Introducing Point Mutations into Human Pluripotent Stem Cells Using Seamless Genome Editing
Summary
Here, we describe a detailed method for seamless gene editing in human pluripotent stem cells using a piggyBac-based donor plasmid and the Cas9 nickase mutant. Two point mutations were introduced into exon 8 of the hepatocyte nuclear factor 4 alpha (HNF4α) locus in human embryonic stem cells (hESCs).
Abstract
Custom designed endonucleases, such as RNA-guided Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-Cas9, enable efficient genome editing in mammalian cells. Here we describe detailed procedures to seamlessly genome edit the hepatocyte nuclear factor 4 alpha (HNF4α) locus as an example in human pluripotent stem cells. Combining a piggyBac-based donor plasmid and the CRISPR-Cas9 nickase mutant in a two-step genetic selection, we demonstrate correct and efficient targeting of the HNF4α locus.
Introduction
Human pluripotent stem cells (hPSCs) represent an unlimited source of somatic cells for research or the clinic1. Gene targeting in hPSCs offers a powerful method to study gene function during cell specification and to understand mechanisms of disease. Although efficient genome editing methods exist, modifying genes in hPSCs remains technically challenging. Standard gene targeting by homologous recombination in hPSCs occurs at low frequency or is even undetectable at some genes2, and DNA double-strand break (DSB) stimulated gene targeting (termed gene editing) is therefore necessary in these cells2,3. Additionally, transfection of hPSCs and subsequent single-cell cloning is not very efficient, even though single cell-associated apoptosis can be reduced through the use of the Rho-associated protein kinase (ROCK) inhibitor4. Finally, the potential on-target and off-target mutations at the gene of interest can also be problematic. Hence, a reliable protocol is essential to make tailored genetic changes in hPSCs.
The RNA-guided CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) and CRISPR associated protein 9 (Cas9) technology is now a well-established tool in gene editing. Transcribed CRISPR RNA (crRNA) and a trans-activating RNA (tracrRNA) form the single guide RNA (sgRNA), which complexes with Cas9 protein allowing gene specific cleavage5. The CRISPR/Cas9 system is programmable, by changing a 20-bp guide sequence of the sgRNA, meaning almost any locus that fulfils the protospacer-adjacent motif (PAM) requirement can be edited. However, the wild type Cas9 can tolerate some mismatches between its guide sequence and a DNA target, which might cause unwanted off-target effects. To improve its specificity, a nickase mutant (Cas9n) has been developed. A Cas9n containing D10A or N863A mutation only possesses one functional nuclease domain and as a result can only nick DNA on one strand. A pair of Cas9n appropriately spaced and oriented can effectively induce DNA DSB and the off-target effects are dramatically reduced6. To ensure efficient Cas9n modification, it has been shown that two Cas9n-sgRNA should ideally be placed with a -4 to 20 bp offset and always create a 5' overhang6.
Cas9(n)-sgRNA induced DSB can be utilized for genome editing in hPSCs7,8. It can create gene knockout via non-homologous end joining repair, or introduce gene modification through homology-directed repair (HDR) if a donor DNA template is present. The protocol described here uses a piggyBac transposon-based donor plasmid (or targeting vector) for HDR, in which a drug resistance marker is flanked by the transposon inverted repeats. The advantage of this approach includes efficient screening and seamless gene editing as firstly demonstrated by Yusa et al.9,10. The drug selection cassette allows enrichment of cells with the integrated vector, which are subsequently screened by junction PCR to identify those derived by HDR. In addition, the drug selection cassette can be positioned to replace the target sequences for Cas9(n) cleavage, so no further DNA breakage occurs after HDR, thus eliminating 'on' target mutations resulting from Cas9(n) re-cleavage. Furthermore, by exploiting the precise excision catalyzed by piggyBac transposase, the selection marker is then excised from the genome without leaving a scar. Only the original endogenous TTAA sequence for piggyBac insertion remains after the removal of the drug selection marker9. Even if the TTAA sites have to be created by introducing substitutions, the chances of disturbing regulatory elements are reduced compared to other methods10.
Here we describe detailed procedures for implementing seamless genome editing in hPSCs. Combining a piggyBac-based donor plasmid and the CRISPR-Cas9 nickase mutant in a two-step genetic selection, we introduced two predetermined point mutations into the hepatocyte nuclear factor 4 alpha (HNF4α) gene11. This approach is reliable and efficient, and has permitted in-depth analyses of the important roles played by HNF4α in endoderm and hepatocyte specification from human pluripotent stem cells11.
Protocol
1. Designing and constructing CRISPR/Cas9n-sgRNA expression plasmids
- Search for 20-bp guide sequences directly upstream of any 5'-NGG near the site to be modified. A pair of single-guide RNAs (sgRNAs) is needed when the Cas9 nickase (Cas9n) from Streptococcus pyogenes is used. The pair of sgRNAs must be able to generate 5' overhangs upon nicking with a -4 to 20 bp offset6.
- Order necessary oligos and pSpCas9n-2A-puro vector.
- Clone sgRNA into the pSpCas9n vector for co-expression with Cas9n following the published protocol12.
2. Designing and constructing a piggyBac-based targeting vector
- Download the target gene sequence and search for a TTAA site near the site to be modified.
NOTE: The distance between the TTAA site and the to-be-modified site should be as small as possible. If within coding sequence and no TTAA site is present within 100-bp distance, it is necessary to introduce one by making synonymous nucleotide substitutions. - Design homology arms (HAs) and incorporate desired point mutations in the HAs. The optimal length of HAs should be around 500 – 1200 bp.
NOTE: It is recommended to introduce synonymous nucleotide substitutions to mutate the PAM sequence in order to avoid potential Cas9n re-cleavage. - Construct the final targeting vector following an established protocol10.
NOTE: In the targeting vector used here, a PGK promoter driven puro-deltaTK selection cassette is flanked by transposon inverted repeats.
3. Genetic editing of human pluripotent stem cells
- Maintain human pluripotent stem cells (hPSCs) in a humidified incubator at 37 °C and 5% CO2 on recombinant laminin 521-coated surfaces (5 µg/mL) in mTeSR1 medium13. The cells should reach 70-85% confluency after 72 hours following a 1:3 split ratio.
NOTE: If present, remove spontaneously differentiated cells before daily medium change by gently aspirating them away. - On the day of transfection, coat enough wells of a 24-well plate with 300 µL of the laminin 521 solution (5 µg/mL) at 37 °C for at least 2 h.
- Remove the coating solution gently and add 300 µL of fresh mTeSR1 medium supplemented with 10 µM ROCK inhibitor to each well, and then put the plate back to the incubator to receive cells.
NOTE: Do not allow the plates to dry at any point when the laminin coating solution is removed. - Move the stock hPSCs from the incubator, remove spent medium, and then wash the cells once using 1 mL of sterile 1x DPBS.
- Add 1 mL of gentle cell dissociation reagent to each well of a 6-well plate and incubate at 37 °C for 6-8 min to dissociate the cells.
NOTE: Gently tap the plate and check whether the cells can detach easily to decide whether the digestion is long enough. - Gently pipette up and down using a P1000 tip to lift off all the hPSCs.
- Terminate the dissociation by adding 2 mL of fresh mTeSR1 medium supplemented with 10 µM ROCK inhibitor (Y-27632) to the cell suspension.
- Mix well and transfer the single-cell suspension to a 50 mL tube, and centrifuge at 200 x g for 3 min at room temperature.
- Remove the supernatant, and resuspend the cells well in 2 mL fresh mTeSR1 medium supplemented with 10 µM ROCK inhibitor.
- Count the viable cells using a hemocytometer. Use Trypan Blue to stain and exclude dead cells.
- Transfer 8 x 105 to 1 x 106 live cells to a 1.5 mL tube for each nucleofection reaction and centrifuge at 200 x g for 3 min. Then carefully aspirate the supernatant.
- Mix 3 µg of the paired Cas9n-sgRNA expression plasmids and 5 µg of targeting vector plasmid in 100 µL of mixed nucleofection solution/supplement from the human stem cell nucleofection kit. Use the GFP control plasmid from the kit and prepare a GFP control plasmid mix as well.
NOTE: It is important to make a maxi-prep of all plasmids to reduce endotoxin contamination before nucleofection. The concentration of the plasmids should be approximately 1 µg/µL. - Use the DNA mix (volume ~111 µL) to resuspend the prepared cells and transfer it to an electroporation cuvette (provided with the nucleofection kit), avoiding any bubbles.
- Electroporate the cells using the nucleofection device by selecting the optimized conditions for human pluripotent stem cells.
NOTE: If using the nucleofection kit for human pluripotent stem cells for the first time, it is necessary to test the electroporation program and choose the most efficient one. - Immediately add 500 µL of fresh and warm to 37 °C mTeSR1 medium supplemented with 10 µM ROCK inhibitor to the electroporated cells. Transfer the mix to 2 wells of a 24-well plate prepared from steps 3.2 and 3.3.
- Quickly put the plate back to the 37 °C/5% CO2 incubator and allow the cells to recover.
- 12-16 h later, change cell maintenance medium. If the cells established cell-cell contact, withdraw the ROCK inhibitor, if not, continue supplementing the medium with the inhibitor.
- Between 24-48 h, check the nucleofection efficiency by examining GFP expression in the control cells. GFP positive cells should be at least 30 percent.
- 48 h after nucleofection, start selecting cells by supplementing the mTeSR1 medium with 1 µg/mL puromycin.
- 72 h after nucleofection, supplement the mTeSR1 medium with 0.5 µg/mL puromycin. If the cell confluency is lower than 30%, also supplement the medium with 10 µM ROCK inhibitor.
- 4-6 days post-nucleofection, passage the puromycin-resistant cells to 10-15 x 96-well plates at the concentration of 0.8 cells/well.
NOTE: It is essential to supplement the medium with 10 µM ROCK inhibitor and 0.5 µg/mL puromycin. - Maintain those cells at 37 °C/10% CO2 for 10-12 days to form single cell-derived colonies. Top up medium once 7 days after seeding.
NOTE: Increased CO2 level helps single hPSCs to form colonies from our experience. - Mark wells containing a single colony and replace the medium with fresh mTeSR1 medium containing 0.5 µg/mL puromycin but no ROCK inhibitor.
NOTE: From this point on, the cells will be grown under 37 °C/5% CO2. - Two days later, change medium for wells containing undifferentiated colonies. Supplement the medium with 10 µM ROCK inhibitor and 0.5 µg/mL puromycin.
NOTE: In some wells, the cells might have differentiated and need to be discarded. - Use P2 pipette tips to scrape off the colonies gently. Transfer the cell suspension derived from one colony into 2 new wells on separate 96-well plates, and use one for genotyping and one for maintaining.
NOTE: It is important to carefully maintain the colonies and keep the spontaneous differentiation level to the minimum. - Take the plate containing cells for genotyping out once the confluency in most wells reached 50% or above. Dump the spent medium and then wash the cells once with DPBS.
- Lyse the cells in well using Bradley lysis buffer and isolate genomic DNA from each well in the plate following associated protocol (https://mcmanuslab.ucsf.edu/protocol/dna-isolation-es-cells-96-well-plate).
- Use a three-primer junction PCR method to genotype both left and right homology arms independently10.
NOTE: A scheme showing the PCR method is presented in Figure 2. - Send the junction PCR products for both homology arms from 4-5 colonies for Sanger sequencing and get the sequences.
NOTE: The sequencing result of 2 established colonies is shown in Figure 3A. - Keep colonies with the correct genotype and discard the rest.
- Expand the correct colonies under continuous puromycin selection and freeze them at the earliest possible passage.
4. Removing transposon from targeted human pluripotent stem cells
- Maintain one colony with correct genotype under 0.5 µg/mL puromycin selection.
- Nucleofect 8 x 105 to 1 x 106 cells with 5 µg hyperactive transposase (pCMV-hyPBase) as described above (Section 3, steps 3.4-3.18). Perform a GFP control nucleofection in parallel.
NOTE: Puromycin should be removed from the mTeSR1 medium immediately after nucleofecting these cells. - Grow and screen for cells with the puro-deltaTK selection cassette removed following published procedures10.
NOTE: It is important to follow the original procedures carefully. FIAU, or 1-(2-deoxy-2-fluoro-β-d-arabinofuranosyl)-5-iodouracil), was used as a thymidine analog for Herpes simplex virus-derived thymidine kinase (HSV-tk)-based negative selection. - Sequence the modified region to confirm the removal of the transposon.
NOTE: The sequencing result of 3 established colonies is shown in Figure 3B. - Keep colonies with the correct genotype and expand for further usage.
- Perform characterization of pluripotency markers in the chosen colonies before using them for further analysis.
NOTE: The characterization of two established colonies is shown in Figure 4.
Representative Results
Targeting vector-based knock-in strategy
The hepatocyte nuclear factor 4 alpha (HNF4α) gene was chosen for targeted genome editing to introduce two point mutations into exon 8. A pair of Cas9n-sgRNA with 11 bp offset was designed close to the site to be modified. The piggyBac-based targeting vector worked as the homology-directed repair template for introducing the desired point mutations. A 967 bp 5'-HA and a 1142 bp 3'-HA incorporating synonymous nucleotide substitutions or the desired point mutations were amplified and cloned into the final targeting vector. The piggyBac insertion site was 16 bp and 22 bp away from the two desired point mutations. Colonies containing the puro-deltaTK selection cassette were selected with puromycin in the first round. Once the selection cassette was removed by transposase excision in the second round, the targeted site was modified seamlessly with only the desired point mutations incorporated in the gene (Figure 1).
Genetic editing in human pluripotent stem cells
To screen for correctly targeted cells, a three primer-based PCR method was used for genotyping (Figure 2). Sanger sequencing was performed to confirm the PCR results (Figure 3A). Post removal of the selection cassette, the modified region was sequenced again to confirm the correct introduction of desired point mutations (Figure 3B).
Colony establishment and characterization of edited human pluripotent stem cells
Colonies with the correct genotype were selected and expanded as needed. The established colonies need to be characterized before being used for further analysis. The edited cells possess the same morphology as the parental cells (Figure 4A). They also express representative human pluripotent stem cell markers, including transcription factors NANOG and OCT4 (Figure 4B), as well as cell surface markers SSEA4 and TRA-1-60 (Figure 4C).
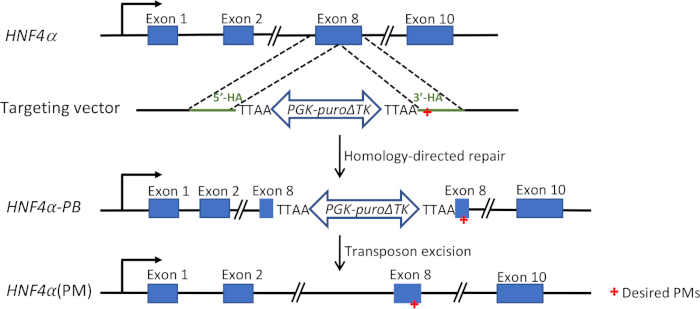
Figure 1: Targeting vector-based knock-in strategy11. A pair of Cas9n-sgRNA expression plasmids were used to induce DNA double-strand break in exon 8 of the HNF4α gene. A targeting vector with a selection cassette was used to introduce predetermined point mutations located 16 bp and 22 bp away. This selection cassette was contained within the piggyBac transposon, consisting of a positive-negative selection marker (puro-deltaTK) driven by a constitutively active promoter (PGK). Via homology-directed repair pathway, the targeted cells incorporated the selection cassette. Transposon excision mediated by transposase results in a seamless modification with only the point mutations present. Red cross indicates the location of desired point mutations. HA = homology arm; PB = piggyBac; PM = point mutation. Please click here to view a larger version of this figure.
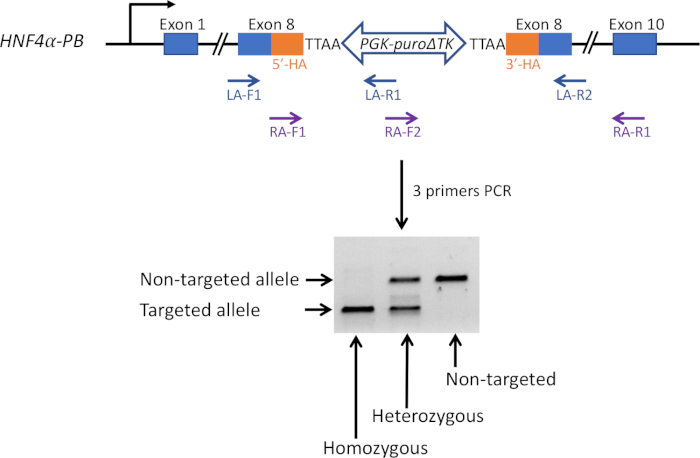
Figure 2: Three primer-based PCR method. To screen gene targeted cells, PCR-based genotyping was used. Three primers, LA-F1, -R1 and -R2 were used to amplify the left homology arm region. Independently, RA-F1, -F2 and -R1 were used to amplify the right homology arm region. Based on the gel electrophoresis result, non-targeted cells, and heterozygous and homozygous cells, were distinguished from each other. Please click here to view a larger version of this figure.
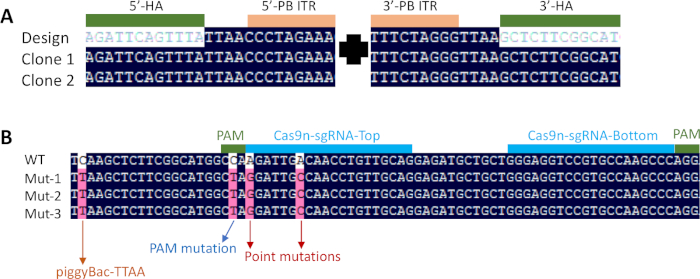
Figure 3: Sequencing results of genetically modified cells11. PCR products from two clones were sequenced and confirmed the correct insertion of the selection cassette at the targeted locus (A). 5' and 3' piggyBac inverted terminal repeats (ITR) were flanked by the TTAA direct repeats. Three clones were sequenced post transposon excision (B). A pair of Cas9n-sgRNA with 11 bp offset was used to introduce DNA double-strand break. Two predetermined point mutations (A to G and A to C) were introduced into the gene. One synonymous mutation was introduced to mutate the protospacer-adjacent motif (PAM), and another one to create the TTAA site necessary for piggyBac excision. Please click here to view a larger version of this figure.
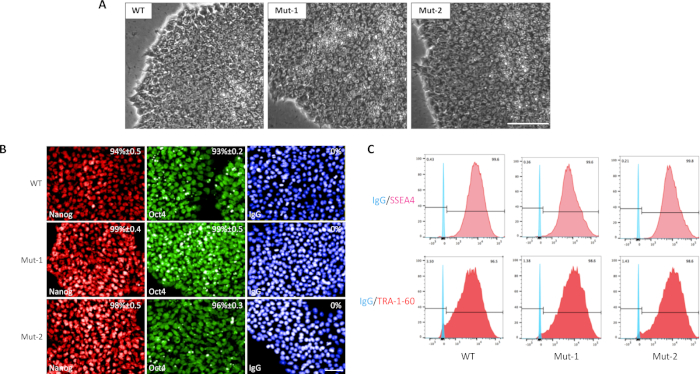
Figure 4: Characterization of edited human pluripotent stem cells. Morphology of two edited cell lines and the parental cells (A), scale bar = 100 µm. The expression of pluripotent stem cell markers NANOG and OCT4 examined by immunostaining (B, scale bar = 50 µm), in addition to SSEA4 and TRA-1-60 by flow cytometry (C) in two modified cell lines and the parental cells. IgG was used as a negative control. DAPI was used to stain the nucleus. The percentages were calculated as the average of three independent experiments. Please click here to view a larger version of this figure.
Discussion
The protocol described herein can be used to introduce predetermined point mutations into an endogenous locus in hPSCs. The combination of a piggyBac-based targeting vector and paired CRISPR/Cas9n expression plasmids proved to be reliable and efficient11. Following HNF4α gene editing, 12 out of 43 analyzed clones were correctly targeted as determined by junction PCR screening. Specifically, the biallelic targeting efficiency was about 21% (9/43) and the monoallelic targeting efficiency was around 7% (3/43).
Following targeting experiments, it is crucial to maintain puromycin selection to keep the targeted locus accessible to the piggyBac transposase during the first round of screening. This will minimize the silencing of the PGK promoter-driven puro-deltaTK selection cassette and reduce the background in the second round of screening10. A recent report showed that CAG promoter is more resistant to silencing than PGK promoter in hPSCs14. Changing the promoter for the selection cassette could therefore improve this system in the future. It is also important to compare the number of resistant colonies from hyPBase- and GFP-transfected cells. Upon successful removal of the transposon, there should be more surviving colonies from the hyPBase- than GFP-transfected cells. In addition to PCR analysis of transposon excision, direct sequencing of the modified region is required to confirm the excision fidelity.
Based on Yusa's piggyBac transposon-based targeting vector strategy9,10, the procedures detailed above focused on improving hPSCs nucleofection and single cell cloning efficiency. Cell seeding density was optimized to reduce the likelihood of heterogeneous colony formation. We also employed 10-12 days culture under 10% CO2 to enhance colony production for genotype screening. In our experience, around 20 colonies could be obtained from each 96-well plate. Although this step might take more time than other methods, it is reliable and highly efficient.
Based on the need, targeting vectors can be designed to introduce or correct point mutations, to create reporter cell lines, as well as knockout gene expression. In conclusion, the combination of the CRISPR/Cas9(n) system and a targeting vector is an efficient way to deliver different types of genetically modified hPSCs.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Kosuke Yusa for sharing the pMCS-AAT_PB-PGKpuroTK and pCMV-hyPBase plasmids. We thank Jia-Yin Yang and Karamjit Singh-Dolt for helpful discussions on genome editing. D.C.H. lab is supported by an award from the Chief Scientist Office (TC/16/37) and the UK Regenerative Medicine Platform (MR/L022974/1). Y.W. was supported by a PhD scholarship funded by the Chinese Scholarship Council and the University of Edinburgh.
Materials
| Anti-Human SSEA4 PE | eBioscience | 12-8843-42 | |
| Anti-Human TRA-1-60 PE | eBioscience | 11-0159-42 | |
| DPBS | Thermo Fisher Scientific | 14190144 | |
| DPBS with calcium and magnesium | Thermo Fisher Scientific | 14040133 | |
| FIAU | Moravek | M251 | |
| Gentle cell dissociation reagent | Stem Cell Technologies | 07174 | |
| H9 human embryonic stem cells | WiCell | WA09 | |
| Human NANOG antibody | R&D systems | AF1997 | |
| Human OCT4 antibody | Abcam | AB19857 | |
| Human stem cell Nucleofector kit 1 | Lonza | VPH-5012 | |
| Mouse IgG3 isotype control PE | eBioscience | 12-4742-41 | |
| mTeSR1 medium | Stem Cell Technologies | 85850 | |
| Nucleofector 2b device | Lonza | AAB-1001 | |
| pCMV-hyPBase | Wellcome Trust Sanger Institute | N/A | |
| pMCS-AAT_PBPGKpuroTK | Wellcome Trust Sanger Institute | N/A | |
| pSpCas9n(BB)-2A-Puro (PX462) | Addgene | PX462 | |
| Puromycin dihydrochloride | Thermo Fisher Scientific | A11138-03 | |
| QIAprep spin maxiprep kit | Qiagen | 12162 | |
| QIAprep spin miniprep kit | Qiagen | 27106 | |
| Recombinant laminin 521 | BioLamina | LN521 | |
| Trypan blue solution, 0.4% | Thermo Fisher Scientific | 15250061 | |
| Y-27632(Dihydrochloride) | Millipore | SCM075 |
References
- Rashidi, H., et al. 3D human liver tissue from pluripotent stem cells displays stable phenotype in vitro and supports compromised liver function in vivo. Archives of Toxicology. 92 (10), 3117-3129 (2018).
- Hockemeyer, D., et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nature Biotechnology. 27 (9), 851-857 (2009).
- Porteus, M. H., Baltimore, D. Chimeric nucleases stimulate gene targeting in human cells. Science. 300 (5620), 763 (2003).
- Watanabe, K., et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nature Biotechnology. 25 (6), 681-686 (2007).
- Jinek, M., et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 337, 816-822 (2012).
- Ran, F. A., et al. Double nicking by RNA-guided CRISPR cas9 for enhanced genome editing specificity. Cell. 154 (6), 1380-1389 (2013).
- Cong, L., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 339 (6121), 819-824 (2013).
- Mali, P., et al. RNA-guided human genome engineering via Cas9. Science. 339 (6121), 823-826 (2013).
- Yusa, K., et al. Targeted gene correction of α1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 478 (7369), 391-394 (2011).
- Yusa, K. Seamless genome editing in human pluripotent stem cells using custom endonuclease-based gene targeting and the piggyBac transposon. Nature Protocols. 8 (10), 2061-2078 (2013).
- Wang, Y., et al. Multiomics Analyses of HNF4α Protein Domain Function during Human Pluripotent Stem Cell Differentiation. iScience. 16, 206-217 (2019).
- Ran, F. A., Hsu, P. D., Wright, J., Agarwala, V., Scott, D. A., Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nature Protocols. 8 (11), 2281-2308 (2013).
- Wang, Y., et al. Defined and Scalable Generation of Hepatocyte-like Cells from Human Pluripotent Stem Cells. Journal of Visualized Experiments. (121), 1-8 (2017).
- Eggenschwiler, R., et al. Improved bi-allelic modification of a transcriptionally silent locus in patient-derived iPSC by Cas9 nickase. Scientific Reports. 6, 1-14 (2016).

