Protein Extract Preparation and Co-immunoprecipitation from Caenorhabditis elegans
Summary
This method describes a protocol for high-throughput protein extract preparation from Caenorhabditis elegans samples and subsequent co-immunoprecipitation.
Abstract
Co-immunoprecipitation methods are frequently used to study protein-protein interactions. Confirmation of hypothesized protein-protein interactions or identification of new ones can provide invaluable information about the function of a protein of interest. Some of the traditional methods for extract preparation frequently require labor-intensive and time-consuming techniques. Here, a modified extract preparation protocol using a bead mill homogenizer and metal beads is described as a rapid alternative to traditional protein preparation methods. This extract preparation method is compatible with downstream co-immunoprecipitation studies. As an example, the method was used to successfully co-immunoprecipitate C. elegans microRNA Argonaute ALG-1 and two known ALG-1 interactors: AIN-1, and HRPK-1. This protocol includes descriptions of animal sample collection, extract preparation, extract clarification, and protein immunoprecipitation. The described protocol can be adapted to test for interactions between any two or more endogenous, endogenously tagged, or overexpressed C. elegans proteins in a variety of genetic backgrounds.
Introduction
Identifying the macromolecular interactions of a protein of interest can be key to learning more about its function. Immunoprecipitation and co-immunoprecipitation experiments can be used to identify the entire interactome of a protein through large-scale proteomic approaches1 or to specifically test a protein’s ability to coprecipitate with a hypothesized interactor. In C. elegans, both methods have been successfully employed to learn more about the activity of a variety of proteins, including those that closely function with microRNAs to regulate gene expression2,3,4. Co-immunoprecipitation experiments have the advantage of testing the protein-protein interactions in their native cellular environment, but extract preparation can be challenging and time-consuming. Efficient lysis of the sample is necessary, but care must be taken to minimize the disruption of protein-protein interactions. Methods such as douncing5, sonication6, Balch homogenization7, and zirconia beads-homogenization8,9 have been used to successfully prepare C. elegans total protein extracts. These methods, with the exception of zirconia bead homogenization, have limitations in terms of the number of samples that can be processed simultaneously. Presented is an alternative method that can be easily scaled up to allow for high-throughput, rapid protein extract preparation from C. elegans samples followed by co-immunoprecipitation. Specifically, the method can prepare up to 24 samples at a time, greatly reducing the time required for extract preparation. By contrast, for example, douncing typically allows for only one sample preparation at a time. This extract method can be used to prepare extracts from any developmental stage of C. elegans.
Described is a step-by-step procedure for animal sample collection, extract preparation, immunoprecipitation, and presentation of Western blotting data to confirm successful protein pulldown and detection of the co-immunoprecipitating protein of interest. To demonstrate the effectiveness of the protocol, two co-immunoprecipitation experiments were performed between 1) microRNA Argonaute ALG-1 and AIN-1, a GW182 homolog; and 2) ALG-1 and HRPK-1, a newly identified ALG-1 interactor2. ALG-1 and AIN-1 are core proteins that comprise the microRNA-induced silencing complex (miRISC) and the interaction between these two proteins is well established10,11. The extract preparation protocol was effective in the ALG-1-AIN-1 co-immunoprecipitation experiment. This protocol also successfully confirmed the interaction between ALG-1 and its newly identified interactor, HRPK-12.
In summary, the manuscript describes a C. elegans extract preparation protocol that can be scaled up to simultaneously process 24 samples along with a co-immunoprecipitation protocol that can be used to identify new or confirm hypothesized interactions between proteins. The extract preparation protocol is compatible with a number of downstream experiments, including protein immunoprecipitation2 and microRNA pulldowns12. Furthermore, the immunoprecipitation protocol can be adapted to test for interactions between any two or more endogenous, endogenously tagged, or overexpressed C. elegans proteins in a variety of genetic backgrounds.
Protocol
1. Worm sample collection
- Seed mixed stage or synchronized13 worms on NGM solid plates at the required temperature and allow the worms to grow until the desired stage. For basic C. elegans growth and maintenance, please see Stiernagle et al. and Porta-de-la-Riva et al.14,13.
- Collect worms in a 15 mL conical centrifuge tube by washing the worm plates with M9 buffer.
- Pellet the worms by centrifuging at 400 x g at room temperature (RT) for 2 min and discard the supernatant.
NOTE: The worm pellet size for extract preparation is between 100 µL and 500 µL. A 300 µL pellet of packed worms is recommended for downstream immunoprecipitation experiments and typically yields ~4.5 mg of total protein, while a 500 µL pellet will yield ~7.5 mg of total protein. - Perform additional 3–5 washes with M9 buffer (see Table 1) or until the supernatant is no longer cloudy.
- Perform one final wash with ddH2O.
- Move the loose worm pellet to a 1.5 mL microcentrifuge tube and spin down at 400 x g at RT for 2 min. Discard the remaining supernatant to obtain a packed worm pellet and proceed to extract preparation.
NOTE: The protocol can be paused here. Worm pellets may be flash frozen in liquid nitrogen immediately and stored at -80 °C or in liquid nitrogen. Please note that worm pellets can only be thawed once and cannot be refrozen.
2. Extract preparation of the worm pellet
NOTE: The extract preparation should be performed on ice or at 4 °C.
- If frozen, thaw worm pellet on ice.
NOTE: If the desired packed worm pellet size of 300 µL was not obtained during sample collection, multiple smaller pellets can be combined until enough material is present for further extraction. - Add an equal volume of ice-cold 2x lysis buffer (60 mM HEPES, pH = 7.4, 100 mM potassium chloride, 0.1% Triton X, 4 mM magnesium chloride, 10% glycerol, 2 mM DTT with RNase inhibitor, protease inhibitor, and phosphatase inhibitors; see Table 1) and vortex or pipette up and down to mix. Spin the tube(s) down to collect the mixture at the bottom of the tube.
- Move the mixture into a 1.5 mL RNase-free tube containing metal beads (see Table of Materials) and put the sample in the bead mill homogenizer (see Table of Materials) at 4 °C. Ensure that the tube caps are tightened and the samples are balanced inside the homogenizer.
- Homogenize the sample at the highest speed (setting 12) for 4 min.
- Remove the sample from the beads and place it into a new 1.5 mL microcentrifuge tube. Alternatively, a magnet provided with the homogenizer can be used to remove the beads from the sample.
- Spin down the extract at 19,000 x g for 20 min at 4 °C to clarify the protein extract.
- Transfer the supernatant into a fresh 1.5 mL tube on ice, while avoiding carry-over of the white, cloudy precipitate that forms on top of the sample. The supernatant is now the clarified extract.
NOTE: Save 10 µL of the clarified extract to determine the total protein concentration. - Use the extract immediately for the following experiments or flash freeze the extract in liquid nitrogen and store at -80 °C.
NOTE: The protocol may be paused here. Extracts may be stored at ultralow temperature (-80 °C freezer or liquid nitrogen for ~6 months). Frozen extracts may be thawed once and cannot be refrozen. - Determine the total protein concentration of the extract using a protein concentration assay kit compatible with detergents (see Table of Materials) according to the manufacturer’s instructions.
3. Immunoprecipitation
NOTE: All the immunoprecipitation steps for extract preparation should be performed on ice or at 4 °C. It is recommended to use 2 mg of total protein for each immunoprecipitation. However, successful immunoprecipitations with 0.8–1 mg of total protein have been performed. Always use fresh or freshly thawed protein extracts. The following protocol is outlined to perform immunoprecipitation from 2 mg of total protein, or a single immunoprecipitation experiment. The amount of beads and antibody may be increased or decreased accordingly for multiple samples or if a different amount of protein extract is used.
- Place or thaw the protein extract on ice. The sample can be diluted to 10 mg/mL or 5 mg/mL with ice-cold 1x lysis buffer (see Table 1).
- Resuspend the magnetic beads by inversion and transfer 150 µL of the 50% beads suspension into a 1.5 mL tube. Magnetize the beads on ice against a magnetic stand for 1 min or until the solution is clear. Discard the supernatant.
- Remove the tube from the magnetic stand and wash the beads in 1x lysis buffer using 2 volumes (i.e., 300 µL) of bead slurry. Repeat the wash 2x for a total of three washes.
- Resuspend the beads in 150 µL of ice-cold lysis buffer.
- Transfer 75 µL of the bead slurry to 2 mg of protein extract and incubate at 4 °C for 1 h with gentle agitation. Save the remaining bead suspension on ice for later use.
NOTE: This step is performed to reduce nonspecific protein binding to beads during the immunoprecipitation step. - Place the tube with sample on the magnetic stand on ice for 1 min or until the beads are fully magnetized and the sample is clear. Transfer the supernatant to a new 1.5 mL tube; do not disturb the beads. This is the precleared protein lysate. Save 10% of the sample for Western blot analysis.
- Add 20 µg of affinity purified antibody to the precleared lysate and incubate at 4 °C for 1 h with gentle agitation.
NOTE: The amount of antibody used for immunoprecipitation is specific to the antibody and protein and should be empirically determined to ensure effective immunoprecipitation of the target protein. - Add the remaining 75 µL of the prewashed beads suspension (step 3.5) to the antibody/lysate mixture and incubate for 1 h at 4 °C with gentle agitation.
- Place the tube in the magnetic stand on ice for 1 min or until the beads are fully magnetized and the sample is clear. Save the supernatant for Western blot analysis (optional).
- Wash the beads containing the immunoprecipitate 3x in 450 µL of wash buffer (30 mM HEPES, pH = 7.4, 100 mM potassium chloride, 0.1% Triton X, 2 mM magnesium chloride, 10% glycerol, 1 mM DTT; see Table 1) on ice.
NOTE: Additional washes may be performed if more stringent washing conditions are preferred. - Resuspend the bead pellet in 20 µL of 2x SDS/BME protein gel loading buffer (see Table of Materials) and denature by boiling at 95 °C for 5 min prior to loading onto an SDS-PAGE gel. Alternatively, denatured samples can be stored at -20 °C for several months.
NOTE: A portion of the bead immunoprecipitate can be saved for downstream RNA isolation, if desired.
4. Western blot detection of IP samples
- Load the IP samples onto the SDS-PAGE gel (see Table of Materials). Avoid transferring the beads by placing the IP tubes on the magnetic stand for 1 min prior to aspiration of the sample.
- Perform the Western blotting15,16 and antibody staining16 with the following modifications: dilute the ALG-1 antibody17 1:500 in 5% non-fat dry milk (NFDM); dilute the HRPK-1 antibody2 1:1,000 in 5% NFDM; and dilute the AIN-1 antibody18 1:10,000 in 5% NFDM. Secondary antibodies (see Table of Materials) were used according to the manufacturer’s instructions.
- Detect the bands with HRP-based chemiluminescence (see Table of Materials).
Representative Results
This protocol (schematized in Figure 1) was used successfully to obtain C. elegans total protein extracts (Figure 2) for downstream immunoprecipitation of several proteins2 (Figure 3 and Figure 4). The presented bead mill homogenizer protocol was comparable in total protein extraction to dounce-based methods (Figure 2) and efficiently extracted nuclear (COL-19::GFP(NLS) (Figure 2) and cytoplasmic proteins (Figure 3 and Figure 4). Multiple samples of various sizes were extracted simultaneously (Figure 2). Argonaute proteins interact with members of the GW182 protein family, forming the miRISCs that bind to the target messenger RNAs and repress their expression10. Figure 3 shows successful co-immunoprecipitation of core miRISC components ALG-1 and AIN-1, consistent with previous reports11,17. More recently, efforts were made to identify additional protein interactors of Argonaute ALG-13 in order to learn more about how microRNA biogenesis and activity might be regulated by auxiliary factors. The RNA-binding protein HRPK-1 was identified in ALG-1 immunoprecipitates3. This interaction was recently confirmed in a reciprocal HRPK-1 immunoprecipitation experiment2. The presented extract and immunoprecipitation protocols successfully recovered ALG-1 in HRPK-1-specific co-immunoprecipitates (Figure 4). In addition, the ALG-1—AIN-1 interaction was tested in a variety of genetic backgrounds and HRPK-1 was shown to be unnecessary for the ALG-1/AIN-1 miRISC assembly2 (Figure 3). Supplemental figures are provided to show the full membrane probed (Supplemental Figure 1).
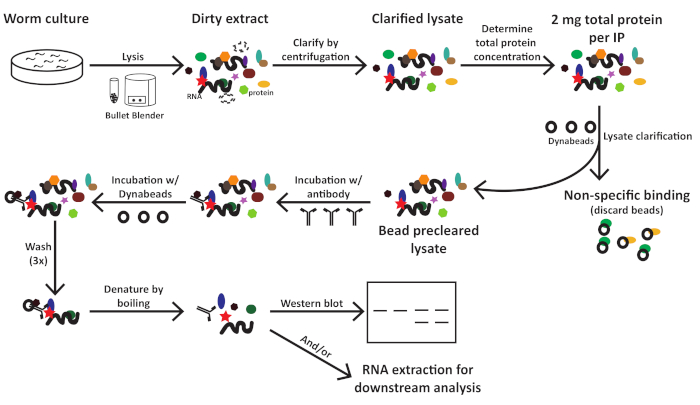
Figure 1: Workflow schematic for C. elegans extract preparation and immunoprecipitation. Please click here to view a larger version of this figure.
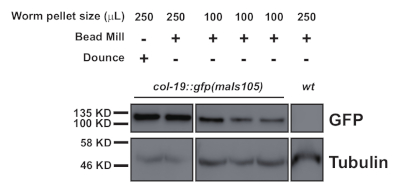
Figure 2: Western blot comparison of a nuclear localized GFP, COL-19::GFP(NLS), levels in dounce-prepared and homogenized samples from 250 µL and 100 µL worm pellets. Please click here to view a larger version of this figure.
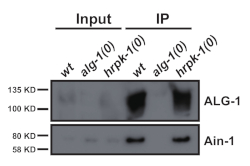
Figure 3: GW182 homolog AIN-1 co-immunoprecipitates with ALG-1. Western blotting for ALG-1 and AIN-1 proteins in ALG-1 immunoprecipitates. The ALG-1/AIN-1 co-immunoprecipitation was not affected by the absence of hrpk-1. Input = 10% of IP. Please click here to view a larger version of this figure.

Figure 4: ALG-1 co-immunoprecipitates with HRPK-1. Western blotting for HRPK-1 and ALG-1 in HRPK-1 immunoprecipitates is shown. Input = 10% of IP. * indicates antibody heavy chain. Please click here to view a larger version of this figure.
| M9 buffer (1 L) | |
| KH2PO4 | 3 g |
| Na2HPO4 | 6 g |
| NaCl | 5 g |
| 1 M MgSO4 | 1 mL |
| ddH2O | up to 1 L |
| 2x Lysis buffer (5 mL) | |
| HEPES (pH 7.4) | 200 µL |
| 2 M KCl | 250 µL |
| 10% TritonX | 100 µL |
| 1 M MgCl2 | 20 µL |
| 100% glycerol | 1 mL |
| ddH2O | up to 5 mL |
| Add fresh: | |
| 1 M DTT | 20 µL |
| EDTA-free protease inhibitor | 1 tablet |
| phosphatase inhibitor cocktail 2 | 100 µL |
| phosphatase inhibitor cocktail 3 | 100 µL |
| 1x Lysis buffer | |
| Dilute 2x Lysis buffer with an equal volume of ddH20. | |
| 1x Wash buffer 10 mL) | |
| HEPES (pH 7.4) | 300 µL |
| 2 M KCl | 500 µL |
| 10% TritonX | 100 µL |
| 1 M MgCl2 | 20 µL |
| 100% glycerol | 1 mL |
| ddH2O | up to 10 mL |
| 1 M DTT | 20 µL (add fresh) |
Table 1: Recipes
Supplemental Figure 1. Full probed Western blot membranes used to generate Figures 2-4 are shown. (A) Probed membrane for Figure 2. Note that membrane was cut to allow for simultaneous probing for GFP and Tubulin, reducing the overall blot size. (B) Probed membrane for Figure 3. (C) Probed membrane for Figure 3. *denotes antibody heavy chain. Please click here to view a larger version of this figure.
Discussion
C. elegans is an excellent model for studying fundamental questions in cell, molecular, and developmental biology19. In addition to its power as a genetic model system, C. elegans is amenable to biochemical approaches, including, but not limited to, protein immunoprecipitation and co-immunoprecipitation. One potential hurdle when conducting immunoprecipitation experiments is lack of antibodies specific to the proteins of interest. If no antibody is available, custom polyclonal or monoclonal antibodies can be generated. However, recent innovations in genome editing technology have allowed researchers to rapidly introduce mutations or tag endogenous C. elegans genes20,21, facilitating studies that unravel the genetic, functional, and physical interactions among the genes and the encoded proteins. Specifically, CRISPR/Cas9-mediated tagging of C. elegans genes at the endogenous loci has reduced the dependence of immunoprecipitation experiments on antibody availability, making co-immunoprecipitation experiments much more feasible. C. elegans genes can be tagged with a variety of tags ranging from fluorescent tags such as GFP or mCherry to small tags such as FLAG and HA. Antibodies recognizing these tags are readily available commercially, facilitating the studies of protein-protein interactions via immunoprecipitation approaches.
The presented protocol, outlined in Figure 1, can be performed for a small number of samples or scaled up, allowing for up to 24 sample preparations at a time. While the initial characterizations of protein-protein interactions via immunoprecipitation are typically done in wild type backgrounds under normal growing conditions, follow-up studies frequently necessitate testing the protein-protein interactions in a variety of genetic backgrounds or under different growth conditions. The ability to simultaneously prepare multiple extracts saves time and, importantly, ensures extract preparation consistency among the different samples. A negative control is always required, with the ideal control being a null mutation in the gene encoding for the immunoprecipitated protein of interest (See Figure 3 and Figure 4 for examples).
This extract protocol allows for rapid protein extract preparation from C. elegans samples and is comparable to zirconium bead-based homogenization8. Bead homogenization in general can be scaled up to multiple simultaneous sample preparations using a variety of bead mill homogenizers or similar equipment. Some more economical bead mill homogenizers may reduce the number of samples that can be processed simultaneously, however. Alternatively, the presented extract protocol is compatible with dounce-based extract preparation, which represents an economical alternative. While different bead mill homogenizers were not tested, most are likely to be compatible with this protein extract protocol, as long as complete disruption of the C. elegans samples is achieved.
As presented, this extract preparation protocol is compatible with multiple downstream experiments, including protein immunoprecipitation2 and microRNA pull-down12 and allows for downstream collection of both protein and RNA components. It also efficiently extracts both nuclear and cytoplasmic proteins (Figure 2, Figure 3, and Figure 4). Similarly, the presented immunoprecipitation protocol permits RNA isolation from protein-associated immunoprecipitates. While the immunoprecipitation protocol was originally developed to identify ALG-1 protein interactors, the method can be adapted to test for interactions between any proteins of interest. In fact, the immunoprecipitation conditions used worked equally well for immunoprecipitating ALG-1 (Figure 3) and HRPK-1 (Figure 4). This protocol is an excellent starting point for immunopurification of RNA-binding proteins. It should be noted, however, that some changes in buffer composition may be required for other proteins of interest. The changes may depend on the physical and biochemical properties of the protein of interest and must be implemented on a case-by-case basis.
Once the target protein (here, ALG-1 or HRPK-1) is immunoprecipitated, Western blotting can be used to test the co-immunoprecipitate for specific protein interactors.
Alternatively, the copurified immunoprecipitate can be subjected to mass spectrometry analysis to identify all the putative interacting proteins. Confirmed co-immunoprecipitation interactions can then be examined in a variety of genetic backgrounds or conditions to identify potential regulation of the specific interaction. For example, to determine whether hrpk-1 plays a role in ALG-1/AIN-1 miRISC assembly, ALG-1-AIN-1 coprecipitation was assessed both in a wild type background and in the absence of HRPK-1 (Figure 3). hrpk-1 was found to be dispensable for ALG-1/AIN-1 interaction2 (Figure 3). In addition, CRISPR/Cas9 genome editing technology can be employed to generate single point or domain deletion mutations in the proteins of interest. Retesting the ability of the generated mutants to coprecipitate with their protein interactors can reveal which domains or residues mediate the physical interaction. Such future studies can yield invaluable information about the mechanism of protein function and regulation. These approaches, combined with the power of C. elegans genetics, can provide important insights into the fundamental molecular processes that govern animal development and cellular function.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was in part supported by Kansas INBRE, P20GM103418 to Li and Zinovyeva and R35GM124828 to Zinovyeva. We thank Min Han for generously sharing the anti-AIN-1 antibody. Some of the strains used in the course of this work were provided by the Caenorhabditis Genetics Center (CGC), funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Materials
| 15 mL tube | VWR | 89039-664 | STEP 1.2 |
| 2x Laemmli Sample Buffer | BioRed | 1610737 | STEP 3.11 |
| 4–20% Mini-PROTEAN TGX Precast Protein Gels | BioRed | 4561096 | STEP 4.1 |
| anti-AIN-1 monoclonal antibody | custom generated | n/a | STEP 4.2, see ref. Zhang et al. 2007 |
| anti-ALG-1 monoclonal antibody | custom generated by PRF&L | n/a | STEP 4.2 |
| anti-HRPK-1 monoclonal antibody | custom generated by PRF&L | n/a | STEP 4.2 |
| Bullet Blender Storm Homogenizer | MidSci | BBY24M | STEP 2.3 |
| DL-Dithiothreitol (DTT) | Sigma | D9779-5G | Table 1 |
| Dynabeads Protein A for Immunoprecipitation | Thermo Fisher | 10002D | STEP 3.2 |
| DynaMag-2 Magnet | Thermo Fisher | 12321D | STEP 3.2 |
| EDTA-free protease inhibitors | Roche | 11836170001 | Table 1 |
| GFP antibody (FL) | Santa Cruz Biotechnology | sc-8334 | Figure 2 |
| Glycerol | Thermo Fisher | G33-500 | Table 1 |
| Goat Anti-Rabbit Secondary Antibody, HRP | BioRed | 1662408 | STEP 4.2 |
| Goat anti-Rat IgG (H+L) Secondary Antibody, HRP | Thermo Fisher | 31470 | STEP 4.2 |
| HEPES | Sigma | H4034-500G | Table 1 |
| LICOR WesternSure PREMIUM Chemiluminescent Substrate, 100 mL Kit | LI-COR | 926-95000 | STEP 4.3 |
| Magnesium chloride hexahydrate ACS | VWR | VWRV0288-500G | Table 1 |
| Magnesium Sulfate Anhydrous | Thermo Fisher | M65-500 | Table 1 |
| Microcentrifuge Tubes, 1.5 mL | VWR | 20170-333 | STEP 1.6 |
| N2 wild type | CGC | ||
| Navy RINO RNA Lysis Kit 50 pack (1.5 mL) | MidSci | NAVYR1-RNA | STEP 2.3 |
| Phosphatase inhibitor cocktail 2 | Sigma | P5726-1ML | Table 1 |
| Phosphatase inhibitor cocktail 3 | Sigma | P0044-1ML | Table 1 |
| Potassium Chloride | Thermo Fisher | P217-500 | Table 1 |
| Potassium phosphate monobasic | Thermo Fisher | P285-3 | Table 1 |
| RC DC Protein Assay Kit I | BioRed | 5000121 | STEP 2.9 |
| RNaseOUT Recombinant Ribonuclease Inhibitor | Thermo Fisher | 10777019 | Table 1 |
| Sodium Chloride | Thermo Fisher | S271-500 | Table 1 |
| Sodium Phosphate Dibasic Anhydrous | Thermo Fisher | S374-500 | Table 1 |
| TritionX-100 | Sigma | X100-500ML | Table 1 |
| UY38 hrpk-1(zen17) | available upon request | ||
| VT1367 col-19::gfp(maIS105) | available upon request | ||
| VT3841 alg-1(tm492) | available upon request |
References
- Liu, X., et al. An AP-MS- and BioID-compatible MAC-tag enables comprehensive mapping of protein interactions and subcellular localizations. Nature Communications. 9 (1), 1188-1216 (2018).
- Li, L., Veksler-Lublinsky, I., Zinovyeva, A. HRPK-1, a conserved KH-domain protein, modulates microRNA activity during Caenorhabditis elegans development. PLoS Genetics. 15 (10), 1008067 (2019).
- Zinovyeva, A. Y., Veksler-Lublinsky, I., Vashisht, A. A., Wohlschlegel, J. A., Ambros, V. R. Caenorhabditis elegans ALG-1 antimorphic mutations uncover functions for Argonaute in microRNA guide strand selection and passenger strand disposal. Proceedings of the National Academy of Sciences of the United States of America. 112 (38), 5271-5280 (2015).
- Hammell, C. M., Lubin, I., Boag, P. R., Blackwell, T. K., Ambros, V. nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell. 136 (5), 926-938 (2009).
- Zou, Y., et al. Developmental decline in neuronal regeneration by the progressive change of two intrinsic timers. Science. 340 (6130), 372-376 (2013).
- Zanin, E., Dumont, J., et al. Affinity purification of protein complexes in C. elegans. Methods in Cell Biology. 106, 289-322 (2011).
- Bhaskaran, S., et al. Breaking Caenorhabditis elegans the easy way using the Balch homogenizer: an old tool for a new application. Analytical Biochemistry. 413 (2), 123-132 (2011).
- Kohl, K., et al. Plate-based Large-scale Cultivation of Caenorhabditis elegans: Sample Preparation for the Study of Metabolic Alterations in Diabetes. Journal of Visualized Experiments. (138), e58117 (2018).
- Larance, M., Bailly, A. P., et al. Stable-isotope labeling with amino acids in nematodes. Nature Methods. 8 (10), 849-851 (2011).
- Ding, L., Han, M. GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends in Cell Biology. 17 (8), 411-416 (2007).
- Ding, L., Spencer, A., Morita, K., Han, M. The Developmental Timing Regulator AIN-1 Interacts with miRISCs and May Target the Argonaute Protein ALG-1 to Cytoplasmic P Bodies in C. elegans. Molecular Cell. 19 (4), 437-447 (2005).
- Jannot, G., Vasquez-Rifo, A., Simard, M. J. Argonaute Pull-Down and RISC Analysis Using 2’-O-Methylated Oligonucleotides Affinity Matrices. Methods in Molecular Biology. 725, 233-249 (2011).
- Porta-de-la-Riva, M., Fontrodona, L., Villanueva, A., Ceron, J. Basic Caenorhabditis elegans methods: synchronization and observation. Journal of Visualized Experiments. (64), e4019 (2012).
- Stiernagle, T. Maintenance of C. elegans. WormBook: the Online Review of C. elegans Biology. , 1-11 (2006).
- Gallagher, S., Chakavarti, D. Immunoblot analysis. Journal of Visualized Experiments. (16), e759 (2008).
- Eslami, A., Lujan, J. Western blotting: sample preparation to detection. Journal of Visualized Experiments. (44), e2359 (2010).
- Zinovyeva, A. Y., Bouasker, S., Simard, M. J., Hammell, C. M., Ambros, V. Mutations in conserved residues of the C. elegans microRNA Argonaute ALG-1 identify separable functions in ALG-1 miRISC loading and target repression. PLoS Genetics. 10 (4), 1004286 (2014).
- Zhang, L., et al. Systematic Identification of C. miRISC Proteins, miRNAs, and mRNA Targets by Their Interactions with GW182 Proteins AIN-1 and AIN-2. Molecular Cell. 28 (4), 598-613 (2007).
- Corsi, A. K., Wightman, B., Chalfie, M. A Transparent Window into Biology: A Primer on Caenorhabditis elegans. Genetics. 200 (2), 387-407 (2015).
- Farboud, B., et al. Enhanced Genome Editing with Cas9 Ribonucleoprotein in Diverse Cells and Organisms. Journal of Visualized Experiments. (135), e57350 (2018).
- Dickinson, D. J., Goldstein, B. CRISPR-Based Methods for Caenorhabditis elegans Genome Engineering. Genetics. 202 (3), 885-901 (2016).

