PCR, Western blot, etc.).
- Optional immunofluorescence of dental pulp cells:
- Gently lift coverslips from Section 1.1 with forceps and transfer them to different wells with 4% PFA for 1 h with rocking.
- Aspirate PFA and rinse the coverslips twice with 1x PBS. Follow this with permeabilization, blocking and immunofluorescence for markers of interest with standard immunofluorescence techniques.
Representative Results
These results show that TG neurite outgrowth was increased in the presence of primary DP cells in the underlying well compared to the control of TG neurite monoculture (Figure 2A,C). There is some assay-to-assay variability in neurite outgrowth. Thus, a TG neuron monoculture should be included in all assays as a control to detect the basal levels of neurite outgrowth. Primary cells from the Tgfbr2f/f mouse were used in this protocol after infection of Ad-Cre-GFP and Ad-eGFP was confirmed in equivalent numbers of cells (Figure 2D). The Ad-eGFP served as a control viral vector. The Ad-Cre-GFP deleted the flanked gene, Tgfbr2, as demonstrated by semi-quantitative PCR (Figure 2E). In the cultures with Transforming growth factor beta receptor 2 (Tgfbr2) deletion, neurite outgrowth was decreased (Figure 2A-C).
We utilized the Thy1-YFP mouse TG neurons and stained them with an anti-GFP antibody that produced very specific and bright images of axonal structures well above this background, as shown in Figure 2. This allowed the specific staining of neuronal markers without non-specific staining of non-neuronal cells by utilizing previously reported methods such as crystal violet7. The large pores in the filters can autofluoresce and/or accumulate secondary antibodies and decrease the precision of axonal imaging (Figure 3). While the Thy1-YFP neurons with immunofluorescence drastically improves the imaging, further background can be removed with auto-thresholding software and then quantified. We also recommend performing immunofluorescence for Neurofilament 200 based on our preliminary findings (not shown) as well as others8,9 if Thy1-YFP mice are not available.
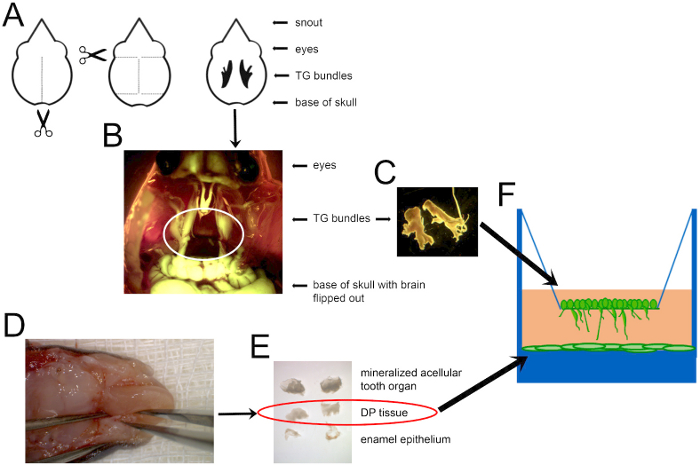
Figure 1: A schematic of the mouse dissection to obtain cells for co-culture. (A) A diagram of where to cut to open the mouse skull and locate TG nerves, shown in black in the last depiction. Scissors indicate where to insert the scissor tips to cut along the dotted lines. (B) A combined darkfield and GFP image showing Thy1-YFP+ TG nerves circled in white. (C) Dissected TG ganglia can then be dispersed and cultured, as shown in F. (D) The mandible of a P7 mouse, with forceps holding the mandible on the left and alveolar bone ridges containing unerupted teeth on each side of the tongue. (E) DP tissue (circled) extracted from the mineralized structure (top), and the enamel outer epithelium (bottom) that was removed to disperse and plate in a tissue culture-treated plate, as shown in F. Images are not shown to scale. DP cells were dispersed and grown to confluence before adding TG neurons. Please click here to view a larger version of this figure.
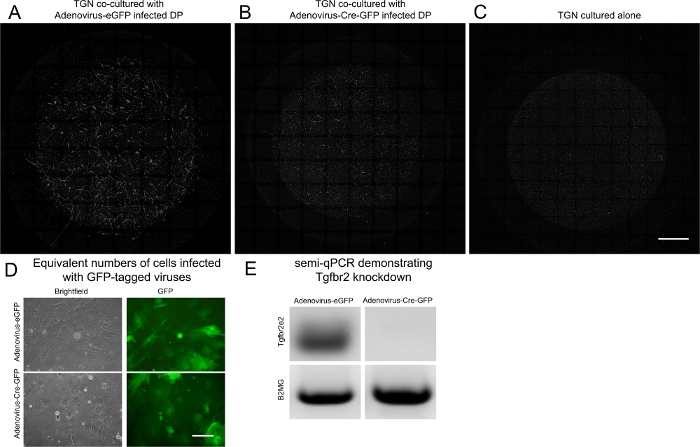
Figure 2: Representative results from co-culture. (A-C) Thy1-YFP TG neurons were cultured in transwell filters with 3 μm pores atop primary Tgfbr2f/f DP cells. Immunofluorescent staining was performed for the YFP protein using an anti-GFP antibody to provide highly specific staining of neuronal structures over the entire filter. The maximum projections of 100 μm z-stack confocal microscopy images at 10x were collected and stitched with stitching software. TG neurons demonstrated significantly more outgrowth when co-cultured with DP cells (A) than when cultured alone (C). Neurite outgrowth was not induced when neurons were co-cultured with DP cells infected with Ad-Cre-GFP to knock down Tgfbr2 (B). Scale bar = 1,000 μm. Equivalent numbers of cells infected with Ad-eGFP and Ad-Cre-GFP are shown in (D). Scale bar = 125 μm. Semi-quantitative PCR confirmed the Tgfbr2 KD (E). Please click here to view a larger version of this figure.
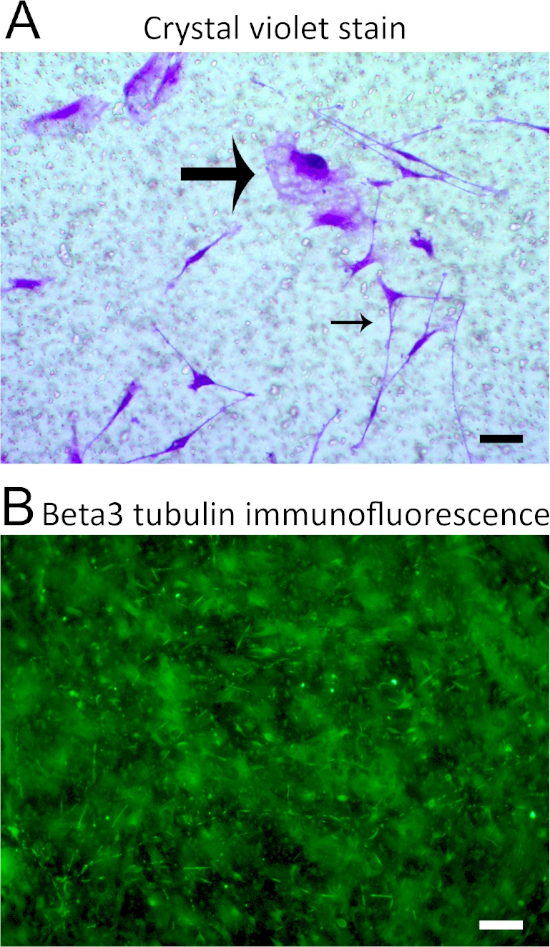
Figure 3: Technical difficulties presented in afferent imaging. (A) Brightfield imaging of transwell filters after crystal violet staining of cell populations. Large pores are prevalent. The large arrow points out a cell that exhibits mesenchymal morphology, whereas the small arrow points to a cell of neuronal morphology. Crystal violet stained both cells without bias. (B) Immunofluorescent staining of β3 tubulin with an Alexa-488 secondary antibody showed non-specific staining of multiple cells, making imaging of afferent structures difficult. Images are representative and were repeated over multiple assays to optimize the imaging shown in Figure 2. Scale bar = 50 μm. Please click here to view a larger version of this figure.
| Component | Volume | Concentration | |
| MEM α | 440 mL | ||
| Heat inactivated fetal bovine serume | 50 mL | 10% | |
| 100x L-glutamine | 5 mL | 1x | |
| Penicillin-streptomycin 100 x | 5 mL | 1x | |
| Change media on day 2 with mitotic inhibitors at these final concentrations | |||
| Uridine | 1 μM | ||
| 5'-Fluor-2'deoxyuridine | 15 μM | ||
Table 1: Co-culture media.
Discussion
The daily activities of the oral cavity require that teeth sense external stimuli and internal inflammation in order to permit proper usage and maintenance. However, only limited information is available regarding the signals that drive the developmental processes of tooth innervation. This protocol provides a method to isolate and co-culture primary DP cells and TG neurons in order to study the cross-communication between the two populations. Several variables were optimized and leave open further avenues of research, as described below.
Controls are important at every step in this assay. A transwell filter with TG neurons without underlying DP cells should be included in every assay to provide a baseline for TG growth. When deleting a flanked gene of interest with an Ad-Cre-GFP recombinase, a control virus expressing only the fluorescent marker should be used to confirm that equivalent numbers of cells have been infected. While we demonstrated high levels of infection with minimal cell death at 100 and 200 MOI for Ad-eGFP and Ad-Cre-GFP, respectively, each lab should optimize this step. Because fluorescent proteins can be attached to different promoters and therefore cause differential expression equivalent numbers of infected, fluorescing cells should be counted. The overall intensity of the fluorescence is irrelevant because it does not accurately reflect the infection status. It is important to demonstrate deletion of the gene, as shown with semi-quantitative PCR in this protocol (Figure 2). While this protocol did not address this topic, previous research demonstrated that control assays with other cells lines could be included to demonstrate that the neurite outgrowth is specifically induced by co-culture with DP cells8.
Because this protocol utilizes primary cells, there are multiple stages at which contamination can be introduced. To prevent this, all reagents should be sterile filtered. Additionally, it is recommended that experiments for every variable be run in duplicate or triplicate to allow for the removal of a filter and sterilization of a contaminated well without complete failure of the assay.
Coverslips must be coated with poly-D-lysine and/or an extracellular matrix protein to ensure DP cell adhesion. While the cells do initially attach, viral infection causes cell lifting death on uncoated coverslips and prevents the genetic manipulation of the co-culture assay.
It is well established that non-neuronal cells, such as Schwann cells from the TG ganglia, can affect the survival of neuronal cells in culture14,15,16. In this protocol, neuronal survival was optimized by adding 1 μM uridine and 15 μM 5-fluoro-2'deoxyuridine. Without the addition of these antimitotic agents to inhibit Schwann cell proliferation, neurite outgrowth will not occur. It is unknown whether the presence of these senescent Schwann cells in co-culture alter the neuronal response. Isolating murine neurons requires several additional steps, and protocols are available for investigators who want to remove this variable17. In either case, neuron dispersion somewhat mimics an axotomy and could be considered to represent injury/repair18 more than development. Further studies would be required to determine the differences between in vivo axonal growth from fascicles versus axonal growth from individual neurons in vitro, and these are not addressed in this protocol.
This protocol takes 1-3 weeks from start to finish. While it is possible to utilize DP cells that require more than 1 week to reach 85-90% confluence, it is recommended that cells be seeded at a high enough density to reach confluence within a few days since these cells divide very slowly past that point. This generally requires around 5-7 P5-8 mice per row of a 24-well plate. This protocol was optimized for a total of 5 days of co-culture, at which point the media with phenol red began to shift color. The media should be changed if longer assays are desired.
Several co-culture assays have been performed to demonstrate neurite outgrowth in response to factors secreted by the DP secreted factors with standard ECM-coated tissue culture plates3,19,20,21 or microfluidic chambers8,22,23. This protocol offers several advantages over these methods. For instance, TG ganglia and DP tissue co-culture requires a specific spatial relationship for the neurites to sense and respond to short-range paracrine signals. With organ culture, only the neurites in the ganglia closest to the DP tissue are able to respond3, whereas the dispersed TG neurons used in this protocol are cultured at an equal distance from the DP cells underneath. Second, organ cultures can introduce tissue necrosis due to the lack of oxygen and nutrients available in large samples24. The co-culture of dispersed cells removes this possibility. Some co-cultures including neurons require neuronal media3,22 which can play a dominant role in promoting neurite outgrowth. This protocol does not add neuron-specific growth factors, thereby allowing for an evaluation of the direct relationship between paracrine signals from the underlying DP cells and neurite outgrowth responses. It is worth noting that the co-culture media also lacks components to promote mineralization, such as beta-glycerophosphate. This allows investigators to determine how neurites might secrete signals to encourage mineralization. However, it also limits the study by only including less-differentiated DP cells without the mineralizing odontoblasts that would typically be present in vivo.
Colorimetric responses from previous research7,8 do not delineate Schwann cell contributions nor demonstrate neuronal morphology since crystal violet non-specifically stains all cells. Immunofluorescent staining of filters can result in high background levels that make afferent imaging difficult (Figure 2). The present protocol allows for the precise staining of neuronal afferents by utilizing Thy1-YFP TG neurons and an anti-GFP antibody and provides a signal bright enough to generate large images of growth throughout an entire figure (Figure 3). It is possible to utilize other neuronal markers, such as Anti-Neurofilament 200, if Thy1-YFP mice are not available.
Finally, using primary DP cells from mice with genes of interest flanked by loxP sites allows for simple and efficient deletion of these genes with an Ad-Cre-GFP system. In future studies, the Ad-Cre recombinase system could be used on the TG neurons if they have a gene of interest flanked by loxP sites. This would facilitate studies on how paracrine signals from the neuronal population influence DP cells, particularly if the DP cells are seeded atop the coverslips (Section 1.1). Future studies can utilize other manipulations, such as the addition of pharmacological inhibitors and/or growth factors. It is also possible to modify this protocol to include migration studies by using 8 μm porosity transwell filters.
In conclusion, this transwell co-culture assay utilizing neurons and DP cells allows for the investigation of multiple cellular parameters. This makes it possible to broaden the body of knowledge about the mesenchymal-neuronal interactions that promote and support tooth innervation.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by a) the National Institutes of Health/NIAMS (grant numbers R01 AR062507 and R01 AR053860 to RS), b) the University of Alabama at Birmingham Dental Academic Research Training (DART) grant (number T90DE022736 (PI MacDougall)) to SBP from the National Institute of Dental and Craniofacial Research/National Institutes of Health, c) a UAB Global Center for Craniofacial, Oral and Dental Disorders (GC-CODED) Pilot and Feasibility grant to SBP and d) the National Institute of Dental and Craniofacial Research/National Institutes of Health K99 DE024406 grant to SBP.
Materials
| 5-Fluoro-2'-deoxyuridine | Sigma-Aldrich | F0503 | Used as a mitotic Inhibitor at 15 μM concentration in co-culture media, Day 2 |
| 24 Well Cell Culture Plate | Corning | 3524 | Co-culture plate |
| Alexa-546 anti-chicken | Invitrogen | A-11040 | Secondary to stain neurite outgrowth labeled by anti-GFP antibody, 1:500 dilution |
| Anti-GFP Antibody | Aves Lab, Inc | GFP-1010 | Primary antibody to label Thy1-YFP neurons, 1:200 dilution |
| Anti-Neurofilament 200 antibody | Sigma-Aldrich | NO142 | Monoclonal primary antibody to label neurons, 1:1000 dilution, alternative if YFP mice are not available |
| B6;129- Tgfbr2tm1Karl/J | The Jackson Laboratory | 12603 | Tgfbr2f/f mouse model used for dental pulp cells in optimized protocol |
| B6.Cg-Tg(Thy1-YFP)16Jrs/J | The Jackson Laboratory | 3709 | Thy1-YFP mouse model genotype used for trigeminal neurons |
| Collagenase Type II | Millipore | 234155-100MG | Used to disperse trigeminal neurons |
| Fetal Bovine Serum | Gibco | 10437 | Additive to co-culture media |
| Fine forceps | Fine Science Tools | 11413-11 | Fine forceps for TG dissection |
| Laminin | Sigma-Aldrich | L2020 | Coats the transwell inserts at final concentration of 10 μg/ml, stock solution is assumed at 1.5 mg/ml |
| Lysis Buffer (Buffer RLT) | Qiagen | 79216 | Extracts RNA from dental pulp cells post co-culture |
| L-Glutamine | Gibco | 25030081 | Additive to co-culture media |
| Micro-dissecting scissors | Sigma-Aldrich | S3146-1EA | Dissection scissors to open skull |
| Microscope Cover Glass | Fisherbrand | 12-545-81 | Circlular coverslip for optional cell culturing and immunofluorescence processing |
| Minimal Essential Medium a | Gibco | 12571063 | Co-culture media base |
| Penicillin-Streptomycin | Gibco | 15070063 | Antibiotic additive to co-culture media |
| Phosphatase Inhibitor | Sigma-Aldrich | 04 906 837 001 | Additive to RIPA Buffer for extracting protein from dental pulp cells post co-culture |
| Polybrene | Millipore | TR-1003-G | Used to aid in dental pulp cell transfection |
| Poly-D-Lysine | Sigma-Aldrich | P7280 | Coverslip coating to aid dental pulp cellular adhesion |
| Protease Inhibitors | Millipore | 05 892 791 001 | Additive to RIPA Buffer for extracting protein from dental pulp cells post co-culture |
| RNAse/DNAse free eppendorf tubes | Denville | C-2172 | Presterilized 1.7 ml tubes for RNA, DNA or protein collection at the end of assay |
| ThinCert Cell Culture Insert | Greiner Bio-One | 662631 | Transwell inserts for trigeminal neurons in co-culture assays |
| Trypsin-EDTA (0.25%) | Gibco | 25200056 | Used fto disperse dental pulp cells |
| Trypsin Type II | Sigma-Aldrich | T-7409 | Used to disperse trigeminal neurons |
| Ultra Fine Forceps | Fine Science Tools | 11370-40 | Ultra fine forceps for dissection |
| Uridine | Sigma-Aldrich | U3750 | Used as a mitotic Inhibitor at 1 μM concentration in co-culture media, Day 2 |
| Vacuum Filtration System | Millipore | SCNY00060 | Steriflip disposable filter, 50 μm nylon net filter |
| Vial forceps | Fine Science Tools | 110006-15 | Long forceps for tissue transfer to conicals |
References
- Moe, K., Sijaona, A., Shrestha, A., Kettunen, P., Taniguchi, M., Luukko, K. Semaphorin 3A controls timing and patterning of the dental pulp innervation. Differentiation. 84 (5), 371-379 (2012).
- Kollar, E. J., Lumsden, A. G. Tooth morphogenesis: the role of the innervation during induction and pattern formation. Journal de biologie buccale. 7 (1), 49-60 (1979).
- Lillesaar, C., Fried, K. Neurites from trigeminal ganglion explants grown in vitro are repelled or attracted by tooth-related tissues depending on developmental stage. Neuroscience. 125 (1), 149-161 (2004).
- Fried, K., Lillesaar, C., Sime, W., Kaukua, N., Patarroyo, M. Target finding of pain nerve fibers: Neural growth mechanisms in the tooth pulp. Physiology & Behavior. 92 (1-2), 40-45 (2007).
- Pagella, P., Jiménez-Rojo, L., Mitsiadis, T. A. Roles of innervation in developing and regenerating orofacial tissues. Cellular and Molecular Life Sciences. 71 (12), 2241-2251 (2014).
- Luukko, K., Kettunen, P. Integration of tooth morphogenesis and innervation by local tissue interactions, signaling networks, and semaphorin 3A. Cell Adhesion & Migration. , 1-9 (2016).
- Smit, M., Leng, J., Klemke, R. L. Assay for neurite outgrowth quantification. BioTechniques. 35 (2), 254-256 (2003).
- de Almeida, J. F. A., Chen, P., Henry, M. A., Diogenes, A. Stem cells of the apical papilla regulate trigeminal neurite outgrowth and targeting through a BDNF-dependent mechanism. Tissue engineering. Part A. 20 (23-24), 3089-3100 (2014).
- Pagella, P., Miran, S., Mitsiadis, T. Analysis of Developing Tooth Germ Innervation Using Microfluidic Co-culture Devices. Journal of Visualized Experiments. (102), e53114 (2015).
- Coelen, R. J., Jose, D. G., May, J. T. The effect of hexadimethrine bromide (polybrene) on the infection of the primate retroviruses SSV 1/SSAV 1 and BaEV. Archives of Virology. 75 (4), 307-311 (1983).
- Caroni, P. Overexpression of growth-associated proteins in the neurons of adult transgenic mice. Journal of neuroscience methods. 71 (1), 3-9 (1997).
- Alić, I., et al. Neural stem cells from mouse strain Thy1 YFP-16 are a valuable tool to monitor and evaluate neuronal differentiation and morphology. Neuroscience Letters. 634, 32-41 (2016).
- Howroyd, P. C. Dissection of the Trigeminal Ganglion of Nonrodent Species Used in Toxicology Studies. Toxicologic Pathology. , (2019).
- Schwieger, J., Esser, K. H., Lenarz, T., Scheper, V. Establishment of a long-term spiral ganglion neuron culture with reduced glial cell number: Effects of AraC on cell composition and neurons. Journal of Neuroscience Methods. 268, 106-116 (2016).
- Liu, R., Lin, G., Xu, H. An Efficient Method for Dorsal Root Ganglia Neurons Purification with a One-Time Anti-Mitotic Reagent Treatment. PLoS ONE. 8 (4), 60558 (2013).
- Burry, R. W. Antimitotic drugs that enhance neuronal survival in olfactory bulb cell cultures. Brain Research. 261 (2), 261-275 (1983).
- Katzenell, S., Cabrera, J. R., North, B. J., Leib, D. A. Isolation, Purification, and Culture of Primary Murine Sensory Neurons. Methods in molecular biology. 1656, 229-251 (2017).
- Dussor, G. O., Price, T. J., Flores, C. M. Activating transcription factor 3 mRNA is upregulated in primary cultures of trigeminal ganglion neurons. Molecular Brain Research. 118 (1-2), 156-159 (2003).
- Lillesaar, C., Arenas, E., Hildebrand, C., Fried, K. Responses of rat trigeminal neurones to dental pulp cells or fibroblasts overexpressing neurotrophic factors in vitro. Neuroscience. 119 (2), 443-451 (2003).
- Lillesaar, C., Eriksson, C., Fried, K. Rat tooth pulp cells elicit neurite growth from trigeminal neurones and express mRNAs for neurotrophic factors in vitro. Neuroscience Letters. 308 (3), (2001).
- Lillesaar, C., Eriksson, C., Johansson, C. S., Fried, K., Hildebrand, C. Tooth pulp tissue promotes neurite outgrowth from rat trigeminal ganglia in vitro. Journal of neurocytology. 28 (8), 663-670 (1999).
- Chmilewsky, F., Ayaz, W., Appiah, J., About, I., Chung, S. H. Nerve Growth Factor Secretion From Pulp Fibroblasts is Modulated by Complement C5a Receptor and Implied in Neurite Outgrowth. Scientific reports. 6, 31799 (2016).
- Pagella, P., Neto, E., Jiménez-Rojo, L., Lamghari, M., Mitsiadis, T. A. Microfluidics co-culture systems for studying tooth innervation. Frontiers in Physiology. 5, 326 (2014).
- Miura, T., Yokokawa, R. Tissue culture on a chip: Developmental biology applications of self-organized capillary networks in microfluidic devices. Development, Growth & Differentiation. 58 (6), 505-515 (2016).

