Co-Culturing Microglia and Cortical Neurons Differentiated from Human Induced Pluripotent Stem Cells
Summary
This protocol describes a methodology to differentiate microglia from human iPSCs and maintain them in co-culture with iPSC-derived cortical neurons in order to study mechanistic underpinnings of neuroimmune interactions using human neurons and microglia.
Abstract
The ability to generate microglia from human induced pluripotent stem cells (iPSCs) provides new tools and avenues for investigating the role of microglia in health and disease. Furthermore, iPSC-derived microglia can be maintained in co-culture with iPSC-derived cortical neurons, which enable investigations of microglia-neuron interactions that are hypothesized to be dysregulated in a number of neuropsychiatric disorders. Human iPSCs were differentiated to generate microglia using an adapted version of a protocol developed by the Fossati group, and the iPSC-derived microglia were validated with marker analysis and real-time PCR. Human microglia generated using this protocol were positive for the markers CD11C, IBA1, P2RY12, and TMEM119, and expressed the microglial-related genes AIF1, CX3CR1, ITGAM, ITGAX, P2RY12, and TMEM119. Human iPSC-derived cortical neurons that had been differentiated for 30 days were plated with microglia and maintained in co-culture until day 60, when experiments were undertaken. The density of dendritic spines in cortical neurons in co-culture with microglia was quantified under baseline conditions and in the presence of pro-inflammatory cytokines. In order to examine how microglia modulate neuronal function, calcium imaging experiments of the cortical neurons were undertaken using the calcium indicator Fluo-4 AM. Live calcium activity of cortical neurons was obtained using a confocal microscope, and fluorescence intensity was quantified using ImageJ. This report describes how co-culturing human iPSC-derived microglia and cortical neurons provide new approaches to interrogate the effects of microglia on cortical neurons.
Introduction
In the human brain, microglia are the primary innate immune cells1. Brain development is regulated by microglia via two routes: release of diffusible factors and phagocytosis1. Microglia-derived diffusible factors help support myelination, neurogenesis, synaptic formation, maturation, cell death, and cell survival1. Microglia also phagocytize various elements in brain synapses, axons and in both living and dead cells2,3,4,5,6,7,8. Receptors on microglia recognize tags such as calreticulin, ATP, and sialic acid and regulate cellular phagocytosis9,10. In the hippocampus, microglia maintain the homeostasis of neurogenesis through its phagocytic role11.
Synaptic phagocytosis in the dorsolateral geniculate nucleus (dLGN) of the rodent brain has been shown to be regulated by microglia1. In rodents, it has been shown that there are two periods during the development when intense microglial synaptic phagocytosis is observed. The first period occurs during initial synapse formation and the second period occurs when connections are being fine-tuned and pruned12. Other factors that are involved in synaptic pruning are inflammatory proteins and the Class I major histocompatibility complex (MHC1, H2-Kb and Db)13,14. It has been suggested that C1q (complement component 1q) on the microglia colocalizes with MHC1, which triggers synaptic pruning15. Furthermore, mouse studies show that interleukin-33 (IL-33) secreted by astrocytes regulates synapse homeostasis in the thalamus and the spinal cord through its effects on microglia, though this has yet to be investigated in humans13. Microglia secrete a variety of cytokines that help maintain neuronal health, such as tumor necrosis factor α (TNFα), IL-1β, IL-6, IL-10 and interferon-γ (IFN-γ) and these cytokines can modulate dendritic spine and synapse formation16,17,18. There are significant gaps in our knowledge of neuron-microglia interactions during human brain development. Most of our knowledge comes from studies from rodent models, while there is a paucity of information on the temporal and mechanistic aspects of synaptic pruning in the human cortex. Microglia support neuronal survival in the neo-cortex, and other cell types contribute as well1. It is not clear how microglia contribute to this preservation and what the interplay between microglia and the other cell types are. Microglia release several cytokines that affect neuronal and synaptic development but the mechanistic basis of their effects of these cytokines in neurons are largely unknown19,20. In order to develop a more complete understanding of the function of microglia in the human brain, it is critical to explore its interactions with different cell types found in the human brain. This report describes a method to co-culture human iPSC-derived neurons and microglia generated from the same individual. Establishing this methodology will enable well-defined investigations to interrogate the nature of microglia-neuronal interactions and to develop robust in vitro cellular models to study neuroimmune dysfunction in the context of different neurodevelopmental and neuropsychiatric disorders.
The role of microglia in schizophrenia
Synaptic pruning is a major neurodevelopmental process that takes place in the adolescent brain21,22. Multiple lines of evidence suggest that synaptic pruning during this critical period is abnormal in schizophrenia (SCZ)23,24,25,26. SCZ is a chronic, debilitating psychiatric disorder characterized by hallucinations, delusions, disordered thought processes and cognitive deficits23,24. Microglia, the resident macrophages in the brain, play a central role in synaptic pruning25,26. Postmortem and positron emission tomography (PET) studies show evidence for dysfunctional microglial activity in SCZ25,26,27,28,29,30,31,32. Postmortem SCZ brains show well-replicated but subtle differences in the brain – pyramidal neurons in the cortical layer III show decreased dendritic spine density and fewer synapses33,34,35. Synaptic pruning is a process by which superfluous excitatory synaptic connections are eliminated by microglia during adolescence, when SCZ patients usually have their first psychotic break22,36. Postmortem studies show an association between SCZ and microglial activation, with increased density of microglia in SCZ brains, as well as increased expression of proinflammatory genes27. In addition, PET studies of human brains using radioligands for microglial activation show increased levels of activated microglia in the cortex25,26,27,28. Recent genome-wide association studies (GWAS) show that the strongest genetic association for SCZ resides in the major histocompatibility complex (MHC) locus, and this association results from alleles of the complement component 4 (C4) genes that are involved in mediating postnatal synaptic pruning in rodents37. This association has provided additional support for the hypothesis that aberrant pruning by microglia may result in the decreased dendritic spine density seen in SCZ postmortem brains. Investigations of microglial involvement in synaptic pruning in SCZ have so far been limited to indirect studies with PET imaging or inferences from investigations of postmortem brains.
Generating human microglia in the laboratory
Cultured primary mouse microglia have been frequently used in studying microglia, though there are several indications that rodent microglia may not be representative of human microglial anatomy and gene expression (Table 1)38. Several studies have also differentiated microglia directly from blood monocytes through transdifferentiation39,40,41,42. Blood monocyte-derived microglia-like cells exhibit major differences from human microglia in gene and protein expression profile pro-inflammatory responses, and they appear to be more macrophage-like in their biology43. Recent methodological advances now enable the generation of microglia from human iPSCs, which provide opportunities to study live microglia that more accurately resemble the biology of microglia found in the human brain (Table 2). These iPSC-derived microglial cells have been shown to recapitulate the phenotype, gene expression profiles, and functional properties of primary human microglia44,45,46,47,48. This paper provides a method to co-culture human iPSC-derived neurons and microglia generated from the same individual in order to develop personalized in vitro models of neuron-microglia interactions. For this in vitro co-culture model, a microglial differentiation protocol from the Fossati group was adapted (Table 3) and combined with an adapted version of a cortical neuronal generation protocol from the Livesey group (Table 4)49,50.
Protocol
The human iPSCs used in this study were reprogrammed from fibroblasts that had been obtained through informed consent from healthy control subjects, with approval from the institutional review board (IRB). The reprogramming and characterization of iPSCs used in this study (ML15, ML27, ML40, ML56, ML141, ML 250, ML292) were described in a prior study51.
1. Maintenance of iPSCs
- Prepare a 1:50 dilution of LDEV-free reduced growth factor basement membrane matrix in DMEM/F12 without phenol red and pre-coat a 6-well plate with 1 mL of the diluted solution for at least 2 h at 37 °C prior to thawing cell stocks.
- Thaw cryopreserved iPSC stocks in a 37 °C water bath for 2 min. Add the cells to a 15 mL centrifuge tube containing 5 mL of DMEM/F12. Spin the cells down at 300 x g for 5 min.
- Remove the coating solution from the pre-coated LDEV-free reduced growth factor basement membrane matrix plate and add 1 mL of stem cell medium (SCM) with 10 µM Rock inhibitor (Y-27632).
- Resuspend the cell pellet in SCM with 10 µM Y-27632 and add to the pre-coated plate for a final volume of 2 mL of SCM plus Y-27632. Maintain iPSC cell cultures in this medium for 24 h in a 37 °C incubator.
- Replace the medium with 2 mL of fresh SCM without Y-27632 24 h after thawing.
- After iPSCs reach 80-90% confluence, passage them onto 75 mL flasks coated with LDEV-free reduced growth factor basement membrane matrix.
- Passage cells by first rinsing cells with HBSS and remove after letting sit for 30 s. Add 1 mL of non-enzymatic cell dissociation reagent and incubate for 4 min at 37 °C. Prepare the plate with SCM containing 10 µM Y-27632.
- Aspirate non-enzymatic cell dissociation reagent and add 1 mL of SCM + 10 µM Y-27632 to each well. Gently scrape the well with a cell lifter and obtain cells with a 1000 µL pipette.
- Deposit cells onto LDEV-free reduced growth factor basement membrane matrix-coated 75 mm flask in a total volume of 8 mL of SCM + Y-27632.
2. Microglia differentiation
NOTE: A schematic outlining the microglia differentiation protocol is depicted in Figure 1A. Media were warmed to room temperature before use.
- Day 0: Perform a complete medium change with SCM medium supplemented with 80 ng/mL BMP-4. Perform daily medium changes with this same medium during Days 1-3, without any washing between medium changes.
- Day 4: Prepare Day 4-5 medium: Hematopoietic medium (HM), supplemented with 25 ng/mL FGF, 100 ng/mL SCF, and 80 ng/mL VEGF. Remove the medium and replace with Day 4-5 medium containing 5 µM Y-27632.
NOTE: Cells begin to float at this point – about half the cells are floating, and half are adherent. - Day 6: Prepare Day 6-13 medium: HM supplemented with 50 ng/mL SCF, 50 ng/mL IL-3, 5 ng/mL TPO, 50 ng/mL m-CSF, and 50 ng/mL Flt3-L. Collect the supernatant, add to a 15-mL conical tube and spin down for 8 min at 300 x g. Resuspend the pellet in a flask with 8 mL of Day 6-13 medium supplemented with 5 µM Y-27632.
- Day 10: Add 8 mL of Day 6-13 medium on top of the existing medium.
- Day 14: Prepare Day 14+ medium: HM supplemented with 50 ng/mL m-CSF, 50 ng/mL Flt3-L, 50 ng/mL GM-CSF. Collect the supernatant, add to a 50 mL conical tube and spin down for 8 min at 300 x g. Resuspend the pellet in 8 mL of Day 14+ medium containing 5 µM Y-27632 and continue to culture in this medium.
- Day 18: Add 8 mL of Day14+ medium.
- Day 22: Add 8 mL of fresh Day14+ medium without removing the existing medium.
- Day 25: Move cells to step 2.9 or continue to maintain in Day14+ medium until day 50 of differentiation.
- After Day 25, collect the supernatant in a 50-mL conical tube and spin down for 8 min at 300 x g.
- Prepare adherent medium: RPMI supplemented with 1% of 200 mM L-alanyl-L-glutamine dipeptide in 0.85% NaCl solution, 25 ng/mL GM-SCF and 100 ng/mL IL-34.
- Resuspend the pellet in the adherent medium containing 5 µM Y-27632.
- Plate cells at a density of 50,000 cells per cm2 on 24-well plates for different experiments: LDEV-free reduced growth factor basement membrane matrix-coated plates for microglial monoculture, 10 µg/mL poly-L-ornithine and 10 µg/mL laminin coated glass imaging plates for imaging experiments.
- Dilute poly-L-ornithine and laminin in DPBS and add 250 µL/well for a 24 well imaging plate.
NOTE: Cells in culture are now adherent in nature (Figure 1C).
- Maintain cells in culture for at least 14 days, with bi-weekly medium changes. After day 14 post-adherence, cells have reached maturation and may be used for experiments.
3. Cortical neuron differentiation
NOTE: A schematic outlining the cortical neuron differentiation protocol is depicted in Figure 1G.
- Day 0:
- Once iPSCs are confluent on LDEV-free reduced growth factor basement membrane matrix-coated plates, switch from SCM to a 50/50 mix of N2/B27 medium supplemented with 10 µM SB431542, 1 µM dorsomorphin, 100 nM LDN193189.
NOTE: N2 medium consists of basal medium supplemented with 1% N-2 supplement, 1% 200 mM L-alanyl-L-glutamine dipeptide in 0.85% NaCl solution, 1% pen/strep. B27 medium consists of DMEM/F12 supplemented with 2% B-27 supplement, 1% 200 mM L-alanyl-L-glutamine dipeptide in 0.85% NaCl solution, 1% pen/strep. - Change the medium with the above supplements added daily for 7 days.
- Once iPSCs are confluent on LDEV-free reduced growth factor basement membrane matrix-coated plates, switch from SCM to a 50/50 mix of N2/B27 medium supplemented with 10 µM SB431542, 1 µM dorsomorphin, 100 nM LDN193189.
- Day 7:
- Pre-coat plates in LDEV-free reduced growth factor basement membrane matrix for at least 2 h.
- Passage cells 1:1 onto pre-coated plates. Rinse cells with 1 mL/well HBSS and remove after letting it sit for 30 s. Add 1 mL/well of cell detachment medium (e.g., Accutase) and incubate for 4-5 min at 37 °C. While incubating, prepare 15 mL conical tubes with 5 mL of DMEM.
- Gently pipette enzymatic dissociation agent to remove cells from the plate with a P1000 pipettor. Collect enzymatic dissociation agent and cell mixture in the 15 mL conical tube containing 5 mL DMEM.
- Centrifuge the tubes for 5 min at 300 x g. Resuspend the pellet in 1 mL of N2/B27 medium containing 10 µM Y-27632. Continue daily feedings with N2/B27 medium without any supplements.
- Day 12: Passage cells 1:1 using methods described in 3.2.2. Continue daily feedings with N2/B27 medium.
- Day 15/16: Passage cells 1:2 using methods described in 3.2.2. Continue daily feedings with N2/B27 medium.
- Day 18/19: Passage cells 1:3 using methods described in 3.2.2. Continue daily feedings with N2/B27 medium until day 25.
- Day 25: Feed cells with N2/B27 medium supplemented with 10 µM of DAPT.
- Day 28: Feed cells with fresh, untreated N2/B27 medium.
- Day 30: Passage cells using methods described in 3.2 onto the microglia culture plates and maintain in BrainPhys medium supplemented with 1% B-27 supplement.
4. Microglia/neuron co-cultures
- Plate Day 30 cortical neurons on top of microglial cultures at a density of 50,000 cells per cm2. Supplement medium with laminin 1 µg/mL to improve cell adherence.
- Maintain cultures in a mix of 50% adherent medium and 50% Brainohys medium supplemented with 1% B-27 supplement. Perform half-medium change bi-weekly until experimentation.
- Perform experiments after neurons reach day 60.
5. Interferon-γ treatment
- Prepare fresh medium supplemented with 100 ng/mL IFN-γ. Add the medium and let it incubate for 24 h. Remove the medium and proceed to experimentation.
6. Immunocytochemistry
- Fix cells in the culture dish with 100 μL of 4% paraformaldehyde at room temperature for 20 min.
- Rinse cells in 1 mL of PBS thrice for 5 min each.
- Add 1 mL of PBST (PBS + 0.1% Triton X) for 10 min.
- Add blocking buffer: 1 mL of PBS plus 5% goat serum for 1 h.
- Add primary antibody diluted in 100 μL of PBS + 1% goat serum – overnight at 4 °C. Optimize all primary antibodies accordingly.
- Rinse cells in 1 mL of PBS thrice for 5 min each.
- Add secondary antibody, diluted in 100 μL of PBS plus 1% goat serum – for 1 h at room temperature.
7. Spine analysis
- Obtain images on a confocal microscope at 60x magnification.
- Use ImageJ function NeuronJ52 to analyze images.
- Obtain measurements for neurite length, spine length, and spine count through NeuronJ.
8. Calcium imaging
- Prepare fresh medium with 3 µM Fluo-4AM dye. Incubate co-cultures in this medium for 30 min at 37 °C. Then rinse the cells with PBS, add live-cell imaging solution to the cells, and proceed to the imaging.
- Using a confocal microscope equipped for live-cell imaging, obtain time-lapse images at 40x for 2 min. Activity can be recorded at baseline, with exposure to 15 mM glutamate, or in the setting of depolarization with 5 mM potassium chloride.
- Using ImageJ, measure fluorescence intensity over time for individual cell bodies. Using the selection tool, select individual region of interest (ROI) surrounding each cell body. Open the ROI Manager and press Add to select. Continue to add new selections to the ROI manager.
- Use the Set Measurements tool to measure Mean Gray Area. When the number of desired cell bodies has been added to the ROI manager, select them all and then use the Multi-Measure tool. This will provide a readout of the mean gray area for each over the time course of the video file. The exported data will give mean fluorescence intensity for the region of interest for each frame.
- Determine fluorescence intensity ratio, F/F0, where F is the fluorescence intensity at a given time and F0 is the initial fluorescence intensity. F/F0 can be graphed over time to examine spontaneous activity in neurons or examined at the maximum fluorescent intensity in the setting of stimulation.
Representative Results
Protocol Validation
The iPSC-derived microglia were generated from seven iPSC lines over three different rounds of differentiation. Control iPSC lines ML27, ML56, ML292, and ML364 and schizophrenia iPSC lines ML40, ML141, and ML250 were utilized. Characterization of these iPSC lines have been described previously51. These iPSC-derived microglia were validated using ICC and qPCR. Microglia generated from the adapted protocol exhibited typical ramified microglial morphology (Figure 1C), and expressed microglial markers CD11c, TMEM119, and IBA1, as examined by immunocytochemistry (Figure 1D,E). Cells with nuclei expressing microglial markers CD11c, P2RY12, and IBA1 were quantified. CD11c, P2RY12, IBA1 and TMEM119 were present in 63%, 60%, 65%, and 44% of the cells respectively, which is consistent with data described in the original differentiation protocol paper49. These experiments were performed with iPSC lines ML27, ML40, ML141, and ML250. Expression of specific genes was examined using qPCR to confirm the expression of microglial genes AIF1, CX3CR1, ITGAM, ITGAX, P2RY12, and TMEM119 (Figure 1F). This data was obtained from iPSC-derived microglia from two lines and normalized to an iPSC line. The SYBR Green real-time PCR protocol was used.
Dendritic Spines
Cortical neurons and microglia in co-culture were visualized using confocal microscopy (Figure 2A). Dendritic spines were quantified in the co-cultured cortical neurons (Figure 2B). Co-cultures were analyzed to determine the proportion of microglia and cortical neurons by marker analysis using P2RY12 and MAP2 to identify microglia and neurons respectively. In these co-cultures, 32.5% of the cells were positive for P2RY12 and 37.7% of the cells were positive for MAP2. Cortical neurons co-cultured with microglia treated with IFN-γ exhibited no significant differences in spine count, spine length, and neurite length when compared to cortical neurons co-cultured with untreated microglia (Figure 2D). This data was collected from the four control iPSC lines ML27, ML56, ML292, ML364, with two separate wells per experimental condition and ten images obtained from each well.
Calcium Imaging
Neurons co-cultured with microglia were stained with calcium fluorescence indicator in order to examine differences in neuronal firing with stimulation from glutamate with and without IFN-γ induced microglial activation (Figure 2E). Cortical neurons co-cultured with microglia treated with IFN-γ showed significant reduction in fluorescence intensity after stimulation with glutamate compared to cortical neurons co-cultured with untreated microglia (Figure 2F). This data was collected for three healthy control iPSC lines ML27, ML56, and ML292, with two wells per experimental condition and five images obtained from each well.
Supplementary Figure 1 further validates antibodies and staining protocol.
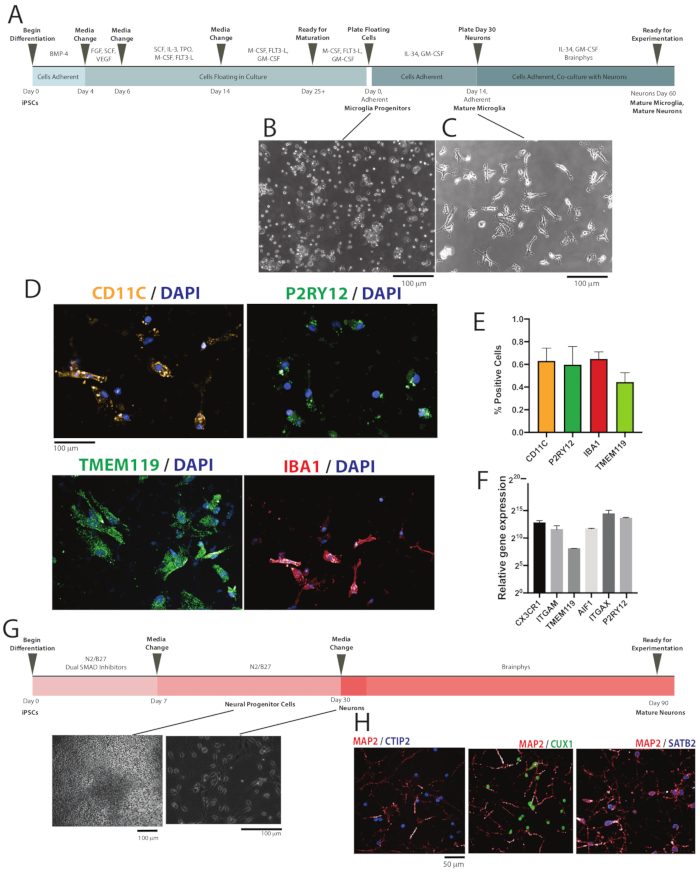
Figure 1: Differentiation and validation of iPSC-derived microglia. (A) Schematic depiction of microglial differentiation from iPSCs through microglial maturity and co-culture with cortical neurons. (B) Representative image of microglial progenitor cells after re-plating following day 25 of differentiation at 10x magnification. (C) Representative image of microglia in monoculture at day 14 at 10x magnification. (D) Immunocytochemistry staining of CD11c, P2RY12, IBA1, and TMEM119 to confirm expression of microglial markers, shown at 20x magnification. (E) Percentage of cells positively stained for microglial markers CD11c, P2RY12, IBA1, and TMEM119. (F) qPCR validation showing microglia exhibiting microglial-signature genes including AIF1, CX3CR1, ITGAM, ITGAX, P2RY12, and TMEM119. (G) Schematic depicting cortical neuron differentiation from iPSCs. (H) Immunocytochemistry staining of CTIP2, CUX1, SATB2, and MAP2 to confirm generation of cortical neurons, shown at 63X magnification. Please click here to view a larger version of this figure.
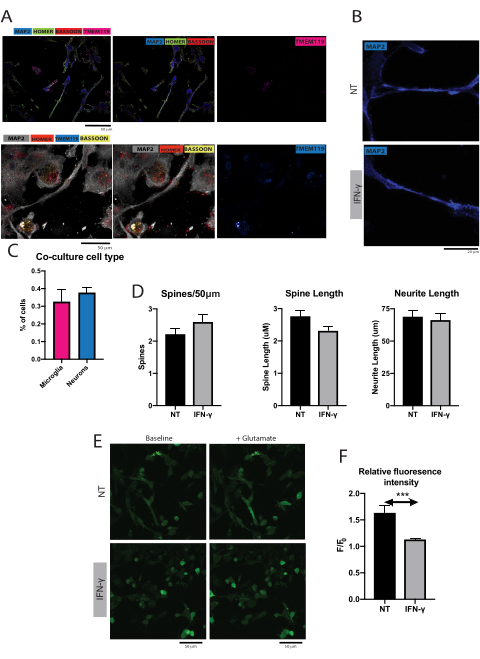
Figure 2. Functional changes in microglia and neuron co-cultures with and without interferon-gamma treatment. (A) Representative image of microglia and cortical neuron co-culture, with neurons stained for MAP2, pre-synaptic marker bassoon and post-synaptic marker homer, and microglia stained for TMEM119. (B) Representative image of dendritic spines in microglia/neuron co-cultures, with and without IFN-γ treatment. Scale bar = 50 μm. (C) Percentage of cells positive for P2RY12 or MAP2 in the co-cultures (mean + SEM). (D) Cortical neurons co-cultured with microglia treated with IFN-γ showed no significant differences when compared to untreated microglia in terms of spine count (Mann-Whitney test, P > 0.05, mean + SEM), spine length (Mann-Whitney test, P > 0.05, mean + SEM), or neurite length (unpaired t test, P > 0.05, mean + SEM). (E) Representative images of cortical neurons co-cultured with microglia in the presence of the calcium indicator Fluo-4AM, before and after glutamate stimulation and with and without IFN-γ treatment. (F) Cortical neurons co-cultured in the presence of microglia treated with IFN-γ had significantly lower fluorescence intensity with glutamate stimulation when compared to neurons co-cultured with untreated microglia (Mann-Whitney test, P = 0.0003, mean + SEM). Please click here to view a larger version of this figure.
| Protocol | Supplements used | Sorting stage | Length of Differentiation | Protein markers expiressed | Characterization | |||
| Abud et al. | FGF, BMP4, Activin A, LiCl, FGF, VEGF, TPO, SCF, IL-3, IL-6, M-CSF, IL-34, TGFb1, insulin, CD200, CX3CL1 | At day 10, isolate CD34+ cells | 38 days | CD45, CX3CR1, ITGB5, MERTK, PROS1, TGFbR1, P2RY12, TREM2 | Cytospin/Giemsa staining, transcriptomic profiling, RNA seq, flow cytometry, RT-QPCR, cell type analysis, flow cytometry, motility assay, inflamation response assay, phagocytosis assay, transplantation | |||
| Douvaras et al. | BMP4, FGF, SCF, VEGF, IL-3, TPO, M-CSF, Flt-3L, GM-CSF, IL-34 | At day 25, for CD14+/ CX3CR1+ cells | 40 days | CD11b, CD11c, CX3CR1, IBA1, P2RY12, TMEM119 | ICC, RT-QPCR, cell type analysis, flow cytometry, RNA seq, calcium assay, motility assay | |||
| Haenseler et al. | BMP4, SCF, VEGF, M-CSF, IL-3, GM-CSF | None | 42 days | CD11b, CD14, CD45, IBA1, MERTK | ICC, RT-QPCR, cell type analysis, flow cytometry, motility assay, inflamation response assay | |||
| Mcquade et al. | IL-34, TGF-β1, M-CSF, CLxCL1, CD200 | None | 38 days | P2RY12, TMEM119 | ICC, RNA seq, phagocytosis assay, transplantation assay | |||
| Muffat et al. | M-CSF, IL-34 | None | 74 days | CD11B, IBA1, P2RY12 TMEM119 | ICC, RNA seq, flow cytometry, cell size comparison Endotoxin response, motility assay | |||
Table 1: Overview of current protocols for differentiation of iPSCs to microglial cells.
| Day | Medium | Cytokines | Concentration |
| 0-3 | Stem Cell Medium | BMP-4 | 80ng/mL |
| 4-5 | Hematopoietic Medium | FGF | 25ng/mL |
| SCF | 100ng/mL | ||
| VEGF | 80ng/mL | ||
| 6-13 | Hematopoietic Medium | SCF | 50ng/mL |
| IL-3 | 50ng/mL | ||
| TPO | 5ng/mL | ||
| M-SCF | 50ng/mL | ||
| Flt3-Ligand | 50ng/mL | ||
| 14-25+ | Hematopoietic Medium | M-SCF | 50ng/mL |
| Flt3-Ligand | 50ng/mL | ||
| GM-SCF | 50ng/mL | ||
| Adherent | RPMI 1640 | GM-SCF | 25ng/mL |
| IL-34 | 100ng/mL |
Table 2: Overview of media used for microglial differentiation, listed with the concentration of cytokines used.
| Protocol | Day 0-3 Medium | Feeding method | Sorting | Supplements |
| Douvaras et al. | Custom medium; medium without Lithium Chloride, GABA, Pipecolic Acid, bFGF and TGFβ1 supplemented with 80ng/mL BMP4 | Every four days, cells pelleted and resuspended in fresh medium | Isolation of CD14+ or CD14+CX3CR1+ progenitors via FACS sorting | |
| This protocol | iPSC medium supplemented with 80ng/mL BMP4 | Every four days fresh medium added on top of existing medium | None | Rock inhibitor (10μM) used after centrifugation |
Table 3: Brief overview of microglial differentiation protocol from Douvaras et al.49 and adaptations made in this study.
| Protocol | Neural Maintenance Medium | Neural Induction Medium | Supplements | Notable Differences | ||
| Shi et al. 201250 | N2/B27 medium. N2 medium: Basal medium with 1% N-2 supplement, 1% 200mM L-alanyl-L-glutamine dipeptide in 0.85% NaCl solution, 1% pen/strep, 5μg/mL insulin, 1mM ;-glutamine, 100μM non-essential amino acids, 100μM 2-mercaptoethanol. B27 medium: DMEM/F12 supplemented with 2% B-27 supplement, 200mM L-glutamine, 1% pen/strep. | N2/B27 medium supplemented with 10μM SB431542 and 100ng/mL noggin OR 10μM SB431542 and 1μM dorsomorphin. | 20ng/mL FGF2 upon appearance of rosettes | Use of Insulin, NEAA, 2-mercaptoethanol | ||
| This protocol | N2/B27 medium. N2 medium: Basal medium with 1% N-2 supplement, 1% 200mM L-alanyl-L-glutamine dipeptide in 0.85% NaCl solution, 1% pen/strep. B27 medium: DMEM/F12 supplemented with 2% B-27 supplement, 1% 200mM L-alanyl-L-glutamine dipeptide in 0.85% NaCl solution, 1% pen/strep. | N2/B27 medium supplemented with 10μM SB431542, 1μM dorsomorphin, 100nM LDN193189. | Use of 200mM L-alanyl-L-glutamine dipeptide in 0.85% NaCl solution in B27 medium, use of 100nM LDN193189 in Neural Induction Medium | |||
Table 4: Brief overview of neuronal differentiation protocol from Shi et al.50 and adaptations made in this study.
Supplementary Figure 1: (A) Control iPSCs stained Microglia specific antibodies CD11c, P2RY12 and (B) TMEM119 and Iba1 to ensure these antibodies do not stain non-microglia cells. Control iPSC line did not exhibit any positive staining for these markers. (C) Co-cultures analyzed by cell type visible by specific line. Co-cultures were stained with a neuronal marker MAP2 or microglial marker P2RY12. Please click here to download this File.
Supplementary Table 1: List of primary antibodies used in this protocol and their optimal dilutions. Please click here to download this Table.
Supplementary Table 2: List of primers used for RT PCR experiments with forward and reverse sequences. Please click here to download this Table.
Discussion
The development of differentiation methods along different trajectories for pluripotent stem cells have opened many avenues for the investigation of brain function and disease processes53,54,55. Initial studies had focused on the development of specific neuronal cell types hypothesized to be important in specific brain disorders56,57. Recently, brain organoids have also provided new ways to study disease biology using patient-specific three-dimensional models51,58. The two-dimensional and three-dimensional cellular models provide specific advantages when trying to tackle different scientific questions59. While the early studies focused on cells along the neuronal lineage, recent developments now enable generation of other cell types in the brain, i.e. microglial cells and brain microvascular endothelial cells60,61. While studying the development of these cell types have provided valuable information and knowledge, it is important understand the interplay between these cell types to fully understand the role of neuroimmune and neurovascular interactions on brain function and development. This paper provides a detailed protocol on generating and co-culturing cortical neurons and microglia derived from the same iPSC line in order to develop personalized in vitro co-culture models to study the effects of different cell types on neuronal biology. By optimizing a method to efficiently differentiate microglia from iPSCs, we are now poised to examine disease-specific phenotypes using microglia and cortical neurons differentiated from iPSCs from disease subjects. Furthermore, this experimental approach allows for cross-culturing of microglia and neurons from control and disease subjects that can be leveraged to delineate specific contributions of microglia and neurons in disease-related phenotypes.
A number of rodent studies have examined co-cultures of microglia and neurons. A tri-culture with astrocytes, microglia and primary neurons reported significant improvement in neuronal health and reduced caspase 3/7 activity in the co-cultures62. An additional co-culture protocol for primary cerebellar granule neurons and primary cortical microglia from mice revealed that the use of co-culture methodology helps prevent the negative effect of toxicants on neuronal function and survival by mediating the release of cytokines63. These findings suggest that it is important to study co-cultures of different cell types to accurately depict relevant neurobiology and highlight the need for these experiments to be undertaken using human cells to understand the role neuro-immune interactions in psychiatric disease biology.
Only one previous study has investigated co-cultures of human iPSC-derived microglia or precursor macrophages with cortical neurons48. Co-cultures of embryonic MYB-independent iPSC-derived macrophages with iPSC-derived cortical neurons found that co-culture with microglia led to downregulation in pathogen-response pathways, upregulation in homeostatic function pathways, and promoted an anti-inflammatory and pro-remodeling cytokine response than corresponding monocultures, further suggesting the important crosstalk between microglia with neurons that can be recapitulated in in vitro co-culture models48. This study used iPSC-derived neurons to help mature MYB-independent iPSC-derived macrophages into microglial cells whereas the current protocol utilizes iPSC-derived microglia that had been differentiated to maturity separately. Given the significant interplay between these cell types, using a methodology where the maturation of microglia is dependent on the health of the neurons may present challenges and confounding factors when studying disease-related phenotypes.
There are three critical steps in this protocol that should be followed to ensure success. First, spinning cells for medium change for the floating microglial cultures should be kept to a minimum, which is why it is outlined that media should be added on top of the existing media during this stage instead of exchanging media. This step helps prevent loss and death of differentiating microglial cells. Second, when plating cortical neurons onto microglial cultures, it is crucial to add laminin to the medium in order to ensure that neurons will adhere to the plate. Neurons have a tendency to lift off of the plate and adding laminin helps prevent this. Third, for calcium imaging experiments, ensure that the confocal microscope used is equipped with an incubation chamber for live-cell imaging. This allows the cells to remain healthy throughout the experiment and prevents variability in data due to the timing of when the imaging data was collected.
This protocol adapted an aspect of the Fossati group protocol49 that had led to significant loss and death of differentiating microglial cells. Rather than collecting the supernatant and floating cell mixture and pelleting the cells every four days as originally described, fresh media was added on top to prevent cell loss and death in the centrifugation process. Cells were grown in 75mm flasks rather than in 6-well plates in order to maximize volume of media that could be added during this stage.
This protocol also removed a sorting step described in the Fossatti group protocol49. The sorting step resulted in a much lower cellular yield and high cell death in our hands. Hence, floating cells at day 25+ were plated without using this sorting step. Cells differentiated using this modified approach had ramified morphology characteristic of microglial cells, expressed microglial genes and showed robust staining of microglial markers. The percentage of cells expressing microglial markers is similar to the data in the original protocol49, suggesting that the purity of these cells is not significantly affected by removal of this sorting step.
Though the specific protocols for generating co-culturing iPSC-derived microglia and cortical neurons involved adapting from two well-established protocols, the microglia generated with this adapted approach have not been as extensively characterized as the original protocol. The downside of this protocol relates to length and cost of differentiation. The differentiation process takes at least 40 days and requires the use of a number of expensive reagents, especially the cytokines. Also, in this report, IFN-γ was used as an activator of microglia, but it should also be noted that this pro-inflammatory cytokine can elicit direct changes in cortical neurons as well64 and there is a need to undertake further studies to delineate the effect of cytokines on microglia and neurons.
These experiments provide proof-of-concept approaches to examine the effect of microglia on neuronal biology, which sets the stage for interesting explorations of various facets of neuro-immune interactions in the context of disease biology using microglia and neurons generated from patients with specific neurodevelopmental and neuropsychiatric disorders. The ability to cross-culture microglia and neurons from healthy subjects and disease subjects provide interesting avenues to dissect the specific roles of these cell types in the manifestation of disease-related phenotypes. Furthermore, co-culture models can be expanded to include astrocytes, oligodendrocytes and endothelial cells in order to develop novel in vitro models that reflect the different niches found in the brain.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by a National Institute of Mental Health Biobehavioral Research Awards for Innovative New Scientists (BRAINS) Award R01MH113858 (to R.K.), National Institute of Mental Health Clinical Scientist Development Award K08MH086846 (to R.K.), the Doris Duke Charitable Foundation Clinical Scientist Development Award (to R.K.), the Ryan Licht Sang Bipolar Foundation (to R.K.), the Jeanne Marie Lee-Osterhaus Family Foundation and the NARSAD Young Investigator Award from the Brain & Behavior Research Foundation (to A.K.), the Phyllis & Jerome Lyle Rappaport Foundation (to R.K.), the Harvard Stem Cell Institute (to R.K.) and by Steve Willis and Elissa Freud (to R.K.). We would like to thank Dr. Bruce M. Cohen and Dr. Donna McPhie from Harvard Medical School and McLean Hospital for providing us with the fibroblasts used in the study.
Materials
| Accutase | Sigma-Aldrich | A6964 | |
| B-27 supplement | Gibco | 17504044 | |
| b-FGF | Peprotech | 100-18B | |
| BMP-4 | Peprotech | 120-05ET | |
| Brainphys | StemCell Technologies | 5790 | |
| CD11C antibody | Biolegend | 337207 | Dilution 1:200 |
| Costar Flat Bottom Cell Culture Plates | Corning | 07-200-83 | |
| Ctip2 antibody | Abcam | ab18465 | |
| CUTL1 monoclonal antibody | Abnova | H00001523-M01 | |
| DMEM/F-12, no phenol red | Gibco | 21041025 | |
| dorsomorphin | Sigma-Aldrich | P5499 | |
| DPBS, no calcium, no magnesium | Gibco | 14190144 | |
| Dulbecco's Modified Eagle Medium (DMEM) | Sigma-Aldrich | D6421 | |
| EasYFlask Cell Culture Flasks | Nunc | 156499 | |
| Fisherbrand Cell Lifters | Fisher Scientific | 08-100-240 | |
| Flt3-Ligand | Peprotech | 300-19 | |
| Fluo4-AM | Life Technologies | F-14201 | |
| Geltrex LDEV Free RGF BME 1 ML | ThermoFisher Scientific | A1413201 | |
| Glutamax | ThermoFisher Scientific | 35050061 | |
| GM-CSF | Peprotech | 300-03 | |
| Goat Anti Chicken- IgG H&L (Alexa Fluor 488) | Abcam | ab150169 | Dilution 1:1000 |
| Goat Anti mouse- IgG H&L (Alexa Fluor 568) | Invitrogen | A-11004 | Dilution 1:1000 |
| Goat Anti Rat- IgG H&L (Alexa Fluor 405) | Abcam | ab175670 | Dilution 1:1000 |
| Goat Anti-Guinea pig IgG H&L (Alexa Fluor 405) | Abcam | ab175678 | Dilution 1:1000 |
| Goat Serum | Sigma-Aldrich | G9023 | |
| HBSS | Invitrogen | 14170120 | |
| IBA1 antibody | Abcam | ab5076 | Dilution 1:500 |
| IL-34 | Peprotech | 200-34 | |
| INF-y | Peprotech | 300-02 | |
| KiCqStart SYBR Green Primers | Sigma-Aldrich | KSPQ12012 | |
| Laminin | Sigma-Aldrich | L2020 | |
| LDN193189 | Sigma-Aldrich | SML0599 | |
| Live Cell Imaging Solution | Invitrogen | A14291DJ | |
| MAP2 antibody | Synaptic Systems | 188 004 | |
| M-CSF | Peprotech | 300-25 | |
| N-2 supplement | Gibco | 17502001 | |
| Neurobasal medium | Life Technologies | 21103049 | |
| NutriStem hPSC XF Medium | Biological Industries | 01-0005 | |
| P2RY12 antibody | Biolegend | 848002 | |
| Paraformaldehyde 16% | Fisher Scientific | 50-980-488 | |
| Penicillin-streptomycin | Gibco | 15140122 | |
| Poly-L-Orthinine | Sigma-Aldrich | P3655 | |
| SATB2 antibody | Abcam | ab51502 | |
| SB431542 | Sigma-Aldrich | S4317 | |
| SCF | Stemcell Technologies | 78062 | |
| SensoPlate 24-Well Glass-Bottom Plate | Greiner-Bio | 662892 | |
| StemPro-34 SFM (1X) | Gibco | 10639011 | |
| TMEM119 antibody | Abcam | ab185333 | Dilution 1:1000 |
| TPO | Peprotech | 300-18 | |
| Triton-X | Sigma-Aldrich | 9002-93-1 | |
| VEGF | Peprotech | 100-20 | |
| Versene | ThermoFisher Scientific | 15040066 | |
| Y-27632 dihydrochloride (ROCK inhibitor) | Tocris | 1254 |
References
- Lenz, K. M., Nelson, L. H. Microglia and beyond: Innate immune cells as regulators of brain development and behavioral function. Frontiers in Immunology. 9, 698 (2018).
- Hoshiko, M., Arnoux, I., Avignone, E., Yamamoto, N., Audinat, E. Deficiency of the microglial receptor CX3CR1 impairs postnatal functional development of thalamocortical synapses in the barrel cortex. Journal of Neuroscience. 32 (43), 15106-15111 (2012).
- Cunningham, C. L., Martínez-Cerdeño, V., Noctor, S. C. Microglia regulate the number of neural precursor cells in the developing cerebral cortex. Journal of Neuroscience. 33 (10), 4216-4233 (2013).
- Hagemeyer, N., et al. Microglia contribute to normal myelinogenesis and to oligodendrocyte progenitor maintenance during adulthood. Acta Neuropathologica. 134 (3), 441-458 (2017).
- Lenz, K. M., Nugent, B. M., Haliyur, R., McCarthy, M. M. Microglia are essential to masculinization of brain and behavior. Journal of Neuroscience. 33 (7), 2761-2772 (2013).
- Miyamoto, A., et al. Microglia contact induces synapse formation in developing somatosensory cortex. Nature Communications. 7 (1), 1-12 (2016).
- Pont-Lezica, L., et al. Microglia shape corpus callosum axon tract fasciculation: Functional impact of prenatal inflammation. European Journal of Neuroscience. 39 (10), 1551-1557 (2014).
- Schafer, D. P., et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent. Neuron. 74 (4), 691-705 (2012).
- Hornik, T. C., Vilalta, A., Brown, G. C. Activated microglia cause reversible apoptosis of pheochromocytoma cells, inducing their cell death by phagocytosis. Journal of Cell Science. 129 (1), 65-79 (2016).
- Brown, G. C., Neher, J. J. Microglial phagocytosis of live neurons. Nature Reviews Neuroscience. 15 (4), 209-216 (2014).
- Sierra, A., et al. Microglia shape adult hippocampal neurogenesis through apoptosis-coupled phagocytosis. Cell Stem Cell. 7 (4), 483-495 (2010).
- Schafer, D. P., et al. Microglia contribute to circuit defects in Mecp2 null mice independent of microglia-specific loss of Mecp2 expression. Elife. 5, 15224 (2016).
- Vainchtein, I. D., et al. Astrocyte-derived interleukin-33 promotes microglial synapse engulfment and neural circuit development. Science. 359 (6381), 1269-1273 (2018).
- Datwani, A., et al. Classical MHCI molecules regulate retinogeniculate refinement and limit ocular dominance plasticity. Neuron. 64 (4), 463-470 (2009).
- Färber, K., et al. C1q, the recognition subcomponent of the classical pathway of complement, drives microglial activation. Journal of Neuroscience Research. 87 (3), 644-652 (2009).
- Wlodarczyk, A., et al. A novel microglial subset plays a key role in myelinogenesis in developing brain. The EMBO Journal. 36 (22), 3292-3308 (2017).
- Lim, S. -. H., et al. Neuronal synapse formation induced by microglia and interleukin 10. PloS One. 8 (11), 81218 (2013).
- Squarzoni, P., et al. Microglia modulate wiring of the embryonic forebrain. Cell Reports. 8 (5), 1271-1279 (2014).
- Shin, W. H., et al. Microglia expressing interleukin-13 undergo cell death and contribute to neuronal survival in vivo. Glia. 46 (2), 142-152 (2004).
- Giulian, D., Li, J., Leara, B., Keenen, C. Phagocytic microglia release cytokines and cytotoxins that regulate the survival of astrocytes and neurons in culture. Neurochemistry International. 25 (3), 227-233 (1994).
- Petanjek, Z., et al. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proceedings of the National Academy of Sciences. 108 (32), 13281-13286 (2011).
- Giedd, J. N., et al. Brain development during childhood and adolescence: A longitudinal MRI study. Nature Neuroscience. 2 (10), 861-863 (1999).
- Lewis, D. A., Lieberman, J. A. Catching up on schizophrenia: Natural history and neurobiology. Neuron. 28 (2), 325-334 (2000).
- Jablensky, A. Epidemiology of schizophrenia: The global burden of disease and disability. European Archives of Psychiatry and Clinical Neuroscience. 250 (6), 274-285 (2000).
- Van Berckel, B. N., et al. Microglia activation in recent-onset schizophrenia: A quantitative (R)-[11C] PK11195 positron emission tomography study. Biological Psychiatry. 64 (9), 820-822 (2008).
- Doorduin, J., et al. Neuroinflammation in schizophrenia-related psychosis: a PET study. Journal of Nuclear Medicine. 50 (11), 1801-1807 (2009).
- Van Kesteren, C., et al. Immune involvement in the pathogenesis of schizophrenia: a meta-analysis on postmortem brain studies. Translational Psychiatry. 7 (3), 1075 (2017).
- Bloomfield, P. S., et al. Microglial activity in people at ultra high risk of psychosis and in schizophrenia: An [11C] PBR28 PET brain imaging study. American Journal of Psychiatry. 173 (1), 44-52 (2016).
- Fillman, S., et al. Increased inflammatory markers identified in the dorsolateral prefrontal cortex of individuals with schizophrenia. Molecular Psychiatry. 18 (2), 206-214 (2013).
- Radewicz, K., Garey, L. J., Gentleman, S. M., Reynolds, R. Increase in HLA-DR immunoreactive microglia in frontal and temporal cortex of chronic schizophrenics. Journal of Neuropathology & Experimental Neurology. 59 (2), 137-150 (2000).
- Steiner, J., et al. Distribution of HLA-DR-positive microglia in schizophrenia reflects impaired cerebral lateralization. Acta Neuropathologica. 112 (3), 305-316 (2006).
- De Picker, L. J., Morrens, M., Chance, S. A., Boche, D. Microglia and brain plasticity in acute psychosis and schizophrenia illness course: a meta-review. Frontiers in Psychiatry. 8, 238 (2017).
- Garey, L., et al. Reduced dendritic spine density on cerebral cortical pyramidal neurons in schizophrenia. Journal of Neurology, Neurosurgery & Psychiatry. 65 (4), 446-453 (1998).
- Glantz, L. A., Lewis, D. A. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Archives of General Psychiatry. 57 (1), 65-73 (2000).
- Konopaske, G. T., Lange, N., Coyle, J. T., Benes, F. M. Prefrontal cortical dendritic spine pathology in schizophrenia and bipolar disorder. JAMA Psychiatry. 71 (12), 1323-1331 (2014).
- Petanjek, Z., et al. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proceedings of the National Academy of Sciences U. S. A. 108 (32), 13281-13286 (2011).
- Sekar, A., et al. Schizophrenia risk from complex variation of complement component 4. Nature. 530 (7589), 177-183 (2016).
- Landry, R. P., Jacobs, V. L., Romero-Sandoval, E. A., DeLeo, J. A. Propentofylline, a CNS glial modulator does not decrease pain in post-herpetic neuralgia patients: In vitro evidence for differential responses in human and rodent microglia and macrophages. Experimental Neurology. 234 (2), 340-350 (2012).
- Sievers, J., Parwaresch, R., Wottge, H. U. Blood monocytes and spleen macrophages differentiate into microglia-like cells on monolayers of astrocytes: morphology. Glia. 12 (4), 245-258 (1994).
- Simard, A. R., Rivest, S. Bone marrow stem cells have the ability to populate the entire central nervous system into fully differentiated parenchymal microglia. The FASEB Journal. 18 (9), 998-1000 (2004).
- Asheuer, M., et al. Human CD34+ cells differentiate into microglia and express recombinant therapeutic protein. Proceedings of the National Academy of Sciences U. S. A. 101 (10), 3557-3562 (2004).
- Ohgidani, M., et al. Direct induction of ramified microglia-like cells from human monocytes: Dynamic microglial dysfunction in Nasu-Hakola disease. Scientific Reports. 4 (1), 1-7 (2014).
- Melief, J., et al. Characterizing primary human microglia: A comparative study with myeloid subsets and culture models: Characterizing primary human microglia. Glia. 64 (11), 1857-1868 (2016).
- Muffat, J., et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nature Medicine. 22 (11), 1358-1367 (2016).
- Abud, E. M., et al. iPSC-derived human microglia-like cells to study neurological diseases. Neuron. 94 (2), 278-293 (2017).
- Pandya, H., et al. Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nature Neuroscience. 20 (5), 753-759 (2017).
- Pocock, J. M., Piers, T. M. Modelling microglial function with induced pluripotent stem cells: an update. Nature Reviews Neuroscience. 19 (8), 445-452 (2018).
- Haenseler, W., et al. A highly efficient human pluripotent stem cell microglia model displays a neuronal-co-culture-specific expression profile and inflammatory response. Stem Cell Reports. 8 (6), 1727-1742 (2017).
- Douvaras, P., et al. Directed differentiation of human pluripotent stem cells to microglia. Stem Cell Reports. 8 (6), 1516-1524 (2017).
- Shi, Y., Kirwan, P., Livesey, F. J. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nature Protocols. 7 (10), 1836-1846 (2012).
- Kathuria, A., et al. Transcriptomic landscape and functional characterization of induced pluripotent stem cell-derived cerebral organoids in schizophrenia. JAMA Psychiatry. 77 (7), 745-754 (2020).
- Meijering, E., et al. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry Part A: The journal of the International Society for Analytical Cytology. 58 (2), 167-176 (2004).
- Watmuff, B., et al. Disease signatures for schizophrenia and bipolar disorder using patient-derived induced pluripotent stem cells. Molecular and Cellular Neuroscience. 73, 96-103 (2016).
- Karmacharya, R., Haggarty, S. J. Stem cell models of neuropsychiatric disorders. Molecular and Cellular Neurosciences. 73, 1 (2016).
- Karmacharya, R., Kieling, C., Mondelli, V. Integrating stem cell-based experiments in clinical research. European Psychiatry. 63 (1), (2020).
- Kathuria, A., et al. Synaptic deficits in iPSC-derived cortical interneurons in schizophrenia are mediated by NLGN2 and rescued by N-acetylcysteine. Translational Psychiatry. 9 (1), 1-13 (2019).
- Watmuff, B., Liu, B., Karmacharya, R. Stem cell-derived neurons in the development of targeted treatment for schizophrenia and bipolar disorder. Pharmacogenomics. 18 (5), 471-479 (2017).
- Kathuria, A., et al. Transcriptome analysis and functional characterization of cerebral organoids in bipolar disorder. Genome Medicine. 12, 1-16 (2020).
- Kathuria, A., Lopez-Lengowski, K., Watmuff, B., Karmacharya, R. Comparative transcriptomic analysis of cerebral organoids and cortical neuron cultures derived from human induced pluripotent stem cells. Stem Cells and Development. 29 (21), 1370-1381 (2020).
- Pong, S., Lizano, P., Karmacharya, R. Derivation, expansion, cryopreservation and characterization of brain microvascular endothelial cells from human induced pluripotent stem Cells. Journal of Visualized Experiments: Jove. (165), e61629 (2020).
- Pong, S., Karmacharya, R., Sofman, M., Bishop, J. R., Lizano, P. The Role of Brain Microvascular Endothelial Cell and Blood-Brain Barrier Dysfunction in Schizophrenia. Complex Psychiatry. 6 (1-2), 30-46 (2020).
- Goshi, N., Morgan, R. K., Lein, P. J., Seker, E. A primary neural cell culture model to study neuron, astrocyte, and microglia interactions in neuroinflammation. Journal of Neuroinflammation. 17 (1), 155 (2020).
- Roqué, P. J., Costa, L. G. Co-Culture of Neurons and Microglia. Current Protocols in Toxicology. 74 (1), 11-17 (2017).
- Warre-Cornish, K., et al. Interferon-γ signaling in human iPSC-derived neurons recapitulates neurodevelopmental disorder phenotypes. Science Advances. 6 (34), (2020).

.
