Particle Templated Emulsification enables Microfluidic-Free Droplet Assays
Summary
Water-in-oil droplet assays are useful for analytical chemistry, enzyme evolution, and single cell analysis, but typically require microfluidics to form the droplets. Here, we describe particle templated emulsification, a microfluidic-free approach to perform droplet assays.
Abstract
Reactions performed in monodispersed droplets afford enhanced accuracy and sensitivity compared to equivalent ones performed in bulk. However, the requirement of microfluidics to form controlled droplets imposes a barrier to non-experts, limiting their use. Here, we describe particle templated emulsification, an approach to generate monodisperse droplets without microfluidics. Using templating hydrogel spheres, we encapsulate samples in monodispersed droplets by simple vortexing. We demonstrate the approach by using it to perform microfluidic-free digital PCR.
Introduction
Droplet microfluidics leverages compartmentalization in picoliter droplets to increase the sensitivity and accuracy of assays compared to bulk reactions, and have numerous applications in chemical screening, protein engineering, and next generation sequencing1,2,3. For example, digital droplet polymerase chain reaction (ddPCR) affords increased accuracy compared to bulk quantitative polymerase chain reaction (qPCR), with applications for genetic variation in cancers, detection of disease causing mutations, and prenatal diagnostics4,5,6. A challenge of droplet microfluidics, however, is the requirement of microfluidic devices to partition samples; while microfluidics afford excellent control over droplet properties, they require specialized expertise to build and operate7,8. Consequently, droplet-based methods are largely limited to expert labs or, in rare instances, applications in which a commercial instrument is available9,10. To broaden the use of droplet assays, the requirement for specialized microfluidic instrumentation is a hurdle that must be overcome.
In this article, we describe Particle Templated Emulsification (PTE), a microfluidic-free method for performing reactions in monodispersed droplets. In PTE, templating particles engulf the sample into droplets in carrier oil by simple vortexing (Figure 1). As the system mixes, the aqueous portion fragments into droplets of reducing size until the droplets contain single particles, at which point further fragmentation is not possible because it requires breaking the particles. The engulfed sample surrounds the particles as a shell in the droplets, thereby encapsulating any dispersed cells, reagents, or functional moieties (Figure 1D). Thus, PTE requires no equipment or expertise to perform droplet reactions beyond a common vortexer. Additionally, droplet generation takes seconds compared to minutes or hours with microfluidics, and the amount produced is proportional to the container volume, not device operation time, making it supremely scalable. These benefits make PTE ideal for conducting droplet assays in a variety of circumstances in which microfluidics are impractical. Here, we demonstrate PTE and use it to conduct ddPCR.
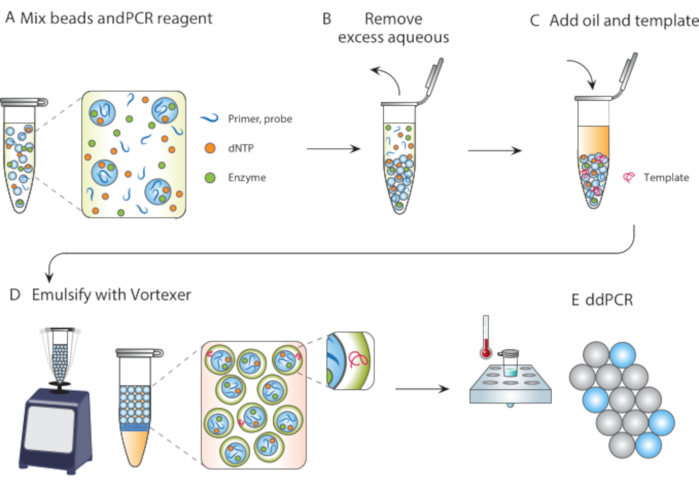
Figure 1. Overview of particle templated emulsification process. (A) Templating particles are mixed with reagents. (B) Excess reagents are removed following centrifugation. (C) The addition of template molecules occurs before the addition of oil. (D) Vortexing produces droplets containing a single template molecule. (E) Subsequent thermocycling and imaging allows for digital droplet analysis of target template. Please click here to view a larger version of this figure.
Protocol
1. Preparation of hydrogel particles for particle templated emulsification.
Hydrogel particles used for particle templated emulsification can be prepared using two different methods.
- Preparation using commercially available particles
- Add 0.5 g of dried polyacrylamide particles compatible with PTE (e.g., Bio-Gel P-60 Gel (Bio-Rad), 45-90 µm diameter) to 30 mL of sterile water in a 50 mL conical tube and mix well. Incubate at room temperature for 30 min.
- Preparation using microfluidic fabrication of particles
NOTE: Polyacrylamide particles compatible with PTE can be prepared using commercially available drop makers (e.g., QX200 Drop Generator (Bio-Rad), RayDrop (Fluigent) etc.), or by custom microfluidic design.- Fabrication of custom master
- Design a soft photolithography mask using computer-aided design (CAD) software. Print the photomask with a 10 µm resolution on circuit board film.
- Pour 1 mL of photoresist onto the center of a 3 in silicon wafer. Use a spin coater to create a 50 µm layer of photoresist by spinning it at 500 rpm for 30 sec followed by 1250 rpm for 30 sec. Place the wafer onto a hotplate set to 95 °C for 15 min to evaporate the solvent.
- Secure the photomask onto the silicon wafer with a cover glass slide and expose the wafer under a collimated 190 mW, 365 µm UV LED for 2.5 min. Place the wafer on a hotplate set to 95 °C for 5 min for post exposure baking.
- Develop the photoresist-silicon wafer by immersing it in a bath of 100% propylene glycol monomethyl ether acetate (PGMEA) for up to 15 min. Rinse the wafer with fresh 100% PGMEA followed by 100% isopropanol. Air dry the wafer.
- Remove any residual isopropanol by drying the wafer on a hotplate set to 95 °C for 1 min. Place the wafer into a clean 3 in Petri dish.
- Fabrication of the custom microfluidic device
- Mix the polydimethylsiloxane (PDMS) silicon base and curing reagent in a 10:1 ratio by mass. Degas the mixed PDMS using a desiccator under house vacuum until no air bubbles are observable.
- Pour the degassed PDMS over the master in the petri dish, ensuring the silicon wafer is completely submerged. Degas the silicon wafer and PDMS to remove any air bubbles that may have formed during pouring.
- Cure the PDMS by placing the silicon wafer and PDMS into an oven set to 65 °C for at least 60 min. Excise a block of PDMS containing the microfluidic features from the petri dish using a scalpel. Take extra care to avoid damaging any features present on the silicon master.
- Punch the inlets and the outlets into the PDMS block corresponding to the inlets and outlets in the microfluidic device using a 0.75 mm biopsy punch. Remove any dust and particulates with the repetitive application and removal of packaging tape to the surface of the PDMS block.
- Clean a 50 mm x 75 mm glass slide by rinsing it with 100% isopropanol and subsequently air drying the surface. Plasma treat both the glass slide and the PDMS (features facing up) using 1 mbar of O2 plasma for 1 min using a plasma bonder.
- Affix the PDMS to the glass slide by placing the plasma treated PDMS with features facing down onto the glass slide, plasma treated side facing up. Place the slide into an oven set to 65 °C for at least 30 min to complete the bonding.
- Treat all microfluidic channels with a fluorinated surface treatment to ensure surface hydrophobicity and prevent wetting. Bake the device at 65 °C for at least 10 min.
- Fabrication of templating particles
- Prepare a polyacrylamide (PAA) solution consisting of 6.2% acrylamide, 0.18% N,N′-methylenebis (acrylamide), and 0.3% ammonium persulfate. Load this solution into a 1 mL syringe with a 28G needle.
- Prepare an insoluble continuous phase consisting of 5% (w/w) fluorosurfactant and 1% N,N,NN-tetramethylethylenediamine (TEMED) in hydrofluoroether (HFE) oil for the generation and stabilization of droplets. Load the solution into new 1 mL syringe.
- Load both PAA and HFE solution containing syringes into syringe pumps (E.g., NE-501). Connect both syringes to the microfluidic device using polyethylene tubing inserted onto the syringe and into the device. Before the connecting, prime the pumps to remove the air from the tubing.
NOTE: Depending on the model, syringe pumps may be controlled with built in input, manufacture software, or a custom script (available at https://github.com/AbateLab/Pump-Control-Program). - Run the drop generation device with PAA and HFE oil inputs at 300 µL/h and 500 µL/h, respectively. Collect 1 mL of the droplets in a 15 mL collection tube and incubate for 3 h at room temperature for polymerization. After the incubation, remove the lower layer of oil by pipetting.
- Add 1 mL of 20% (v/v) perfluoro-1-octanol (PFO) in HFE oil to the 15 mL collection tube as a chemical demulsifier. After mixing, spin down the 15 mL collection tube at 2000 x g for 2 min. Remove the PFO/HFE supernatant by pipetting. Repeat 1x.
- Add 2 mL of 2% sorbitan monooleate in hexane to the 15 mL collection tube and vortex to mix. Spin the tube at 3000 x g for 3 min. Remove the supernatant by pipetting to remove surfactant/hexane solution. Repeat 2x.
- Add 5 mL of TEBST buffer (20 mM Tris-HCl pH 8.0, 274 mM NaCl, 5.4 mM KCl, 20 mM EDTA, 0.2% Triton X-100) and mix well. Spin down at 3,000 x g for 3 min. Remove the supernatant by pipetting. Repeat 3x.
- Resuspend in 5 mL TEBST. This solution may be stored at 4 °C indefinitely.
- Fabrication of custom master
2. Particle templated emulsification.
Following the preparation of templating particles, PTE is used to encapsulate the sample and reagents in droplets.
- Prepare the polyacrylamide particles for particle templated emulsification by centrifuging at 6000 x g for 1 min to pellet the particles, then remove the supernatant by pipetting and resuspend using sterile water. Repeat 3x to ensure removal of any residual TEBST.
- Determine the concentration and diameter of the templating particles using a hemocytometer (or equivalent). Calculate individual particle diameter by measuring the diameters in pixels and converting to microns. Conversion of pixels to microns may be calculated using the hemocytometer (or equivalent) as a calibration slide and measuring the known grid distance in pixels.
- Prepare the disperse phase in a fresh 1.5 mL microcentrifuge tube using a PCR master mix, the appropriate primers, and a fluorescein hydrolysis probe according to Table 1. Incubate at room temperature for 5 min under gentle agitation (10 rpm) using a tube rotator to ensure homogenous distribution of the components.
NOTE: The volume and target concentration of particles is based upon Poisson loading. As general rule, the number of particles should be an order of magnitude more than the number of samples to be encapsulated. For samples of unknown concentrations, a dilution series is necessary to ensure Poisson loading.
| Volume | Reagent |
| 100 μL | Particles (450 particles / μL) |
| 200 μL | 2x PCR master mix |
| 18 μL | 10 μM forward primer |
| 18 μL | 10 μM reverse primer |
| 18 μL | 10 μM probe |
| 0.8 μL | Triton X100 |
| 45.2 μL | Nuclease free water |
Table 1. Preparation of the PCR master mix used with PTE for digital droplet PCR.
- Centrifuge the disperse phase at 6000 x g for 1 min and remove the supernatant. Record the volume of the supernatant extracted and using the total disperse phase volume calculated in 2.3 determine the pellet volume.
NOTE: The amount of supernatant extracted will vary depending on particle packing, diameter, and concentration with a minimum expected volume of 300 µL. - Add 1 µL of 1.62 pg /µL Saccharomyces cerevisiae genomic DNA to the pellet from 2.4 and mix thoroughly by pipetting or vigorous tapping.
NOTE: The presence of excess aqueous content can decrease encapsulation efficiency. If the sample volume exceeds 1% of the pellet volume, concentrate the sample. If the sample cannot be concentrated, scale the PCR master mix and resulting pellet volume according to the sample volume. PTE permits the emulsification of small (10 µL) to large (2 mL) volumes of templating particles. The PCR master mix (2.3) and oil (2.6) can be scaled according to the target (2.3) and measured (2.4) volume of the particle pellet respectively. - Add 200 µL 2% fluorosurfactant in HFE oil to the tube as the insoluble continuous phase for emulsification. Ensure the pellet is dislodged by pipetting or tapping/flicking the tube. Then vortex at 3000 rpm for 30 sec.
NOTE: The setting corresponding to 3000 rpm may vary depending on brand and model. - Allow for the emulsions to settle for 1 min. Remove 100 µL of the bottom oil phase and replace this volume with fresh 2% fluorosurfactant in HFE oil. Gently invert the tube several times to mix. Repeat 3-5x or until small satellite droplets have been removed.
3. Digital droplet PCR and analysis.
- After 2-5 min of settling, remove the bottom oil phase. Replace this volume with 5% fluorosurfactant in fluorocarbon oil (e.g., FC-40).
- Using a wide bore pipette tip, carefully pipette the 100 µL of sample into 200 µL PCR tubes. Place the PCR tubes into a thermocycler and run according to Table 2.
| Step | Temperature | Duration | Notes |
| 1 | 95 °C | 2 min | |
| 2 | 95 °C | 30 s | |
| 3 | 50 °C | 90 s | |
| 4 | 72 °C | 60 s | |
| 5 | Repeat x34 steps 2 to 4 | ||
| 6 | 72 °C | 2 min | |
| 7 | 4 °C | hold |
Table 2. Thermocycling conditions for digital droplet PCR using PTE emulsions.
- Pipette the sample onto a counting slide using a wide bore pipette tip for fluorescent imaging. Image the sample using a fluorescent microscope with 490 nm excitation and 525 nm emission detection wavelengths.
- Quantify the positive fluorescent droplets (Np) and total drops (NT) to verify the presence and calculate the number of template molecules (λ) using the fraction of positive droplets (Np/NT) and Poisson statistics:
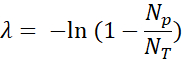
- Calculate the 95% confidence interval (zc = 1.96) by:
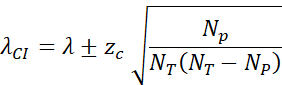
- Calculate the sample concentration (molecules/µL) using the volume (ν in µL) of sample added in step 2.5, using the equation given below. Determine the mean and standard deviation of the sample concentration using technical replicates.
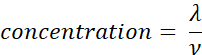
Representative Results
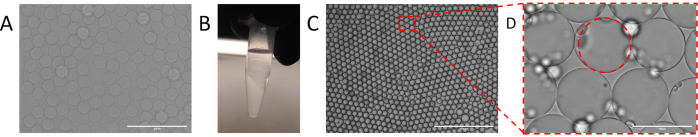
Figure 2. Encapsulation of sample into droplets using particle templated emulsification. (A) templating particles used for particle templating emulsification. (B) Separation of templating particle pellet from supernatant following centrifugation. (C) Droplets resulting from particle templated emulsification with (D) identifiable aqueous shell. Please click here to view a larger version of this figure.
In PTE, the monodispersity of the emulsions is dictated by that of the templating particles, because the droplets have a diameter slightly larger than the particles. Thus, uniform particles are central to controlled PTE encapsulation11. A variety of methods exist for generating uniform templating particles, including chemical (sol-gel, emulsion polymerization), hydrodynamic (membrane emulsification, homogenization), and filtration methods. Microfluidic approaches in particular, afford superb monodispersity (Figure 2A) and allow additional particle engineering to enhance their functionality in PTE12. Alternatively, templating particles can be purchased, although their uniformity, while adequate, is typically less than with microfluidic generation11.
To perform PTE, the particles are mixed with the sample to be encapsulated (Figure 1A), and the excess supernatant is removed by centrifugation and pipetting (Figure 1B), as illustrated by a photograph of a particle pellet at the bottom of a PCR tube (Figure 2B). The encapsulating oil containing a stabilizing surfactant is then added (Figure 1C), and the sample gently pipetted before vortexing for 30 seconds (Figure 1D), to generate the emulsion (Figure 2C). The resultant droplets contain a particle core and aqueous shell comprising the initial sample, within which reside the reagents, target molecules, and cells necessary for the reaction (Figure 2D). Just as in droplet microfluidic encapsulation, discrete entities like small beads or cells are encapsulated randomly and in accordance with a Poisson distribution, although nearly all droplets contain a templating particle due to the nature of PTE physics.
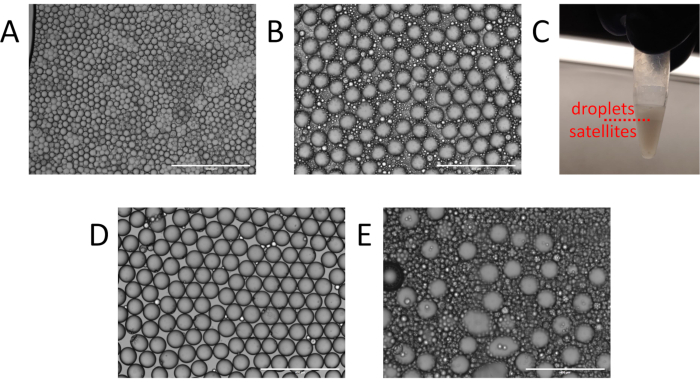
Figure 3. Identification and cleanup of particle templated emulsification droplets. (A) Example of non-uniform droplet generation with multiple particles per droplet from insufficient vortexing. (B) Expected presence of satellites and droplets following particle templated emulsification and (C) the water-in-oil fractionation. (D) Resulting emulsion following oil washing. (E) Excessive satellite generation resulting from residual supernatant during particle templated emulsification. Please click here to view a larger version of this figure.
Even in successful PTE, double or triple core droplets exist, though they generally contribute negligibly to the reaction, provided they are rare. Achieving a low frequency of multicore droplets while retaining adequate shells requires optimization of process parameters, including surface tension, inter-particle adhesion forces, sample viscosity, container size, and vortexing power and time. For example, a poorly optimized emulsification may contain polydispersed droplets with many templating particles (Figure 3A), indicating that the vortexing was insufficient to fully emulsify the sample. In such instances, detergents can be added to reduce inter-particle adhesion and lower surface tension, or vortexing power or time can be increased. Another common issue is generation of excessive satellites, which are small empty droplets (Figure 3B). Satellites can be unavoidable in PTE emulsions depending on the interfacial tension and rheological properties of the sample and carrier oil. However, they often result from not adequately removing excess sample prior to emulsification (Figure 2B), or vortexing with too much power, stripping the shells from the droplets. In a successful PTE emulsification, satellites should comprise no more than ~10% of the total encapsulated sample volume (Figure 3C)11. At this level, they usually contribute negligibly to the reaction and can be ignored. For aesthetic purposes, they can be cleared from the emulsion by washing with fresh oil (Figure 3D).
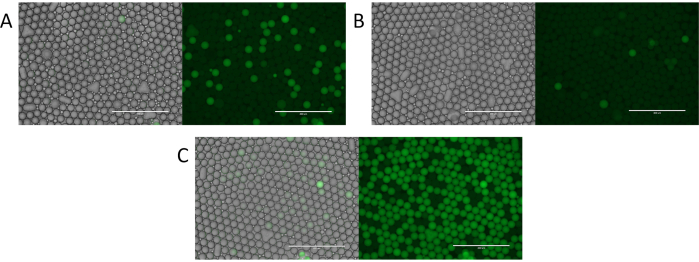
Figure 4. Evaluation of particle templated emulsification digital droplet PCR. (A) Fluorescent imaging of the droplets identifies positive fluorescent droplets and negative non-fluorescent droplets. (B) Identification of rare template or low concentrations of template with digital droplet PCR. (C) Over abundant template encapsulation resulting in a variable number of template molecules per droplet. Please click here to view a larger version of this figure.
To demonstrate the utility of PTE, we used it to perform microfluidic-free digital PCR11. Using the process, we encapsulated a sample comprising S. cerevisiae genomic DNA, and thermocycled it. In digital PCR, droplets containing amplified targets become fluorescent, while those without remain dim. Thus, a fluorescent droplet indicates a target, allowing direct quantitation of targets by counting positive droplets (Figure 4A). The number of fluorescent droplets thus scales with the target molecules, yielding few positives when the target is rare (Figure 4B) and many when it is abundant (Figure 4C). As with encapsulation of other discrete components, target encapsulation follows a Poisson distribution, allowing the positive droplet fraction to be transformed into the target concentration (Figure 4D), thereby demonstrating the ability to perform digital PCR with PTE11.
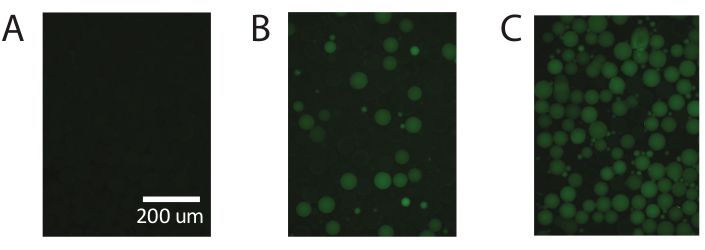
Figure 5. Demonstration of digital droplet PCR using commercially available PAA. (A) Fluorescent imaging of the droplets identifies negative non-fluorescent droplets. (B) Identification of low concentrations of template with digital droplet PCR. (C) Identification of high concentrations of template with digital droplet PCR. Please click here to view a larger version of this figure.
These results are repeatable using commercially available polyacrylamide particles (Figure 5) and demonstrate the ability of PTE to perform standard digital PCR with commercially available polyacrylamide particles, achieving accurate measurements over the same range.
Supplemental File. Please click here to download this file.
Discussion
PTE uses particles to encapsulate samples in monodispersed droplets by vortexing. In addition to its simplicity and accessibility, PTE provides several additional benefits, including allowing large volumes of droplets to be generated instantaneously. Moreover, the process can be conducted in an isolated tube, obviating the need to transfer samples to microfluidic devices, streamlining the overall workflow and limiting opportunities for sample contamination or loss. The templating particles also provide a means by which to engineer the contents of the resultant droplet reactions. For example, particle size, chemistry, and wettability can be engineered for targeted biomolecule or cell capture, while functional moieties such as enzymes, actives, or nucleic acids, can be displayed on particle to facilitate reactions, such as for single cell sequencing or functional characterization. While the approach is flexible, there are nevertheless important constraints to its use. For example, it is not currently possible to perform droplet additions as are often conducted with microfluidics, requiring that all reaction components be introduced before encapsulation; this requires that reagents be compatible and stable until the droplets can be generated and, in the case of troublesome combinations, can often be addressed by quickly mixing and emulsifying the sample on ice. Alternatively, reactive components that can be triggered externally with light or heat can be used13. PTE thus provides a flexible and scalable method for conducting droplet assays accessible to non-experts. This, coupled with its innate simplicity and flexibility, makes PTE ideal for the execution and development of numerous droplet applications.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work developing this protocol was supported by the National Institutes of Health (R01-EB019453-02), the Office of the Director of National Intelligence, Intelligence Advanced Research Projects Activity through Raytheon BBN Technologies Corp (N66001-18-C-4507), the Chan-Zuckerberg Biohub Investigator Program, Defense Advanced Research Projects Agency through Texas A&M University (W911NF1920013), and Centers for Disease Control and Prevention through Johns Hopkins University Applied Physics Laboratory (75D30-11-9C-06818 (CDC3)). The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the above organizations or the U.S. Government. The U.S. Government is authorized to reproduce and distribute reprints for governmental purposes notwithstanding any copyright annotation therein.
Materials
| 0.22 um syringe filter | Milipore Sigma | SLGP033RS | |
| 0.5M EDTA, pH 8.0 | Thermo-Fisher | 15575020 | |
| 0.75 mm biopsy punch | World Precision Instruments | 504529 | |
| 1 mL syringes | BD | 309628 | |
| 1H,1H,2H-Perfluoro-1-Octanol (PFO) | Sigma-Aldrich | 370533 | |
| 1M Tris-HCI, pH 8.0 | Thermo-Fisher | 15568025 | |
| 27 gauge needles | BD | 305109 | |
| 3" silicon wafers, P type, virgin test grade | University Wafers | 447 | |
| 3D-printed centrifuge syringe holder | (custom) | (custom) | |
| Acrylamide solution,40%, for electrophoresis, sterile-filtered | Sigma-Aldrich | A4058-100ML | |
| Ammonium persulfate | Sigma-Aldrich | A3678-25G | |
| Aquapel (fluorinated surface treatment) | Pittsburgh Glass Works | 47100 | |
| Hexane | Sigma-Aldrich | 139386 | |
| FC-40 fluorinated oil | Sigma-Aldrich | F9755 | |
| Isopropanol | Sigma-Aldrich | 109827 | |
| N,N′-Methylenebis(acrylamide) | Sigma-Aldrich | 146072-100G | |
| NaCl | Sigma-Aldrich | S9888 | |
| Novec-7500 Engineering Fluid (HFE oil) | 3M | 98-0212-2928-5 | |
| polyethylene tubing | Scientific Commodities | B31695-PE/2 | |
| fluorosurfactant | Ran Biotechnologies | 008-FluoroSurfactant | |
| PGMEA developer | Sigma-Aldrich | 484431 | |
| Photomasks | CadArt Servcies | (custom) | |
| Platinum Multiplex PCR Master Mix (Taq Master Mix) | Applied Biosystems | 4464263 | |
| Spin coater | Specialty Coating Systems | G3P-8 | |
| Span 80 (sorbitane monooleate) | Sigma-Aldrich | s6760 | |
| SU-8 3025 photoresist | Kayaku | 17030192 | |
| Triton X-100 (octylphenol ethoxylate) | Sigma-Aldrich | t8787 | |
| Tween 20 (polysorbate 20) | Sigma-Aldrich | p2287 | |
| Platinum Multiplex PCR Master Mix (Taq Master Mix) | Applied Biosystems | 4464263 | |
| Yeast FWD | IDT | 5′-GCAGACCAGACCAGAACAAA-3′ | |
| Yeast REV | IDT | 5′-ACACGTATGTATCTAGCCGAATA AC-3 |
|
| Yeast Probe | IDT | 5′-/56-FAM/ATATGTTGT/ZEN/TCACTCGCGCCTGGG/3IABk FQ/-3′ |
|
| EVOS FL AUTO | Life Technologies | ||
| EVOS LED Cube, GFP | Life Technologies | AMEP4651 | |
| SYLGARD 184 KIT 1.1 LB (PDMS base and curing reagents) | Dow Corning | DC4019862 | |
| TEMED | Thermo Fisher | 17919 | |
| Saccharomyces cerevisiae genomic DNA | Milipore | 69240-3 | |
| Expanded plasma cleaner (plasma bonder) | Harrick Plasma | PDC-002 (230V) | |
References
- Mashaghi, S., Abbaspourrad, A., Weitz, D. A., van Oijen, A. M. Droplet microfluidics: A tool for biology, chemistry, and nanotechnology. Trends in Analytical Chemistry. 82, 118-125 (2016).
- Gielen, F., et al. Ultrahigh-throughput-directed enzyme evolution by absorbance-activated droplet sorting (AADS). Proceedings of the National Academy of Sciences of USA. 113 (47), 7383-7389 (2016).
- Mai, S., Murphy, T. W., Lu, C. Microfluidics for genome-wide studies involving next generation sequencing. Biomicrofluidics. 11 (2), 021501 (2017).
- Olmedillas-López, S., García-Arranz, M., García-Olmo, D. Current and emerging Applications of Droplet Digital PCR in Oncology. Molecular Diagnosis and Therapy. 21 (5), 493-510 (2017).
- Tong, Y., Shen, S., Jiang, H., Chen, Z. Application of Digital PCR in Detecting Human Diseases Associated Gene Mutation. Cellular Physiology and Biochemistry. 43 (3), 1718-1730 (2017).
- Yan, Y., et al. Evaluation of droplet digital PCR for non-invasive prenatal diagnosis of phenylketonuria. Analytical and Bioanalytical Chemistry. 411 (27), 7115-7126 (2019).
- Shang, L., Cheng, Y., Zhao, Y. Emerging Droplet Microfluidics. Chemical Reviews. 117 (12), 7964-8040 (2017).
- The, S., Lin, R., Hung, L., Lee, A. P. Droplet Microfluidics. Lab Chip. 8 (2), 198-220 (2008).
- Baker, M. Digital PCR hits its stride. Nature Methods. 9, 541-544 (2012).
- Kolodziejczyk, A. A., Kim, J. K., Svensson, V., Marioni, J. C., Teichmann, S. A. The Technology and Biology of Single-Cell RNA Sequencing. Molecular Cell. 58 (4), 610-620 (2015).
- Hatori, M. N., Kim, S. C., Abate, A. R. Particle-Templated Emulsification for Microfluidics-Free Digital Biology. Analytical Chemistry. 90 (16), 9813-9820 (2018).
- Panda, P., et al. Stop-flow lithography to generate cell-laden microgel particles. Lab Chip. 8 (7), 1056-1061 (2008).
- Yozwiak, C. E., Hirschhorn, T., Stockwell, B. R. Towards a microparticle-based system for pooled assays of small molecules in cellular contexts. ACS Chemical Biology. 13 (3), 761-771 (2018).

