Characterization of In Vitro Differentiation of Human Primary Keratinocytes by RNA-Seq Analysis
Summary
Presented here is a stepwise procedure for in vitro differentiation of human primary keratinocytes by contact inhibition followed by characterization at the molecular level by RNA-seq analysis.
Abstract
Human primary keratinocytes are often used as in vitro models for studies on epidermal differentiation and related diseases. Methods have been reported for in vitro differentiation of keratinocytes cultured in two-dimensional (2D) submerged manners using various induction conditions. Described here is a procedure for 2D in vitro keratinocyte differentiation method by contact inhibition and subsequent molecular characterization by RNA-seq. In brief, keratinocytes are grown in defined keratinocyte medium supplemented with growth factors until they are fully confluent. Differentiation is induced by close contacts between the keratinocytes and further stimulated by excluding growth factors in the medium. Using RNA-seq analyses, it is shown that both 1) differentiated keratinocytes exhibit distinct molecular signatures during differentiation and 2) the dynamic gene expression pattern largely resembles cells during epidermal stratification. As for comparison to normal keratinocyte differentiation, keratinocytes carrying mutations of the transcription factor p63 exhibit altered morphology and molecular signatures, consistent with their differentiation defects. In conclusion, this protocol details the steps for 2D in vitro keratinocyte differentiation and its molecular characterization, with an emphasis on bioinformatic analysis of RNA-seq data. Because RNA extraction and RNA-seq procedures have been well-documented, it is not the focus of this protocol. The experimental procedure of in vitro keratinocyte differentiation and bioinformatic analysis pipeline can be used to study molecular events during epidermal differentiation in healthy and diseased keratinocytes.
Introduction
Human primary keratinocytes derived from the human skin are often used as a cellular model to study the biology of the epidermis1,2,3,4. The stratification of the epidermis can be modeled by keratinocyte differentiation, either in a 2D submerged monolayer fashion or 3D air-lift organotypic model2,3,5,6,7. Although 3D models have become increasingly important to assess the epidermal structure and function, 2D differentiation models are still widely used, due to their convenience and the possibility to generate large numbers of cells for analyses.
Various conditions have been applied for inducing keratinocyte differentiation in 2D, including addition of serum, high concentration of calcium, lower temperature and inhibition of epidermal growth factor receptors2,3. Each of these methods has been validated by a number of keratinocyte differentiation marker genes and shown to be effective in assessing keratinocyte differentiation, including under pathological conditions. However, these induction conditions also show differences in their differentiation efficiency and kinetics when specific panels of marker genes are examined2,3.
One of these methods involves keratinocyte contact inhibition and depletion of growth factors in the culture medium8. It has been shown that keratinocytes can differentiate spontaneously when cells reach full density. Excluding growth factors in the culture medium can further enhance differentiation. The method combining contact inhibition and depleting growth factors has been shown to generate differentiated keratinocytes with gene expression patterns similar to the normal stratified epidermis when using several epidermal markers3, suggesting that this model is suitable for studying normal keratinocyte differentiation. Recently, two comprehensive gene expression analyses of keratinocyte differentiation using this model have been reported9,10. Researchers validated this model at the molecular level and showed that it can be used to study normal and diseased keratinocyte differentiation.
This protocol describes the procedure for the in vitro differentiation method and molecular analysis of differentiated cells using RNA-seq. It also illustrates characterization of the transcriptome of cells on differentiation day 0 (proliferation stage), day 2, day 4, and day 7 (early, middle, and late differentiation, respectively). It is shown that differentiated keratinocytes display gene expression patterns that largely resemble cells during epidermal stratification. To examine whether this method can be used for studying skin pathology, we applied the same experimental and analysis pipeline to investigate keratinocytes carrying mutations of the transcription factor p63 that are derived from patients with ectrodactyly, ectodermal displasia and cleft lip/palate (EEC) syndrome11,12. This protocol focuses on the in vitro differentiation of keratinocytes as well as subsequent bioinformatic analysis of RNA-seq. Other steps in the complete procedure such as RNA extraction, RNA-seq sample preparation and library construction, are well documented and can be easily followed, especially when using many commonly used commercial kits. Therefore, these steps are only briefly described in the protocol. The data show that this pipeline is suitable for studying molecular events during epidermal differentiation in healthy and diseased keratinocytes.
Protocol
Skin biopsies were taken from the trunk of healthy volunteers or patients with p63 mutations, to set up the primary keratinocyte culture. All procedures regarding establishing human primary keratinocytes were approved by the ethical committee of the Radboud University Nijmegen Medical Centre (“Commissie Mensgebonden Onderzoek Arnhem-Nijmegen”). Informed consent was obtained.
1. Human primary keratinocyte differentiation by contact inhibition
- Prepare keratinocyte growth medium (KGM) from Keratinocyte Basal Medium (Table of Materials) when necessary.
- Make 500 mL proliferation medium using pre-prepared stocks (KGM-pro; see Table 1).
- Make 500 mL differentiation medium (KGM-dif; see Table 2).
NOTE: KGM with supplements can be kept at 4 °C for two weeks. For longer storage, it can be aliquoted and stored at -20°C. - Seed primary keratinocytes in the density of 5.0–20 x 103 cells/cm2, depending on the cell proliferative capacity. Add sufficient KGM-pro medium to cover cells.
NOTE: 1) Details of regular cell culture within KGM medium can be found in the website of Lonza13. 2) The seeding density should be tested for each cell line. For example, a normal primary keratinocyte line can be seeded at a density of 5.0 x 103 cells/cm2, whereas a line carrying mutations in the transcription factor p63 that is less proliferative should be seeded at a density of 20 x 103 cells/cm2. 3) Depending on the experimental set up, several dishes or wells of cells should be seeded to allow collecting samples on different differentiation days in replicas. - Refresh cells with the KGM-pro medium for two days after seeding (seeding on day 0, refreshing for the 1st time on day 3). Afterwards, refresh with KGM-pro medium every other day. Check cells regularly, at least every other day.
- Induce cell differentiation when cells are more than 90% confluent by changing the medium to KGM-dif. The day of changing medium to KGM-dif is defined as differentiation day 0. Collect cells for further RNA analyses by first washing 2x with DPBS, afterwards adding in the lysis buffer (from an RNA extraction kit).
NOTE: With the abovementioned seeding density, cells should reach confluency within 7–10 days. Cells can be seeded with a higher initial density to reach confluency sooner. If cells are not proliferative, longer waiting may not give rise to fully confluent cells. A higher seeding density may be required. - Refresh cells with the KGM-dif medium every day and collect cells on differentiation day 2, 4. and 7 for further RNA extraction.
2. RNA extraction
- Isolate total RNA. This can be performed using a commercial RNA isolation kit (such as Zymo Quick-RNA MicroPrep RNA), or using phenol14. However, an isolation kit containing a DNAse treatment is highly recommended for RNA-seq samples to prevent DNA contamination and phenol. An example of a commercial RNA-isolation kit is given in the Table of Materials.
- Measure RNA concentration with a spectrometer. Both DNA and RNA have an absorption peak at 260 nm.
NOTE: 1) The ratio between the absorbances at 260 nm and 280 nm is used to assess the purity of the RNA. A ratio of around 2.0 is generally accepted as ‘pure’ RNA. If the ratio is substantially lower than 2.0, the sample might be contaminated with contaminants that absorb at 280 nm such as proteins, phenols, or other substances. 2) The ratio between the absorbances at 260 nm and 230 nm measures nucleic acid purity. A ratio between 2.0 and 2.2 is expected. If the ratio is substantially lower, the sample might be contaminated with contaminants that absorb at 230 nm such as EDTA, ethanol, or other substances.
3. RNA quality check
- Run RNA either on a bioanalyzer RNA Pico chip to validate RNA Integrity Number (RIN) quantitatively, or on a gel for a qualitative measure. In general, a RIN of at least eight is highly advisable. However, when extracting RNA from tissues the RIN may be lower.
NOTE: Optionally, quality control can be performed by qPCR on the RNA material.
4. RNA-seq library preparation
NOTE: The RNA-seq library preparation is often performed with a commercial kit or under commercial settings. The described protocol is adapted from a commercial kit, KAPA RNA HyperPrep Kit with RiboErase (Illumina), with a brief description of all required steps: rRNA depletion with oligo hybridization to human ribosomal RNAs, RNA fragmentation, first-strand synthesis, second-strand synthesis and A-tailing, and cleanup after each step15. Other library preparation kits can also be used for this purpose. It is recommended to perform this step using a commercially available kit, as the quality of generated cDNA library is often more consistent. 2) The following steps are described for 1x library preparation. If preparing several samples, make master mixes with 10% extra volume.
- Oligo hybridization and rRNA depletion
- Prepare oligo hybridization master mix (total = 11 µL, with 4.4 µL of hybridization buffer, 4.4 µL of hybridization oligos, and 2.2 µL of RNase-free water) and depletion master mix (total = 5.5 µL, with 3.3 µL of depletion buffer and 2.2 µL of RNase H).
- Set up the PCR reaction program on a thermocycler: 95 °C for 2 min; ramp down to 45 °C at -0.1 °C/s; 45 °C pause; 45 °C for 30 min; 4 °C forever.
NOTE: Consider starting with double the amount of RNA than necessary for amplification. In the first attempt, use one-half of the amount to continue with the following steps. This is to make sure that there is still material to repeat these steps with a different number of cycles (see step 4.7.2), if the cycles of amplification turn out to be insufficient or overamplified. - Use between 25 ng and 1 µg of total RNA in 10 µL of RNAse-free water, add 10 μL of oligo hybridization master mix. Place samples in the pre-programmed thermocycler and start the program.
- When the program reaches the pause step at 45 °C, add 5 µL of depletion master mix to the 20 µL hybridization reaction without removing it from the thermocycler. Mix thoroughly by pipetting up and down several times.
- Resume the thermocycler program to continue with the depletion step (45 °C for 30 min).
NOTE: 1) Put the beads for rRNA depletion (e.g., KAPA pure beads) at room temperature (RT) during the depletion step for the next part of the protocol. 2) Consider preparing the DNase digestion master mix (for section 4.2), since the reagents are needed directly after the rRNA depletion cleanup. - Perform a 2.2x bead-based KAPA Pure Beads cleanup: combine the RNA mix (25 μL) and KAPA Pure Beads (55 μL), by thoroughly resuspend the beads in the RNA mix by pipetting up and down several times.
- Incubate at RT for 5 min to allow RNA binding to the beads, and place the tube(s) on a magnet rack to capture the beads until the liquid is clear, and carefully remove and discard 60 µL of supernatant.
- Keeping the tube(s) on the magnet, wash 2x with 200 µL of 80% ethanol by incubating the beads with the ethanol for ≥30 s and discard the ethanol. Try to remove all residual ethanol without disturbing the beads after the second wash.
- Dry the beads at RT for 3–5 min or until all ethanol has evaporated.
NOTE: Over-drying the beads may result in reduced yield.
- DNase digestion
- Prepare DNase digestion master mix (total = 22 µL, with 2.2 µL of DNase buffer, 2 µL of DNase, and 17.8 µL of RNase-free water) .
- Resuspend the beads in DNAse digestion master mix (20 μL) by pipetting up and down several times. Incubate the tube(s) at RT for 3 min to elute the RNA off the beads.
- Place the tube(s) on a magnet rack to capture the beads, until the liquid is clear, and carefully transfer 20 µL of supernatant into a clean tube.
- Incubate the tube(s) with supernatant at 37 °C for 30 min.
NOTE: Consider preparing the RNA elution, fragmentation, and priming master mixes (for section 4.3), since the reagents are directly needed after the DNase digestion cleanup. - Perform a 2.2x bead-based cleanup by following 4.1.6–4.1.9.
- RNA elution, fragmentation, and priming
- Prepare the Fragment, Prime and Elute Buffer (1x) master mix with 11 µL of Fragment, Prime and Elute Buffer (2x) and 11 µL of RNase-free water.
- Thoroughly resuspend the beads with purified, DNase-treated RNA in 22 µL of Fragmentation, Prime and Elute Buffer (1x) by pipetting up and down several times.
- Incubate at room temperature for 3 min to elute RNA off the beads, then place the tube(s) on a magnet to capture the beads until the liquid is clear.
- Carefully transfer 20 µL of supernatant into a tube(s). Discard the tube(s) with beads.
NOTE: This is a safe stopping point, as samples can be stored at -20 °C for ≤24 h. - Place the tube(s) in a thermocycler and carry out the fragmentation and priming at 94 °C for 6 min. Resulting in approximate 200–300 bp long fragments.
NOTE: Incubate for 8 min at 94 °C for 100–200 bp fragments. When using partially degraded RNA, fragment between 1–6 min at 85 °C depending on the severity of degradation. - Place the tube(s) on ice and proceed immediately to first strand synthesis.
- First strand synthesis
- Prepare first strand synthesis master mix (total = 12 µL, with 11 µL of first strand synthesis buffer and 1 µL of KAPA script).
- Set up the PCR reaction program on a thermocycler: 25 °C for 10 min; 42 °C for 15 min; 70 °C for 15 min; 4 °C forever.
- On ice, combine 20 µL of fragmented primed RNA with 10 µL of first strand synthesis master mix. Keep the tube(s) on ice, mix thoroughly by gently pipetting the reaction up and down several times.
- Place the tube(s) in the pre-programmed thermocycler and start the program.
- Second strand synthesis and A-tailing
- Prepare second strand synthesis and A-tailing master mix on ice (total = 33 µL, with 31 µL of second strand synthesis buffer and 2 µL of second strand synthesis & A-tailing enzyme mix).
- Set up the PCR reaction program on a thermocycler: 16 °C for 30 min; 62 °C for 10 min; 4 °C forever.
- Combine 30 µL of first strand synthesis product with 30 µL of second strand synthesis and A-tailing master mix, and mix thoroughly by pipetting up and down several times on ice.
- Place the tube(s) in the pre-programmed thermocycler and start the program.
- Adapter ligation and post-ligation cleanup
- Dilute stock adapters (NEXTflex DNA barcodes, 25 µM) 3.57x in RNase-free water to 7 µM.
- Prepare adapter ligation master mix (total = 50 µL, with 40 µL of ligation buffer and 10 µL of DNA ligase).
- Combine 60 µL of second strand synthesis product, 45 µL of adapter ligation master mix, and 5 µL of diluted adapters. Mix thoroughly by pipetting up and down several times on ice.
- Incubate the tubes at 20 °C for 15 min and continue immediately with the first post ligation cleanup.
- Perform a 0.63x bead-based cleanup by combining adapter-ligated DNA (110 µL) and KAPA pure beads (70 µL), then thoroughly resuspend the beads in the DNA mix by pipetting up and down several times.
- Repeat steps 4.1.7–4.1.9.
- Remove the tubes from the magnet. Thoroughly resuspend the beads in 50 µL of 10 mM Tris HCL (pH = 8.0–8.5) and incubate the beads at RT for 2 min.
- Place the plate on the magnet and wait until the liquid is clear. Transfer 50 μL of the clear supernatant to a new tube.
NOTE: Safe stopping point for less than 24 h at 4 °C. - Perform a 0.7x bead-based cleanup by combining 50 µL of beads with purified adapter-ligated DNA with 35 µL of PEG/NaCL solution. Mix thoroughly by vortexing.
- Perform cleanup steps 4.1.7–4.1.9. Thoroughly resuspend the beads in 20 µL of 10 mM Tris HCL (pH = 8.0–8.5), and incubate the beads at RT for 2 min.
- Place the plate on the magnet; wait until the liquid is clear. Transfer 20 μL of the clear supernatant to a new tube and proceed to section 4.7.
NOTE: Safe stopping point for less than 1 week at 4 °C or less than 1 month at -20 °C.
- Library amplification and cleanup
- Prepare library amplification master mix (total = 33 µL, with 27.5 µL of 2x KAPA HiFi HotStart ReadyMix and 5.5 µL of 10x library amplification primer mix).
- Set up the PCR reaction program on a thermocycler using the following parameters. 98 °C for 45 s; N cycles (see the note below) of: 98 °C for 15 s; 60 °C for 30 s; 72 °C for 30 s; 72 °C for 1 min; and 4 °C forever.
NOTE: The cycles for the library amplification depend on the input material. It can roughly be estimated as such: starting with RNA = 25–100 ng, N = 11–15 cycles; 100–250 ng, N = 9–12 cycles; 250–500 ng, N = 7–10 cycles. - Perform a 0.8x bead-based cleanup by combining amplified library DNA (50 µL) and KAPA pure beads (40 µL), then thoroughly resuspend the beads in the DNA mix by pipetting up and down several times.
- Perform cleanup steps 4.1.7–4.1.9. Thoroughly resuspend the beads in 50 µL of 10 mM Tris HCL (pH = 8.0–8.5), and incubate the beads at RT for 2 min.
- Transfer 50 μL of the clear supernatant to a new tube and proceed to the second library amplification cleanup.
- Perform a 1x bead-based cleanup by combining amplified library DNA (50 µL) and KAPA pure beads (50 µL), then thoroughly resuspend the beads in the DNA mix by pipetting up and down several times.
- Perform cleanup steps 4.1.7–4.1.9. Thoroughly resuspend the beads in 22 µL of 10 mM Tris HCL (pH = 8.0–8.5), and incubate the beads at RT for 2 min.
- Transfer 20 μL of the clear supernatant to a new tube proceed to Qbit and bioanalyzer measurements.
- Concentration and fragmentation size
- Measure DNA concentrations of the samples. Using a highly sensitive fluorescent dye-based kit (e.g., Denovix Qbit, see Table of Materials), or if the concentration seems to be below 0.5 ng/μL, requantify using a more sensitive qPCR approach (e.g., Kappa Quantification, see Table of Materials).
- Determine the fragment size of the library, using high sensitive electrophoresis (e.g., a bioanalyzer).
NOTE: Optionally, quality control by qPCR can be performed before sequencing (Appendix) using a diluted prepared library instead of cDNA.
- Sequencing
- Send the library for sequencing. For RNA-seq differential gene expression analysis, a sequencing depth of reads between 10–25 million per sample is generally sufficient.
5. Data pre-processing
- Quality check Fastq files
- Download and install Trimgalore16 to remove low quality base pairs and empty reads within the Fastq files.
NOTE: 1) Most software mentioned here only works in Linux. If limited to a Windows computer, it is possible to run analyses on a Linux server using the software MobaXterm or Putty. Keep in mind that for genome indexing a lot of random-access memory (RAM) is required (around 64 GB). 2) Installing all required software into a conda environment is highly recommended. This will allow easy software installation and package management. For more information on conda, visit <https://docs.conda.io>. - Create a directory (folder) for the data e.g. the folder ‘RNA_seq_KC_diff’. Set this as the working directory by typing: “cd /home/user/RNA_seq_KC_diff/”.
NOTE: For an overview of the folder structure, see ‘folder_structure.txt’ in the supplementary coding files. - Create several folders in the working directory with the specific names: ‘Fastq’, ‘CRCh38_fasta’, ‘CRCh38’, ‘scripts’, and ‘Mapping’.
- Move the fasta files of the sequenced data into the fastq folder. Alternatively, generate soft links using the command: “ln –s home/user/old_location_fastq/FastqFile home/user/RNAseq_KC_diff/Fastq/Fastqfile” to the Fastq files to save disk space.
- Run Trim galore on the Fastq files, see the TrimGalore.txt file for the code to copy paste into bash. Change folder names and settings within the command where necessary.
- Download and install Trimgalore16 to remove low quality base pairs and empty reads within the Fastq files.
- Mapping
- Download a genome to map the reads to, e.g., hg38 ensembl release 97 (unmasked version): ftp://ftp.ensembl.org/pub/release-97/fasta/homo_sapiens/dna/Homo_sapiens.GRCH38.primary_assembly.fa.gz ; and the corresponding gene annotation file: hg 38 gene annotation ensembl release 97, ftp://ftp.ensembl.org/pub/release-97/gtf/homo_sapiens/Homo_sapiens.GRCh38.97.gtf.gz. Transfer both the files to the CRCh38_fasta folder.
NOTE: Alternatively use a different version of the genome, for example, the UCSC version of hg38, if mapping to transcript IDs rather than gene IDs is preferred. - Install STAR 2.7.117.
- Generate an in-house reference genome using STAR 2.7.1 by the generate genome script (see Supplemental Coding Files). Change the amount of threads to what the processer has (–runThreadN X). Change folder names and settings in this script where necessary. Run it by copy/pasting into bash.
- Install samtools 1.918 for indexing the bam files and gzip to compress the wiggle files.
- Align the Trim Galore validated sequencing reads to the human genome assembly hg38 (ensembl release 97) using STAR 2.7.1 followed by indexing using samtools and gzip. This should be performed using the script map_fastq.txt (see Supplemental Coding Files). Change the folder names and settings in this script where necessary. Run it by copying/pasting into bash.
NOTE: STAR mapping generates several output files, including a bam file, a wiggle file, and read counts table named *_ReadsPerGene.out.tab, which can directly be used concatenated by R to use as input for Deseq2.
- Download a genome to map the reads to, e.g., hg38 ensembl release 97 (unmasked version): ftp://ftp.ensembl.org/pub/release-97/fasta/homo_sapiens/dna/Homo_sapiens.GRCH38.primary_assembly.fa.gz ; and the corresponding gene annotation file: hg 38 gene annotation ensembl release 97, ftp://ftp.ensembl.org/pub/release-97/gtf/homo_sapiens/Homo_sapiens.GRCh38.97.gtf.gz. Transfer both the files to the CRCh38_fasta folder.
- Genome browser visualization
- Install wigToBigWig from the UCSC genome browser tools to generate bigwig files with the big2bw script19.
- Transfer the files ‘convertBigWigChroms.py’ and ‘CRCH38_ensembl2UCSC.txt’ from the supplementary coding files to a folder ‘scripts’ in the working directory.
- Make the convertBigWigChroms.py executable (using the command: ‘chmod +x scripts/convertBigWigChroms.py’ on a Linux machine).
- Generate bigWig files from the Wiggle files, using the script wig2bw.txt (see Supplemental Coding Files). Run it by copy/pasting into bash.
- Input the BigWig files on the UCSC genome browser or Integrative Genomics Viewer (IGV) for visualization.
6. RNA-seq data analysis
- Input sample data into Deseq2
- Download and install Rstudio (version 1.1.456) and R (version 3.6).
- Install all the required R packages (see Supplemental coding files).
NOTE: For a detailed R-markdown file showing all programming code and output, see the attached files: “RNA_seq_kc_differentiation_wt.html” and “RNA_seq_kc_differentiation_patient.html”. A general description of all steps is included below. - Generate a count table from the ReadsPerGene.out.tab files.
- Write a sample data file, containing all filenames, the day of differentiation and other relevant sample data. For an example sample file, see “sample_data_example.csv” in the Supplemental Coding Files.
- Use the count table and sample data to generate a Deseq220 object containing both the countable and sample data.
- Gene expression normalization, sample distance and PCA
- Normalize the count table in the Deseq2 object, using either Deseq2 rld, or vst normalization. Rld normalization is preferred, but with many samples, vst normalization is much faster.
- Plot sample distance based on the normalized read count intensities using the “dist” function in R and subsequently performing “hclust” clustering based on sample distance. Plot the heatmap itself using the pheatmap package.
- Generate a PCA plot of the normalized read count intensities using the plotPCA function of Deseq2.
NOTE: PCA serves as a tool in exploratory data analysis and can be used to visualize distance and relatedness between different samples.
- Differential/highly variable gene expression analysis
- Calculate either the differential genes using the Deseq2 result function. However, when assessing changes over several timepoints in which pairwise comparisons are not applicable, use highly variable genes. In this case, extract the top 500 highly variable genes via ordering on the variance between samples from different time points with the rowVars function.
NOTE: In our analysis, for the control samples, the top 500 highly variable genes (the genes with the highest standard deviation of their normalized intensity across days of differentiation) were used for further analysis. However, for the diseased vs control analysis differentially expressed genes between disease and healthy controls over time were calculated by Deseq2 using a multiple testing corrected p-value of 1 x 10-4 as a cutoff. Another possible filtering step is to exclude differential genes changing less than a certain fold change cutoff. - Perform kmeans clustering on the differential or highly variable genes to cluster them by different expression patterns.
- Visualize differential or highly variable genes in a heatmap using the pheatmap package. The intensity plotted in the heatmap is the Deseq2 normalized intensity with the median value subtracted.
- Calculate either the differential genes using the Deseq2 result function. However, when assessing changes over several timepoints in which pairwise comparisons are not applicable, use highly variable genes. In this case, extract the top 500 highly variable genes via ordering on the variance between samples from different time points with the rowVars function.
- Gene Ontology (GO) annotation enrichment analysis
- Generate a list of expressed background genes by taking all genes with more than 10 counts in a single sample.
- Perform GO analysis using online tools such as GOrilla21. Use the lists of the differential/highly variable genes in clusters as “gene list”, and the background genes as the background for comparison.
NOTE: More advanced R-users can use the package clusterProfiler22 to automate Go-term enrichment analysis.
Representative Results
Normal keratinocyte differentiation and RNA-seq analysis
In this experiment, keratinocyte lines derived from five individuals were used for differentiation and RNA-seq analyses. Figure 1 summarizes the experimental procedure of differentiation and RNA-seq analysis results. An overview of in vitro differentiation procedures of normal keratinocytes and cell morphology changes during differentiation are illustrated in Figure 1A. Principle component analysis (PCA) showed that keratinocytes undergoing differentiation had connected but distinct overall gene expression profiles (Figure 1B). Highly variable genes were clustered by kmeans to visualize gene expression dynamics and patterns during differentiation (Figure 1C).
Each cluster of genes was represented by the keratinocyte differentiation hallmark genes (e.g., KRT5 for proliferation, KRT1 and KRT10 for early and mid-differentiation, and IVL/LOR/FLG for late differentiation). Gene ontology (GO) annotation analysis of highly variable gene clusters (Figure 1C) showed a clear difference in gene functions of these gene clusters (e.g., keratinization in the mid-stages of differentiation; and epidermal cell differentiation, keratinocyte differentiation, and peptide cross-linking in the late stages; Figure 1D). Protein expression of several differentiation markers were measured by western blotting (Figure 1E).
P63 mutant keratinocyte differentiation and RNA-seq analysis:
In the second experiment, cell morphology and gene expression differences were compared between keratinocytes from healthy controls and three lines derived from patients carrying p63 mutations (mutants R204W, R279H, and R304W). Figure 2A shows an overview of differentiation procedures and cell morphology changes. Mutant keratinocytes remained flat on the surface of the dish and did not become crowded or overlap growth as control keratinocytes on day 7.
In PCA analysis, the control cell lines clearly followed a differentiation pattern, as compared to Figure 1B. However, the pattern of gene expression of mutant cells during differentiation stays largely similar to those of proliferating/undifferentiated cells. Among the three mutant lines, differentiating R279 samples moved along PC1 and PC2 to some extent, indicating that its differentiation was less impaired, compared to R204W and R304W.
In the clustering analysis (Figure 2C), genes downregulated in control cells (cluster 1) were partially downregulated in R204W and R279W, but their gene expression was not drastically changed in R304W. These genes likely play roles in cell proliferation, as shown by GO annotation (Figure 2D). In cluster 2 (Figure 2C), genes were first induced and subsequently downregulated in control cells. These genes are likely involved in keratinocyte differentiation, as epidermal differentiation and keratinization functions were highly enriched for this cluster genes (Figure 2D). These genes were not induced in R204W and R304W, whereas in R279H cells, these genes were induced but not downregulated as much as the control cells.
Genes in cluster 3 were only induced at the end of differentiation in control cells (Figure 2D). Consistent with this, these genes may have a role in the outer most layer of the epidermis, as they have been shown to be responsive to external stimuli and inflammation (Figure 2D). The expression pattern of these genes did not change much in all three mutant cell lines. The visible differences in gene expression patterns between control and mutant cells demonstrate that mutant cells cannot differentiate properly in these in vitro differentiation models.
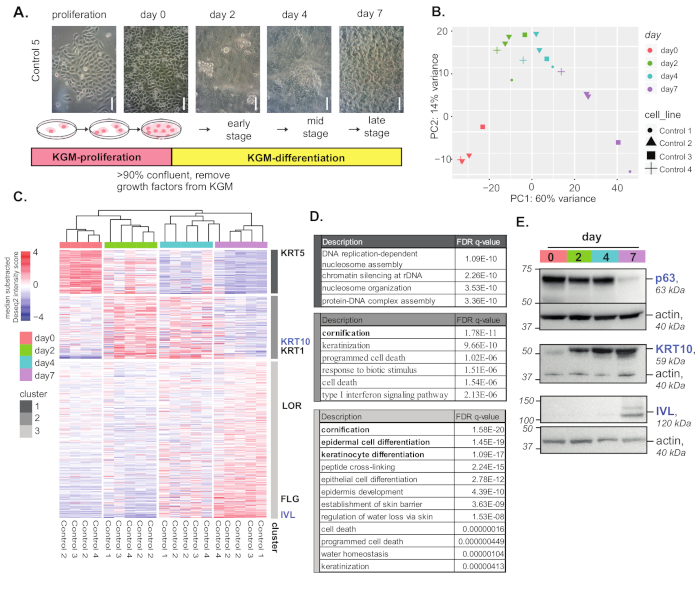
Figure 1: Keratinocyte differentiation and analysis. (A) Overview of the keratinocyte differentiation protocol and cell morphology. Scale bar = 100 µm. (B) Principle component analysis of differentiating control keratinocytes. (C) Heatmap of the top 500 highly variable genes during keratinocyte differentiation. Genes are clustered in three clusters using kmean clustering. Representative differentiation marker genes for each gene cluster are indicated at the side. (D) GO term enrichment analysis of overrepresented functions for genes in the highly variable gene clusters, as compared to a background of all expressed genes (counts of >10). GOrilla was used for the enrichment test. (E) Western blot of keratinocyte differentiation markers during differentiation. Please click here to view a larger version of this figure.
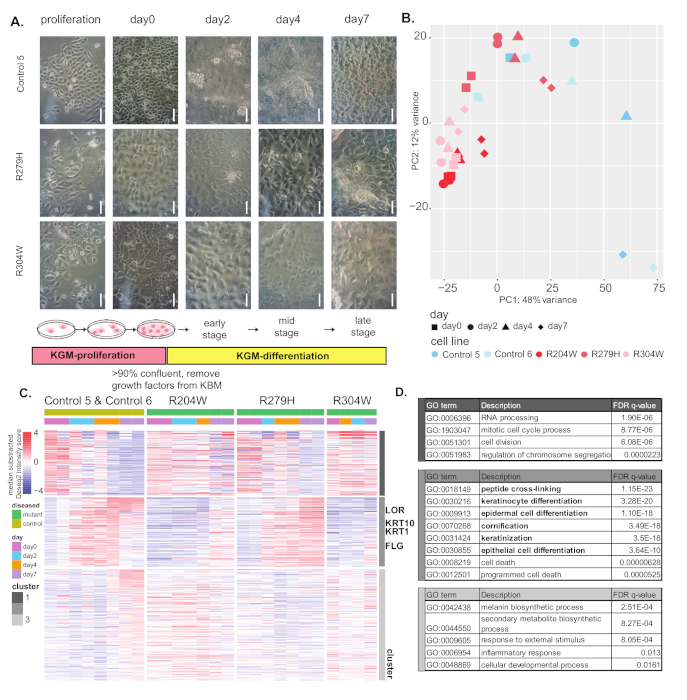
Figure 2: Comparison of control and p63 mutant keratinocytes. (A) Overview of the keratinocyte differentiation protocol and cell morphology of control and p63 mutant keratinocytes. Scale = 100 µm. (B) Principle component analysis of differentiating control and patient (R204W, R279H & R304W) keratinocytes. (C) Heatmap of differential genes between control and mutant keratinocytes. Genes are clustered in three clusters using kmean clustering. Representative differentiation marker genes for each gene cluster are indicated. (D) GO term enrichment analysis of overrepresented functions for genes in the highly variable gene clusters, as compared to a background of all expressed genes (counts of >10). GOrilla was used for the enrichment test. Please click here to view a larger version of this figure.
| KGM component | Stock | Medium | Volume |
| KBM | 500 mL | ||
| Pen/Strep | 100,000 units/mL | 100 units/mL | 5 mL |
| BPE | ~13 mg/mL | 0.4% | 2 mL |
| Ethanolamine | 0.1 M | 0.1 mM | 500 μL |
| o-phosphoethanolamine | 0.1 M | 0.1 mM | 500 μL |
| Hydrocortisone | 0.5 mg/mL | 0.5 µg/mL | 500 μL |
| Insulin | 5 mg/mL | 5 µg/mL | 500 μL |
| EGF | 10 μg/mL | 10 ng/mL | 500 μL |
Table 1. KGM-pro medium supplements.
| KGM component | Stock | Medium | Volume |
| KBM | 500 mL | ||
| Pen/Strep | 100,000 units/mL | 100 units/mL | 5 mL |
| Ethanolamine | 0.1 M | 0.1 mM | 500 μL |
| o-phosphoethanolamine | 0.1 M | 0.1 mM | 500 μL |
Table 2. KGM-diff medium supplements.
Supplemental Coding Files: Folder_structure.txt; Generate_genome.txt; Map_fastq.txt; RNA_seq_kc_differentiation_patient.html; RNA_seq_kc_differentiation_wt.html; Sample_data_example.csv; TrimGalore.txt; and Wig2bw.txt. Please click here to download this file.
Discussion
This work describes a method for inducing human keratinocyte differentiation and subsequent characterization using RNA-seq analyses. In the current literature, many studies on human keratinocyte differentiation use two other methods, with a high calcium concentration or with serum as methods to induce differentiation2,3,23. A previous report carefully compared these three different methods3 and showed that these methods can represent distinct biology of keratinocyte differentiation. In this same report, authors showed that differentiated cells induced by serum have high proliferative potential and express genes such as KRT16 and SKALP/ PI3 that are expressed in the psoriasis skin but not in normal epidermis, and that therefore serum-induced differentiation model can be used to study psoriasis. In contrast, the method of contact inhibition plus growth factor exclusion can induce KRT1 and KRT10, which are normally expressed in the epidermis and resemble normal keratinocyte differentiation. High calcium induction gives rise to a gene expression profile that is between serum induction and contact inhibition, as the least specific method. The analyses using RNA-seq analyses during differentiation confirmed that the method of contact inhibition plus growth factor exclusion results in differentiated keratinocytes with similar gene expression profiles as those expected for epidermal differentiation (e.g., KRT5 in proliferating cells, KRT1 and KRT10 in early differentiation induction, and LOR and FLG in late differentiation; Figure 1C).
These findings demonstrate that this differentiation technique is an easy-to-use and reliable method to study keratinocyte differentiation. Furthermore, this work on keratinocytes derived from p63 mutant keratinocytes also show that the method can be used to study affected keratinocyte differentiation under diseased conditions. In this in vitro differentiation approach, KGM purchased from Lonza is used, as this medium yields consistent results. In principle, other epidermal medium with similar composition such as keratinocyte serum-free medium (KSFM) from Thermo Fisher Scientific is also likely suitable, although this needs to be tested.
It should be noted that this method has some limitations. As it is based on contact inhibition, the confluency of cell density is required. In our experiments with p63 mutant keratinocytes, a higher initial seeding density has been necessary to ensure cells to reach confluency. In addition, when cells do not grow with the same speed, differentiation sometimes must be induced on different days. These considerations should be taken into account when setting up experiments. In experimental settings where cells need to be induced at the same time, other differentiation methods should be considered, including addition of serum, high concentrations of calcium, and inhibition of epidermal growth factor receptors2,3. Nevertheless, all in vitro differentiation methods have pros and cons, as they probably represent partial differentiation induction signals that are present during in vivo skin development2.
A comprehensive molecular analysis that compares these different methods will be highly informative and can instruct the choice of method most suitable for different studies on biological processes. In addition, validating changes measured at the transcriptomic level is highly advised, for example, via western blotting or proteomic analyses. Furthermore, in vitro data should be used with caution, and conclusions from these models should be validated in vivo, preferably in human skin development.
Basic principles of RNA extraction and RNA-seq library preparation have been well described previously24,25,26,27,28. In this protocol, the RNA extraction and RNA-seq library preparation procedures are based on workflows of commercially available kits. In principle, different methods for RNA extraction should not have a major influence on RNA-seq analyses if the RNA quality is good. In cases where RNA quality is poor (i.e., when RNA is extracted formalin-fixed paraffin-embedded tissues), RNA-seq can still be performed; however, the RNA fragmentation step should be adjusted. The RNA-seq library preparation covers from ribosomal RNA (rRNA) removal to the cDNA library construction for sequencing. The major procedures may be performed by using various kits, or using individual enzymes and homemade buffers29,30,31,32. It should be noted that, if RNA-seq analysis is performed using different basic principles (e.g., either ribosomal RNA depletion or polyA mRNA enrichment by hybridization to polyT oligos), the outcome of RNA-seq analyses may differ. Furthermore, the protocol utilizes ribosomal RNA removal by hybridization of DNA oligos to human, mouse, and rat rRNA. When working with different species, an alternative oligo set should be employed.
Sequencing of the generated library can be performed either on both ends of the fragments (paired end) or at one end of the fragment (single end). In general, paired end sequencing vastly improves mappability and provides more information regarding transcript variants. However, for a relatively simple differential gene analysis as described here, single end sequencing can also provide sufficient information.
For the important step of data analysis, a relatively simple method for quality control of the fastq files is described, followed by mapping reads to the genome. Even though the provided bash code does not have ideal scalability, it has the advantage of transparency. The choice of software, which steps it performs, and version of the genome used during data preprocessing are all nontrivial steps that should be well-documented, which is essential for the repeatability.
For more advanced users, an automated pipeline can be used to perform the steps of fastQ file quality check, adapter trimming and mapping, e.g. the ARMOR snakemake workflow33 or the ‘RNA-seq’ Snakemake workflow from the van Heeringen lab34. However, these completely automated pipelines are less transparent and more difficult to change. When using these tools, it is vital to understand the functions within these automated pipelines. Finally, the protocol includes RNA-seq data analysis with an emphasis on variable gene expression over time. Variable gene expression is preferred over pairwise differential testing when looking at processes with multiple timepoints, such as differentiation. In conclusion, this analysis pipeline introduces relevant tools for bioinformatic analysis of RNAseq data. It contains tools that can thoroughly assess keratinocyte differentiation, both under normal and diseased conditions.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This research was supported by Netherlands Organisation for Scientific Research (NWO/ALW/MEERVOUD/836.12.010, H.Z.) (NWO/ALW/Open Competition/ALWOP 376, H.Z., J.G.A.S.); Radboud University fellowship (H.Z.); and Chinese Scholarship Council grant 201406330059 (J.Q.).
Materials
| Bioanalyzer 2100 | Agilent | G2929BA | |
| Bovine pituitary extract (BPE) | Lonza | Part of the bulletKit | |
| CFX96 Real-Time system | Bio-Rad | qPCR machine | |
| Dulbecco's Phosphate-Buffered Saline (DPBS) | Sigma-Aldrich | D8537 | |
| Epidermal Growth Factor (EGF) | Lonza | Part of the bulletKit | |
| Ethanolamine >= 98% | Sigma-Aldrich | E9508 | |
| High Sensitivity DNA chips | Agilent | 5067-4626 | |
| Hydrocortison | Lonza | Part of the bulletKit | |
| Insulin | Lonza | Part of the bulletKit | |
| iQ SYBR Green Kit | BioRad | 170-8886 | |
| iScript cDNA synthesis | Bio rad | 1708890 | |
| KAPA Library Quant Kit | Roche | 07960255001 | Low concentration measure kit |
| KAPA RNA HyperPrep Kit with RiboErase | Roche | KK8540 | RNAseq kit |
| KGM Gold Keratinocyte Growth Medium BulletKit | Lonza | 192060 | |
| Nanodrop | deNovix | DS-11 FX (model) | Nanodrop and Qbit for DNA and RNA measurements |
| NEXTflex DNA barcodes -24 | Illumnia | NOVA-514103 | 6 bp long primers |
| Penicillin-Streptomycin | Gibco | 15140122 | |
| RNA Pico Chip | Agilent | 5067-1513 |
References
- Hardman, J. A. Skin equivalents for studying the secrets of skin: from development to disease. British Journal of Dermatology. 173 (2), 320-321 (2015).
- Borowiec, A. S., Delcourt, P., Dewailly, E., Bidaux, G. Optimal differentiation of in vitro keratinocytes requires multifactorial external control. PLoS One. 8 (10), 77507 (2013).
- Van Ruissen, F., et al. Induction of normal and psoriatic phenotypes in submerged keratinocyte cultures. Journal of Cellular Physiology. 168 (2), 442-452 (1996).
- Ojeh, N., Pastar, I., Tomic-Canic, M., Stojadinovic, O. Stem Cells in Skin Regeneration, Wound Healing, and Their Clinical Applications. International Journal of Molecular Sciences. 16 (10), 25476-25501 (2015).
- Ali, N., et al. Skin equivalents: skin from reconstructions as models to study skin development and diseases. British Journal of Dermatology. 173 (2), 391-403 (2015).
- Luis, N. M., et al. Regulation of human epidermal stem cell proliferation and senescence requires polycomb- dependent and -independent functions of Cbx4. Cell Stem Cell. 9 (3), 233-246 (2011).
- Mulder, K. W., et al. Diverse epigenetic strategies interact to control epidermal differentiation. Nature Cell Biology. 14 (7), 753-763 (2012).
- Poumay, Y., Pittelkow, M. R. Cell density and culture factors regulate keratinocyte commitment to differentiation and expression of suprabasal K1/K10 keratins. Journal of Investigative Dermatology. 104 (2), 271-276 (1995).
- Kouwenhoven, E. N., et al. Transcription factor p63 bookmarks and regulates dynamic enhancers during epidermal differentiation. EMBO Rep. 16 (7), 863-878 (2015).
- Qu, J., et al. Mutant p63 Affects Epidermal Cell Identity through Rewiring the Enhancer Landscape. Cell Reports. 25 (12), 3490-3503 (2018).
- Brunner, H. G., Hamel, B. C., Van Bokhoven, H. The p63 gene in EEC and other syndromes. Journal of Medical Genetics. 39 (6), 377-381 (2002).
- Browne, G., et al. Differential altered stability and transcriptional activity of DeltaNp63 mutants in distinct ectodermal dysplasias. Journal of Cell Science. 124, 2200-2207 (2011).
- . KGM Gold Keratinocyte Growth Medium BulletKit Available from: https://bioscience.lonza.com/Lonza_bs/CH/en/Culture-Media-and-Reagents/p/000000000000192060/KGM-Gold-Keratinocyte-Growth-Medium-BulletKit (2019)
- Doyle, K. a. M. . Protocols and applications guide. , (1996).
- . Hyperprep Kit Available from: https://sequencing.roche.com/en-us/products-solutions/by-category/Library-preparation/rna-library-preparation/kapa-rna-hyperprep-kit-with-riboerasehmr.html (2019)
- . TrimGalore Available from: https://github.com/FelixKrueger/TrimGalore (2019)
- Dobin, A., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29 (1), 15-21 (2013).
- Li, H., et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25 (16), 2078-2079 (2009).
- . Convert chromosome names in a bigWig file Available from: https://gist.github.com/dpryan79/39c70b4429dd4559d88fb079b8669721 (2019)
- Love, M. I., Huber, W., Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550 (2014).
- Eden, E., Navon, R., Steinfeld, I., Lipson, D., Yakhini, Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 10, 48 (2009).
- Yu, G., Wang, L. G., Han, Y., He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 16 (5), 284-287 (2012).
- Kretz, M., et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 493 (7431), 231-235 (2013).
- Mullegama, S. V., et al. Nucleic Acid Extraction from Human Biological Samples. Methods in Molecular Biology. 1897, 359-383 (2019).
- Ma, F., et al. A comparison between whole transcript and 3′ RNA sequencing methods using Kapa and Lexogen library preparation methods. BMC Genomics. 20 (1), 9 (2019).
- Chao, H. P., et al. Systematic evaluation of RNA-Seq preparation protocol performance. BMC Genomics. 20 (1), 571 (2019).
- Podnar, J., Deiderick, H., Huerta, G., Hunicke-Smith, S. Next-Generation Sequencing RNA-Seq Library Construction. Current Protocols in Molecular Biology. 106, 21 (2014).
- Haque, A., Engel, J., Teichmann, S. A., Lonnberg, T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Medicine. 9 (1), 75 (2017).
- Oyola, S. O., et al. Optimizing Illumina next-generation sequencing library preparation for extremely AT-biased genomes. BMC Genomics. 13, 1 (2012).
- Quail, M. A., et al. Optimal enzymes for amplifying sequencing libraries. Nature Methods. 9 (1), 10-11 (2011).
- Quail, M. A., et al. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 13, 341 (2012).
- Ross, M. G., et al. Characterizing and measuring bias in sequence data. Genome Biology. 14 (5), 51 (2013).
- . ARMOR Available from: https://github.com/csoneson/ARMOR (2019)
- . snakemake-workflows Available from: https://github.com/vanheeringen-lab/snakemake-workflows (2019)

