Engineered Lung Tissues Prepared from Decellularized Lung Slices
Summary
This protocol describes a method to generate reproducible, small-scale engineered lung tissues, by repopulating decellularized precision-cut lung slices with alveolar epithelial type 2 cells, fibroblasts, and endothelial cells.
Abstract
There is a need for improved 3-dimensional (3D) lung models that recapitulate the architectural and cellular complexity of the native lung alveolus ex vivo. Recently developed organoid models have facilitated the expansion and study of lung epithelial progenitors in vitro, but these platforms typically rely on mouse tumor-derived matrix and/or serum, and incorporate just one or two cellular lineages. Here, we describe a protocol for generating engineered lung tissues (ELTs) based on the multi-lineage recellularization of decellularized precision-cut lung slices (PCLS). ELTs contain alveolar-like structures comprising alveolar epithelium, mesenchyme, and endothelium, within an extracellular matrix (ECM) substrate closely resembling that of native lung. To generate the tissues, rat lungs are inflated with agarose, sliced into 450 µm-thick slices, cut into strips, and decellularized. The resulting acellular ECM scaffolds are then reseeded with primary endothelial cells, fibroblasts, and alveolar epithelial type 2 cells (AEC2s). AEC2s can be maintained in ELT culture for at least 7 days with a serum-free, chemically-defined growth medium. Throughout the tissue preparation and culture process, the slices are clipped into a cassette system that facilitates handling and standardized cell seeding of multiple ELTs in parallel. These ELTs represent an organotypic culture platform that should facilitate investigations of cell-cell and cell-matrix interactions within the alveolus as well as biochemical signals regulating AEC2s and their niche.
Introduction
Alveoli are the functional units of the distal lung, comprising a meshwork of gas-exchanging airspaces lined by alveolar epithelial type 1 cells (AEC1s) and type 2 cells (AEC2s). Underlying the epithelium is a dense network of capillaries as well as supporting mesenchyme, all buttressed by an extracellular matrix (ECM) scaffold that provides both strength and flexibility to these delicate air sacs1. The alveoli are also the site of injury in numerous lung pathologies, including idiopathic pulmonary fibrosis2, acute respiratory distress syndrome3, and severe coronavirus disease-19 (COVID-19)4. Although work over the past decade has uncovered a remarkable plasticity within the lung epithelium, the mechanisms that enable distal lung repair in some settings – and that preclude repair in others – remain an area of intense investigation5. The development of improved in vitro platforms to model the alveolus would facilitate studies of alveolar biology, regeneration, and therapeutics.
AEC2s self-renew and differentiate into AEC1s, and thus are considered the primary stem cell of the distal lung6,7,8. However, these cells pose a particular challenge to in vitro study given the difficulties associated with culturing primary AEC2s without a loss of phenotype9. In conventional 2-dimensional (2D) culture, AEC2s flatten and adopt some features of AEC1-like cells10. In contrast, 3D culture strategies, most commonly organoids, support the maintenance of differentiated features in primary AEC2s6,11,12 and permit long-term culture of pluripotent stem cell (PSC)-derived AEC2s13,14. Organoids have been used to model distal lung development15, viral infection11,15, and AEC2-related genetic disease13,16,17, enabling important insights into AEC2 biology and regeneration. However, these culture models typically comprise just one or two cellular lineages, and embed the cells in gel-type matrices that fail to recapitulate either the architecture or the ECM substrate of the native lung alveolus.
The ECM is a critical regulator of cell phenotype and behavior via molecular, topological, and mechanical cues; comprises a key component of tissue-specific niches regulating stem cell fate; and serves as a reservoir that modulates the availability of locally-secreted growth factors18,19,20,21. Culturing cells on native ECM may thus increase the predictive capacity of in vitro systems to model the biology of in vivo tissues. Decellularization, a process that removes cellular material from tissues via detergents, enzymes, or physical or other methods, can preserve in large part the ECM scaffolding of a native organ, when carefully performed22,23. Such scaffolds can be repopulated with cells for 3D biomimetic culture. However, while decellularized scaffolds are widely used for tissue engineering applications, their use for routine cell culture has been limited. Several previous studies have reported the decellularization and recellularization of lung slices or small lung tissue segments. In addition to proof-of-concept studies24,25,26, repopulated lung slices have been used to study fibroblast-matrix adhesion27,28 and to investigate the effect of diseased lung matrices on fibroblast phenotype27,29. With improved technologies available for generating precision-cut tissue slices, decellularized lung slices could offer a convenient and small-scale platform with which to culture cells, while preserving alveolar, airway, and vascular substructures. Incorporating multiple cell types would enable studies of cell-cell interactions within a physiologically relevant 3D environment. However, improved strategies are needed to facilitate the handling of tissues throughout the culture process, and to ensure controlled and reproducible seeding of tissues with known numbers of cells.
Here, we present a protocol to generate engineered lung tissues (ELTs) by repopulating decellularized precision-cut lung slices (PCLS) with primary endothelial cells, AEC2s, and fibroblasts. In an adaptation of our previously described engineered heart tissue system30 and whole lung decellularization-recellularization strategies22,31, we describe procedures to cut PCLS from rat lungs and to clip the slices into reusable tissue culture cassettes that simplify and standardize downstream manipulations. Clipped slices are decellularized to form acellular ECM scaffolds, which are repopulated in customized seeding baths. Lung slice scaffolds preserve critical ECM components and architecture, and support the growth of AEC2s within multi-lineage alveolar-like structures for at least 7 days. ELTs represent a novel alveolar co-culture system within a physiologically relevant 3D matrix, which should support the development of lung tissue engineering strategies, while facilitating basic biological studies of AEC2s and the alveolus.
Protocol
All animal experimental procedures described in this paper were approved by the Yale Institutional Animal Care and Use Committee.
1. Creation of Tissue Culture Cassettes and Seeding Baths
NOTE: Once made, tissue culture cassettes and seeding baths may be autoclaved and reused for repeated rounds of ELT culture.
- Tissue Culture Cassettes
- Use a laser cutter to cut tissue culture cassette frames and clips out of 3/32 inch thick polytetrafluoroethylene (PTFE) according to the designs provided in Supplementary File 1 and Supplementary File 2, respectively. Use a laser cutter to cut tissue culture cassette tabs out of 1/16 inch thick PTFE according to Supplementary File 3. Cut outlines 3x using 80% power and 15% speed (for a 30 W laser cutter).
- Seeding Baths
- Use the seeding bath CAD files (Supplementary File 4 and Supplementary File 5) to 3D print the base and ring of the seeding bath mold, respectively, using clear resin.
- Soak the molds in a solution of 10% poloxamer 407 in distilled water overnight before use to aid in PDMS release. Let air dry, then fit the ring over the base of the mold and wrap in flexible plastic film to help prevent leaking.
- Prepare at least 60 g per mold of polydimethylsiloxane (PDMS) by mixing PDMS elastomer in a 10:1 ratio with curing agent, and pour into the 3D printed mold. Degas the PDMS in a vacuum desiccator for 30 min to remove any air bubbles.
- Bake seeding baths at 60 °C for 8 h.
2. Preparation of Precision-Cut Lung Slices from Rat Lungs
- Organ Harvest
- Prepare a split perfusion system comprising gravity- and pump-driven limbs, as pictured in Figure 1. Connect a pulmonary artery (PA) cannula to the end of the tubing, comprising a 1/16 inch barbed Y-connector attached to a ½ inch length of LS 14 silicone tubing and a 3/32 inch female luer-lock connector (see Figure 1). Do not attach a check valve to the cannula at this time.
- Prime the lines with PBS containing 100 U/mL heparin and 0.01 mg/mL sodium nitroprusside (SNP) for anticoagulation and vasodilation, respectively. Pre-set the perfusion pump to 30 mL/min.
NOTE: Add SNP fresh to the heparin solution and keep protected from light. - Dose an adult (8-12 week-old, approximately 300-350 g) Sprague-Dawley rat with an intraperitoneal (IP) injection of 400 U/kg heparin for anti-coagulation, followed by an IP injection of ketamine (75 mg/kg) and xylazine (5 mg/kg) for anesthesia. Confirm a surgical plane of anesthesia via lack of response to noxious stimulus (toe pinch).
- Trim the chest and abdomen of fur using hair clippers. Then spray with 70% ethanol and wipe 3x with 10% povidone-iodine.
- Grasp the skin below the level of the diaphragm with rat tooth forceps. Then make a ½ inch transverse incision in the skin with fine-pointed scissors. Grasp the exposed abdominal fascia with the forceps, make a ½ inch transverse incision in the fascia, and then extend the incision through the skin and fascia across the width of the upper abdomen.
- Use the tip of the fine scissors to make a small incision (not more than 1/8 inch) in the center of the anterior diaphragm, causing the lungs to retract in the thorax. Extend the incision in the diaphragm across the full width of the chest.
- Make two vertical incisions through the full height of the ribcage toward the neck, being careful not to damage the lungs. Extend the incision through the left ribs to cut through the collarbone and along the side of the neck to the level of the larynx, exposing the trachea.
- Dissect the trachea free from surrounding connective tissue and from the esophagus. Make a transverse incision across the anterior half of the trachea between two cartilage rings, close to the larynx. Thread a 4-0 polypropylene suture behind the trachea, below the level of the incision, and loosely pre-tie the first half of a surgeon's knot with two twists.
- Place a cannula comprising a 1/16 inch barbed Y-connecter connected to a one-way check valve and a ½ inch length of LS 14 silicone tubing with a 3/32 inch female luer-lock connector (see Figure 1) into the trachea by inserting one limb of the Y-connector into the tracheal incision toward the direction of the lungs.
- Position the pre-tied suture loop around the trachea at the level of the inserted cannula and tighten around the inserted Y-connector to secure the cannula in place. Add two single-twist throws of the suture to complete the knot.
- Fill a 10 mL syringe with air and connect to the luer-lock of the tracheal cannula.
- Clamp the inferior vena cava close to the diaphragm using a curved hemostat, then inject the heart with 150 U heparin (1000 U/mL) via the right ventricle (RV).
- Partially open the gravity line stopcock, to produce a slow but steady dripping of PBS/heparin/SNP from the PA cannula prepared in step 2.1.1.
- Thread the needle of a 4-0 polypropylene suture behind the base of the PA where it exits the RV. Use the first half of a surgeon's knot to pre-tie a loose loop of suture around the base of the PA.
- Make a small incision (not more than 1/8 inch) in the RV just below and perpendicular to the PA using fine scissors, then insert one limb of the PA cannula Y-connector into the base of the PA. Secure the suture around the PA and the inserted connector and add a single-twist throw to complete the surgeon's knot.
NOTE: Cannulating the PA under flow prevents the introduction of air bubbles into the vasculature that can preclude adequate clearing of the lungs. - Attach a one-way valve to the other end of the PA catheter Y-connector, then cut off the apex of the heart to allow blood flow efflux via the left ventricle.
NOTE: Failure to cut off the apex of the heart prior to perfusing via the pump can cause damage to the blood-gas barrier, leading to leakage of fluid into the airspaces. - Switch the perfusion line to the pump side using the stopcock connecting the two lines, then turn on the pump at 30 mL/min. While perfusing the lungs via the PA, manually ventilate the lungs via the 10 mL tracheal syringe at approximately 10-15 breaths/min, to facilitate clearing the lungs of blood. Perfuse the lungs until they turn mostly white, usually requiring 40 mL of PBS/heparin/SNP or less.
NOTE: Inadequate clearing of the lungs of blood may impair downstream decellularization. - Cut the posterior trachea just above the level of the tracheal cannula, and then dissect the lungs and heart free of all remaining connective tissue and extract the lungs and heart en bloc.
- Fill a 10 mL syringe with 2% low melting point agarose in Hank's balanced salt solution (HBSS) without phenol red, prewarmed to 42 °C.
NOTE: The exact volume of agarose required will vary by lung size. Larger lungs (i.e., from rats larger than 400 g) will require more than 10 mL agarose. - Manually inflate the extracted lungs 3x with 10 mL of air (i.e., to approximately total lung capacity) via the trachea cannula to help recruit collapsed parenchyma.
- Immediately inflate the lungs with the prepared syringe of agarose by manually injecting the agarose via the trachea cannula at a rate of approximately 40 mL/min, just until the most distal tips of the lung lobes are inflated. If distal regions of lung remain collapsed, inject an additional 1-2 mL of agarose.
- Cap the trachea by attaching the white cap from a 4-way stopcock to the female luer-lock of the tracheal cannula. Place the lung in a 150 mm Petri dish on ice to allow the agarose to solidify.
NOTE: Inflation of the lung shortly after extraction is critical to ensure uniform filling of the lung parenchyma, and subsequent successful tissue slicing. If the lung inflates very unevenly, do not proceed with lung slicing as slice quality will be poor.
- Lung Slicing
NOTE: Exact slicing procedure may need to be adapted based on the vibratory microtome (vibratome) being used; additional examples of PCLS preparation with various tissue slicers have been published previously32,33,34.- Pre-chill the metal chilling block at -20 °C and keep on ice when not using throughout the slicing procedure.
- Use a small drop of cyanoacrylate glue to attach a blade to the blade holder. Carefully attach the blade holder to the vibratome using an Allen wrench so that it just lines up with the end of a specimen tube inserted into the buffer tray.
- Prepare 6-well plates with 3 mL per well sterile ice-cold HBSS without phenol red to collect the slices.
- Using a scalpel, cut a piece of lung tissue approximately 1-1.5 cm3.
NOTE: Lung tissue from the lower and middle portions of the left lobe, as well as from the right middle and bottom lobes, most readily yields larger tissue slices that maximize alveolar area. If uninflated tissue regions or areas of connective tissue are present, either trim off this tissue with scissors or orient downward toward the plunger; such tissue tends not to cut cleanly. - Place a small drop of cyanoacrylate glue onto the plunger of the specimen tube. Dab lung tissue on a paper wipe to remove excess moisture, and then immediately place the lung tissue on top of the plunger using a pair of forceps.
- Slide the metal tube of the specimen tube up to the level of the top of the tissue and hold in place, with the plunger receded. Pipette pre-warmed 2% agarose in HBSS into the top of the tube to completely surround the tissue.
- Place the ice-cold chilling block around the tissue for about 1 min to allow the agarose to solidify.
- Insert the specimen tube into the buffer tray. Fill the tray with ice-cold PBS to mid-way up the tissue block. Turn the motor box switch to fast forward (FF) to advance the motor box plunger so that it is just touching the base of the specimen tube.
- Set the desired settings for slice thickness, cutting speed, and oscillation frequency, for example, 450 µm thickness, speed 4 and oscillation frequency 5. Select Continuous mode, and then flip the switch to On to begin slicing.
- As tissue slices fall into the buffer tray, transfer them to the prepared 6-well plates using an inoculating loop or spatula.
- Stop slicing when ~2 mm thickness of tissue remains in the specimen tube, to avoid damaging the blade or cutting tissue containing glue.
- Repeat the above steps to slice additional lung tissue, as desired.
- Decellularize slices immediately for scaffold preparation, or snap-freeze and store at -80 °C for up to 2 months. To freeze, transfer 4-6 slices to a 35 mm Petri dish and carefully aspirate any excess fluid from around the slices. Place the dishes in a bath of dry ice and 100% ethanol to snap-freeze, then wrap in foil, seal in a plastic bag, and transfer to -80 °C.
NOTE: Do not place fresh slices directly into a -80 °C freezer as the relatively slow rate of freezing can cause ice crystals to form that can damage the tissue.
3. Preparation of Lung Tissue Scaffolds
- Preparation of Materials and Decellularization Solutions
- Autoclave frames, clips, and tabs.
- Prepare decellularization solutions as outlined in Table 1.
NOTE: Add benzonase nuclease to pre-warmed buffer immediately before use and sterile filter. Prepare Triton X-100 and sodium deoxycholate (SDC) solutions within 24-48 h of the decellularization procedure. Prepare antibiotic/antimycotic solutions and benzonase buffer up to 30 d in advance and store at 4 °C.
- Cutting and Clipping Lung Slices
NOTE: While cutting and clipping may be done non-sterilely on the benchtop, the decellularization steps in section 3.3 and all subsequent handling of the tissue scaffolds must be performed in a laminar flow hood.- Fill a 100 mm Petri dish about one-third full with PBS. Transfer cassettes (frames containing two clips each) and tabs to the dish using forceps.
- If using frozen slices, thaw one dish at a time by pouring room temperature PBS into the dish to cover the slices. Keep remaining dishes on dry ice.
- Transfer a thawed slice to a 150 mm Petri dish. Gently unfold the slice using fine forceps, if necessary, so that it lies flat, then carefully aspirate excess PBS from around the tissue.
- Use a razor blade, with a ruler as a guide, to cut a 3 mm wide strip from the slice by pressing the full length of the blade firmly against the dish and rocking it slightly side to side with the blade edge held in place. Alternatively, use a rotary cutter retrofitted with 2 parallel blades separated by a 3 mm custom-made spacer (e.g., made of acetal [polyoxymethylene]) to cut tissue strips. Avoid any tears, holes, large airways or vessels, or thick connective tissue.
NOTE: For successful clipping, the strip must be at least 9 mm long. - Using forceps, transfer the tissue strip to the prepared 100 mm Petri dish.
- Clip the tissue strip into the cassette: float the tissue above the cassette, centering the tissue to overhang the holes in the clips at either end. With fine forceps, place a tab partly into the hole at one end, gently straighten the tissue, and then place the second tab. Using forceps in each hand, press each tab in completely to secure the tissue.
NOTE: If having difficulty keeping the tissue in place prior to clipping, aspirate some PBS from the dish to lower the fluid level. Be careful not to stretch the tissue in placing the second clip, as this may lead to tearing. - Repeat the thawing, cutting, and clipping procedure in steps 3.2.2-3.2.6 for as many tissues as desired.
- Slice Decellularization
- Once all slices are clipped, transfer the 100 mm dish containing the cassettes to a laminar flow hood.
- Begin step 1 of the decellularization protocol (see Table 2): using a curved hemostat to grasp the notched sides of each cassette, transfer cassettes to 6-well plates (2 tissues/well) filled with 3 mL of PBS + ions + antibiotics/antimycotics per well (see solution recipe in Table 1).
- Place well plates on an orbital shaker at 30 rpm for 10 min.
- Continue with step 2 of the decellularization protocol (see Table 2): aspirate the fluid from each well, then replace with 3 mL/well PBS + ions, place plate on orbital shaker at 30 rpm, and incubate for 5 min.
- Repeat step 3.3.4 for each of the solutions and corresponding durations as outlined in the decellularization protocol in Table 2.
- After the final rinse step with PBS + antibiotics/antimycotics (step 20 of Table 2), transfer tissues to sterile 6-well plates with fresh PBS + antibiotics/antimycotics, and incubate at 37 °C for 48 h.
NOTE: After sterilization with antibiotics/antimycotics, lung tissue scaffolds can be seeded immediately, or stored at 4 °C for up to 30 d.
4. Slice Recellularization and Culture
NOTE: Figure 2 shows a proposed timeline for tissue seeding and culture, in which slices are seeded first with rat lung microvascular endothelial cells and cultured in low-serum endothelial medium; then seeded with rat AEC2s and rat lung fibroblasts with a serum-free AEC2 growth medium (adapted from Jacob et al.13 and You et al.35); see additional notes on cell sources used in Results and culture media details in Table 3. This strategy yields alveolar-like structures containing AEC2 monolayers.
- Preparing Tissue Scaffolds for Seeding (Day -4 or -3)
- If using tissue scaffolds stored at 4 °C, incubate scaffolds overnight at 37 °C with fresh PBS + antibiotics/antimycotics (10% penicillin/streptomycin, 4% amphotericin B, 0.4% gentamicin in PBS) prior to seeding.
- Rinse scaffolds 3x with sterile PBS (5 mL/well), 5 min each.
- Examine scaffolds under a phase contrast microscope at 5x magnification to select tissues for seeding.
NOTE: The best scaffolds for seeding have no tears or holes and do not contain large airways or vessels. While scaffolds with the features may be seeded successfully, repopulation patterns may differ from those observed in alveolar areas.
- Endothelial Cell Seeding (Day -3)
- Count endothelial cells using a hemocytometer and prepare the endothelial cell suspension in endothelial medium (see Table 3) at 5 x 106 cells/mL, with sufficient cells to seed 500,000 endothelial cells per slice (e.g., for 12 slices, resuspend 6 x 106 cells in 1.2 mL medium).
- Place autoclaved seeding baths in 100 mm Petri dishes. Transfer rinsed scaffolds upside down to seeding baths: use a fine curved hemostat to grasp a cassette by the notched sides, use a straight hemostat or forceps to grasp one end of the cassette (being careful not to touch the tissue itself) and flip, and then grasp the cassette again with the tips of the fine curved hemostat via the holes along the notched sides, and place in a seeding bath well. Repeat for remaining cassettes.
NOTE: When correctly placed, scaffolds will be centered, upside down, in the bottom of each well. If needed, press down gently on the corner of the cassette with the tips of a hemostat to ensure that the cassette is seated flat in the well. Improper seating of the cassette can lead to poor tissue seeding. It is acceptable if the well contains a small amount of PBS. - Swirl the prepared cell suspension gently to mix, then use a manual pipette to pipette 100 µL cells directly on top of each tissue at the base of the well, being careful not to damage the tissue with the pipette tip.
- Transfer seeded tissues to the cell culture incubator at 37 °C/5% CO2.
- After 2 h, add 900 µL pre-warmed culture medium to each well using a manual pipette, then return to incubator. If a cassette becomes unseated (floats) upon adding medium, gently press down on the corner of the cassette with the pipette tip so that it lies flat in the well.
- Change medium on day -2. Remove medium by tilting the Petri dish and manually pipetting with a pipette tip lightly placed in the corner of the well, so as not to disturb the cassette. Replace with 1 mL of fresh endothelial medium per well.
- AEC2 and Fibroblast Seeding and Tissue Culture (Day 0)
- Count AEC2s and fibroblasts using a hemocytometer. Prepare a 1:1 cell suspension of AEC2s and fibroblasts in AEC2 growth medium (epithelial base medium + AEC2 supplements; see Table 3) at 5 x 106 total cells/mL, with sufficient cells to seed 500,000 cells (250,000 AEC2s and 250,000 fibroblasts) per slice (e.g., for 12 slices, resuspend 3 x 106 AEC2s + 3 x 106 fibroblasts together in 1.2 mL medium).
- Pipette out the medium from each well of the seeding bath as described in step 4.2.6. Swirl the prepared cell suspension gently to mix, then pipette 100 µL cells directly on top of each tissue at the base of the well.
NOTE: It is acceptable if a small amount of endothelial medium remains in the well prior to AEC2/fibroblast seeding. - Transfer seeded tissues to the cell culture incubator at 37 °C/5% CO2.
- After 2 h, add 900 µL pre-warmed AEC2 growth medium to each well, then return to incubator.
- After 24 h of culture (day 1), prepare a 12-well plate with 1 mL of pre-warmed AEC2 growth medium per well per cassette.
- Pipette 800 µL of medium from each well of the seeding bath. Remove cassettes from the seeding bath: grasp each with a fine curved hemostat via the holes along the notched sides, transfer to a straight hemostat or forceps to grasp the cassette at one end and flip, then use the curved hemostat to grasp the cassette via the notched sides and transfer right-side-up, one per well, to the prepared 12-well plate.
- Change culture medium in the 12-well plate every other day until day 7 or for the desired length of culture: use a glass Pasteur pipette to aspirate the medium from each well, being careful not to touch the tissue; pipette in 1 mL fresh AEC2 growth medium per well.
NOTE: The degree of tissue repopulation may be monitored via phase contrast microscopy at 5x magnification throughout the duration of culture.
5. Tissue Harvest and Sample Analysis
- To fix ELTs for histology and immunofluorescent staining, transfer tissue culture cassettes to 10% neutral-buffered formalin and incubate for 3-4 h at room temperature on a rocker. Remove tissues from cassettes by using the tip of a fine-pointed forceps to cut the tissue where it meets the tabs. Process tissue according to routine methods for paraffin embedding and histology; no specialized techniques are required.
- To process ELTs for qRT-PCR, rinse tissues in cassettes in PBS 2x, then remove tissues and snap freeze or proceed with lysis for RNA extraction.
NOTE: Pooling at least 2 slices seeded with 1 x 106 cells and cultured for 7 days should yield ample RNA for downstream PCR analysis.
Representative Results
An overview of the process to generate ELTs – comprising lung slicing, slice clipping and decellularization, and scaffold repopulation – is presented in Figure 3. The ELTs presented here were cultured using primary rat lung microvascular endothelial cells (see Table of Materials), neonatal rat AEC2s, and lipofibroblast-enriched neonatal rat lung fibroblasts36. AEC2s were freshly isolated via magnetic bead-based sorting as previously described37; alternative isolation protocols have been detailed and discussed elsewhere38,39,40. Purity of isolated rat AEC2s can be assessed via flow cytometry for rat-specific AEC2 surface marker RTII-7041, or via staining of a cytocentrifuged cell sample for RTII-70 or pro-surfactant protein C (pSPC). Rat lung fibroblasts were isolated from postnatal day 7-9 rat pups according to an adaptation of a published protocol42 and used at passage 1-2; alternative isolation protocols have been described elsewhere43,44. Purity of isolated fibroblasts can be assessed via staining of cultured or cytocentrifuged cells for mesenchymal marker vimentin, and lipofibroblast enrichment can be assessed via staining for Oil Red O45.
When the lung tissue is uniformly inflated with agarose, and tissue pieces strategically selected and oriented for slicing so as to maximize total and parenchymal tissue area, one rat lung may yield tissue for >100 alveolar ELTs. Strips of PCLS exhibit sufficient mechanical integrity to be clipped into tissue cassettes with few (<5%) instances of tearing (Figure 3B).
The protocol for decellularizing lung slices is closely based on our previously published whole lung decellularization protocol, which by quantitative proteomics was demonstrated to preserve many ECM components at levels not significantly different from those in native lung22. Decellularized slice scaffolds preserve the native architecture of the alveoli, as viewed by hematoxylin and eosin (H&E) staining (Figure 4A,B) and by phase contrast microscopy (Figure 4C). We typically exclude scaffolds containing large airways or vessels (Figure 4D) or tears, although the former can be included if they are of interest to the researcher. Decellularization leads to a 96% reduction in tissue DNA content as measured by an assay for double-stranded DNA (see Table of Materials; 0.50 µg/mg ± 0.073 µg/mg vs 0.018 µg/mg ± 0.0035 µg/mg in native vs decellularized tissue, respectively, mean ± SEM) (Figure 5A), with no DNA visible by hematoxylin staining (Figure 4B). Histological and immunofluorescent staining of decellularized scaffolds reveals maintenance of ECM proteins collagen, elastin, collagen IV, and laminin with architecture and quantity similar to that in native lung slices (Figure 5B-E). Note that the nuclei of native tissues stain blue/black with trichome (for collagen) and EVG (for elastin) stains. Immunofluorescent staining was performed as described previously, using standard methods for staining tissues37. The antibodies used and their respective concentrations are listed in Table 4.
Successful scaffold repopulation leads to highly cellular ELTs after 7 days, with an alveolar-like repopulation pattern visible by light microscopy (Figure 6A-C). In some cases, with very high cellularity, organoid-type structures may be visible (Figure 6A,B). Unsuccessful tissue seeding can be visualized by phase-contrast microscopy during culture (Figure 6C). After culturing tissue scaffolds with AEC2s, fibroblasts, and endothelial cells, ELTs are densely repopulated with alveolar-like structures comprising all three cell lineages (Figure 6D,E). At day 7 or 8, AEC2s maintain cuboidal morphology and express surfactant protein-B (SPB) and lamellar body protein ABCA3, without evidence of significant differentiation to AEC1s (Figure 6E,F). AEC2s are highly proliferative in ELTs, as demonstrated by 5-ethynyl-2'-deoxyuridine (EdU) incorporation following a 2 h pulse at 10 µM (Figure 6G).
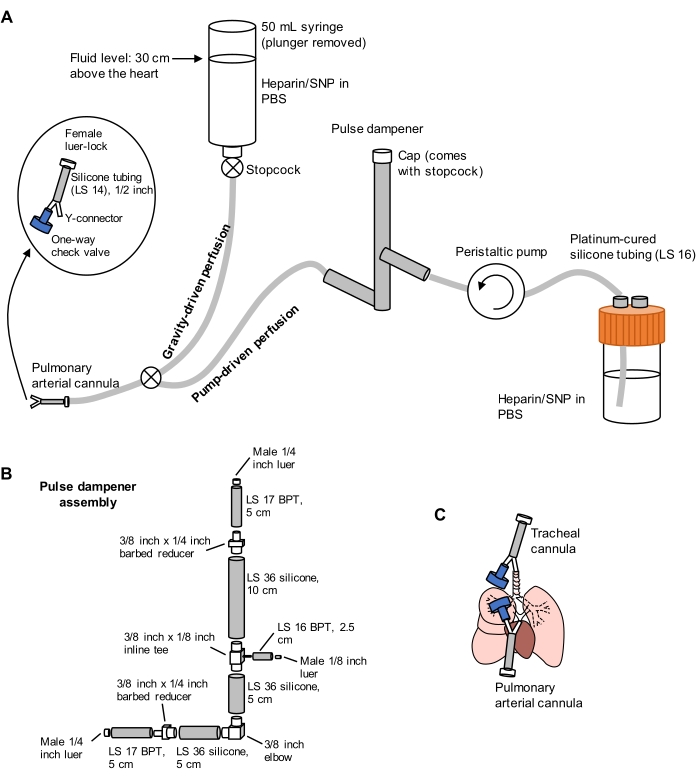
Figure 1: Schematic of perfusion system for lung extraction and clearing. (A) The perfusion system comprises a gravity-driven limb, used for initial cannulation of the pulmonary artery under flow; and a pump-driven limb, used to clear the lungs efficiently after initial cannulation. The pump line includes a "pulse dampener" that dampens the spikes in pressure caused by the pump. The design of the tracheal and pulmonary arterial cannulas is detailed at left. SNP = sodium nitroprusside. (B) Details of pulse dampener assembly. BPT and silicone refer to types of tubing. (C) Positions of tracheal and pulmonary arterial cannulas placed during lung extraction. Please click here to view a larger version of this figure.
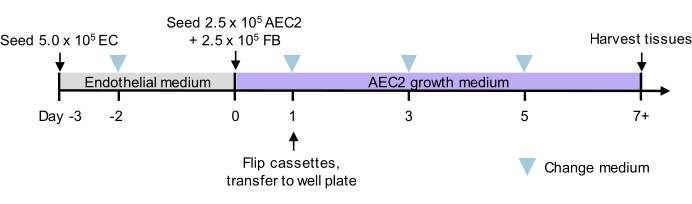
Figure 2: Culture timeline for tri-lineage recellularization. Proposed timeline for tri-lineage ELT seeding and culture, including timing of two-phase seeding. Cell numbers for seeding and culture medium for each phase are indicated. See culture media details in Table 3. AEC2 = alveolar epithelial type 2 cell. EC = endothelial cell. FB = fibroblast. Please click here to view a larger version of this figure.
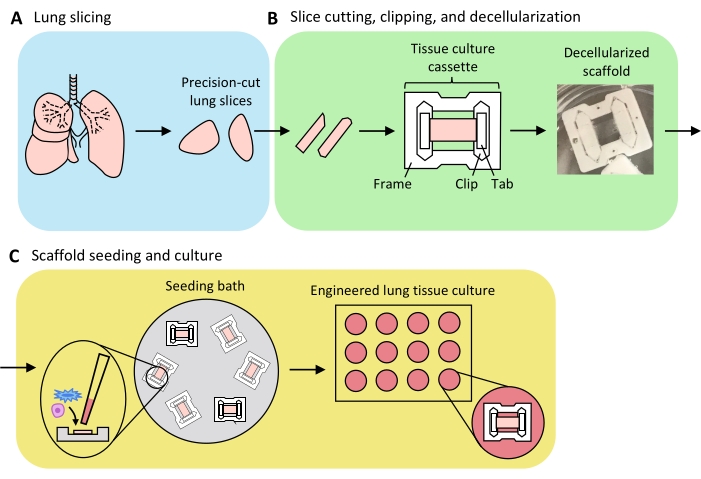
Figure 3: Schematic of engineered lung tissue preparation. (A) Native lung tissue is cut into slices using a vibratome. (B) Precision-cut lung slices are cut into standardized 3 mm wide strips, clipped into polytetrafluorethylene (PTFE) tissue-culture cassettes, and detergent-decellularized to yield acellular extracellular matrix scaffolds. (C) Scaffolds are reseeded in specialized seeding baths that confine the seeding area to the area of the tissue, and then cultured in a standard well plate. Please click here to view a larger version of this figure.
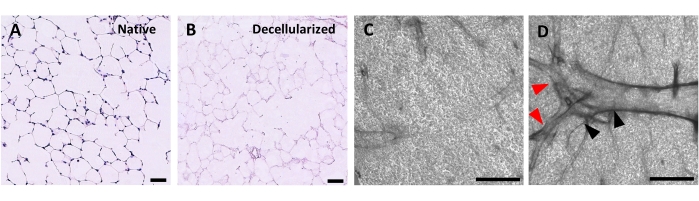
Figure 4: Structure of decellularized lung scaffolds. H&E staining of native (A) and decellularized (B) lung slices showing preservation of alveolar architecture after decellularization. (C,D) Examples of decellularized ECM scaffolds viewed at 5x magnification by phase contrast microscopy, comprising predominately alveolar tissue (C) or containing large branching airways and vessels (D, black and red arrowheads). Scale bars, 50 µm (A,B); 500 µm (C,D). Please click here to view a larger version of this figure.
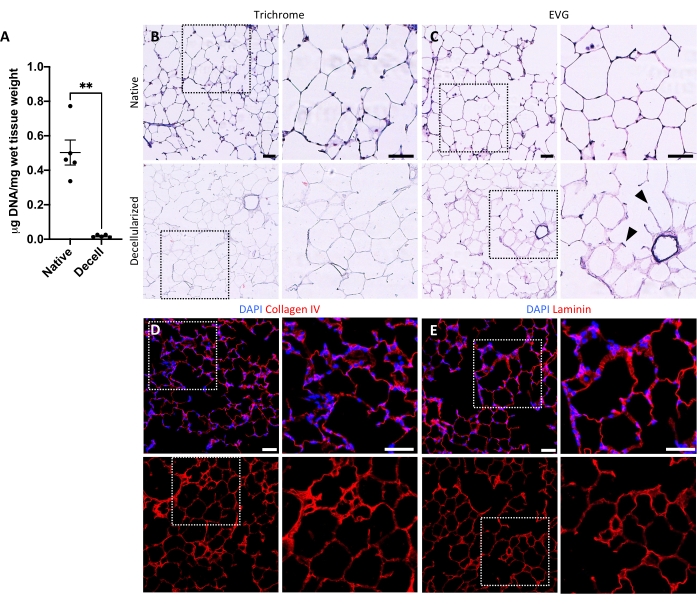
Figure 5: DNA removal and matrix preservation in decellularized lung scaffolds. (A) Quantification of DNA in native and decellularized lung slices (mean ± SEM, n = 5). Welch's t test, **P < 0.01. Decell = decellularized. (B,C) Histological staining of native and decellularized lung slices for collagen (B) and elastin (C). Arrowheads, elastin preserved in alveolar entrance rings of decellularized tissue. (D,E) Immunofluorescent staining of native and decellularized lung slices for collagen IV (D) and laminin (E). Scale bars, 50 µm. In all panels, dotted boxes outline the image region that is magnified to the right in each respective panel. Please click here to view a larger version of this figure.
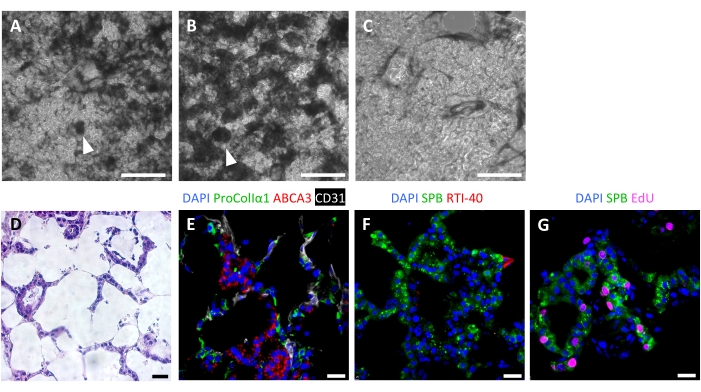
Figure 6: Cellular repopulation of engineered lung tissues. (A-C) Examples of recellularized ELTs on day 7 of culture, as visualized during culture by phase contrast microscopy. The recellularization pattern mirrors the alveolar structure of the tissue. In some areas of high cellularity, organoid-like structures may form (arrowheads). (A) and (B) represent successful cell repopulation, whereas (C) represents a poor level of recellularization after 7 days of culture. (D-G) Staining of recellularized ELTs on day 7 or 8 of culture. (D) H&E staining showing cellular repopulation of the alveolar septa. (E) Immunofluorescent staining labels engrafted proCollagenIα1+ fibroblasts, ABCA3+ AEC2s, and CD31+ endothelial cells. (F) Tissues contain abundant SPB+ AEC2s but few RTI-40 (podoplanin)+ AEC1s under these conditions. (G) Many AEC2s are proliferating in ELTs, as measured by EdU incorporation. Scale bars, 500 µm (A-C); 25 µm (D-G). Please click here to view a larger version of this figure.
Table 1: Decellularization solutions. Preparation details for decellularization solutions. Please click here to download this Table.
Table 2: Decellularization protocol. Details of protocol for decellularizing lung slices. Please click here to download this Table.
Table 3: Culture media. Preparation details for endothelial and AEC2 growth media. Please click here to download this Table.
Table 4: Antibodies used for immunostaining. Details of antibodies and their concentrations used for immunostaining. Please click here to download this Table.
Supplementary File 1: Design for laser cutting tissue culture cassette frames. Please click here to download this File.
Supplementary File 2: Design for laser cutting tissue culture cassette clips. Please click here to download this File.
Supplementary File 3: Design for laser cutting tissue culture cassette tabs. Please click here to download this File.
Supplementary File 4: CAD file for seeding bath mold base. Please click here to download this File.
Supplementary File 5: CAD file for seeding bath mold ring. Please click here to download this File.
Discussion
This paper describes the use of decellularized precision-cut lung slices as a platform to generate engineered lung tissues in vitro, which contain multi-lineage alveolar-like structures. By combining strategies that we previously developed to repopulate high-fidelity acellular ECM lung scaffolds for whole lung engineering22,31, with our robust system for culturing small-scale engineered heart tissues30, this protocol enables the use of physiologically relevant lung ECM as a tissue culture substrate, in a repeatable and moderate-throughput manner.
The methods presented here detail ELT scaffold preparation from rat lungs, which are readily attainable, may be extracted en bloc with direct access to intact airways for agarose inflation, and are of larger size than mouse lung. However, any lung tissue that can be inflated with agarose and yield slices at least 9 mm in length may be used within this system. Regardless of tissue source, uniform inflation of the lung tissue with agarose is the most critical step for ensuring success of downstream tissue slicing, clipping, and tissue handling. Underinflated lung tissue tends not to slice cleanly, while overinflated tissue may tear during clipping. Following agarose gelation, appropriately inflated tissue regions are firm but provide a little give when gently pressed with forceps. For intact rat lungs, we found that pre-inflating the extracted lungs with air several times, followed by agarose inflation as soon as possible after extraction, results in the best slicing outcomes and best quality of resulting tissue scaffolds. The appropriate volume of agarose needs to be optimized empirically; for a rat lung the volume required to inflate the lung to total lung capacity is approximately 30 mL/kg animal mass (e.g., 10.5 mL agarose for lungs from a 350 g rat). For larger resected lung tissues with less straightforward airway access (such as those from human donors), some additional troubleshooting may be necessary to inflate the tissue via a bronchus32. During subsequent lung slicing, the selection and orientation of the tissue on the plunger is another important step to 1) ensure that the slices are large enough to generate tissue strips that can be clipped into tissue culture cassettes and 2) maximize parenchymal (alveolar) tissue area, excluding large airways or vessels.
Clipping the PCLS into tissue culture cassettes can be a challenging step initially, but the cassettes greatly simplify tissue handling during decellularization and seeding. Two potential problems that may arise are tissue tearing (either during the process of clipping, or during decellularization), or tissue positioning in the clips that results in poor downstream seeding (e.g., no seeding, or seeding just at the ends). Tearing may be the result of agarose overinflation, overstretching of the tissue during tab insertion, or leaving too little overhang to provide adequate tissue grip when inserting the tabs. Note that slices that tear at one clip end may be successfully seeded, however, they are difficult to visualize under the microscope during culture as the tissue is not flat. Poor tissue seeding (such as that in Figure 6C) is likely the result of the slice not lying flat between the two clips, and thus making poor contact with the base of the seeding bath well when flipped upside-down. Another possible cause is improper seating of the cassette in the bottom of the seeding bath well. In terms of clipping, apply slightly more tension in the tissue when placing the second clip to help it lie flat. Some slices have a slight concavity; in these cases clip the slice with the convex side up. With practice, we typically experience failed seeding with fewer than 2% of slices.
One limitation of this protocol is the requirement for some specialized equipment – a laser cutter and a 3D printer – to generate the initial materials for ELT preparation. However, once the tissue culture cassettes and seeding baths are created, no additional special materials are required. The lung slicing and decellularization steps of ELT scaffold preparation are moderately time-consuming; however, these steps may be performed in advance, or in numbers sufficient to prepare for multiple experiments at the same time. Many PCLS (>100 if optimizing for parenchymal regions) can be cut from a single lung and snap-frozen for later use. While a single freeze-thaw cycle may cause minor ultrastructural damage to the ECM46, even multiple freeze-thaw cycles have been demonstrated not to cause a significant loss in ECM23,47. PCLS may also be clipped and decellularized in advance of an experiment, to be used within one month. (Notably, the described decellularization protocol can be accomplished in approximately 6 hours, which represents a significant advantage over previously described methods that require a day or more27,28.) Once the scaffolds are prepared, the cell seeding process is simple and fast, and the culture of ELTs does not require specialized techniques.
A caveat of the described ELT method is the lack of region-specific seeding, i.e., the delivery of AEC2s specifically to the alveolar space, or endothelial cells specifically to the vascular space. Nevertheless, although cells are simply seeded on top of the tissue scaffolds, the pattern of recellularization is non-random, with some semblance of alveolar-like organization, including epithelial rings. We suspect that cell-cell interactions, as well as local differences in ECM composition and geometry20,21, likely contribute to the observed recellularization patterns. In support of this hypothesis, a previously published study, in which fibroblasts were seeded non-specifically onto decellularized lung slices, demonstrated that the pattern of tissue repopulation and associated cellular phenotypes varied significantly by microscopic tissue region and ECM scaffold source (e.g., healthy versus diseased)27. Fibroblasts were also observed to invade into the interstitium – the location in which they reside in native lung tissue1,27. The primary alternative method that we can imagine to culture cells on lung slices in a truly region-specific manner would entail seeding intact decellularized lungs via the airway31,48 and vascular compartments49,50, and then slicing the recellularized tissue. However, this alternative 1) is significantly more cost-, time-, and resource-intensive; 2) is lower throughput; 3) requires increased numbers of animals; and 4) is associated with an increased risk of contamination due to the challenges of whole lung culture and subsequent slicing of the seeded lung. While not recapitulating all aspects of native cellular organization, the ELT platform enables lung cell culture on a physiologically relevant ECM substrate, in a manner that is accessible to many more labs.
The flexibility of the ELT system is a major advantage of this platform, and should allow small-scale lung tissue culture with any number of tissue scaffolds, cells, or culture media of interest. The use of scaffolds derived from diseased tissue or from injury models may permit the study of cell-cell or cell-matrix interactions in the setting of disease-altered ECM27,29,51. However, note that the decellularization protocol may need to be adapted to account for matrix differences among species52. The described seeding strategy can be used for any cell type, and the culture timeline adapted to fit the needs of the researcher. As a starting point, 1 x 106 cells per scaffold should yield a highly cellular tissue within 7 days of culture, whereas 1 x 105 total cells result in poor cellularity. In any adaptation of the timeline, the tissue culture cassettes should be removed from the seeding bath 24 h following the last tissue seeding. Here, with the goal of modeling some of the cellular complexity of the lung alveolus, we describe a tri-culture recellularization strategy that supports the maintenance of well-differentiated neonatal AEC2s in alveolar-like structures for at least 7 days. Our results also demonstrate the successful engraftment of both fibroblasts and endothelial cells within ELTs, emphasizing the broad applicability of the culture substrate and its suitability for co-culture studies. Seeding adult cells in ELTs may facilitate the modeling of more quiescent alveolar structures, while seeding human PSC-derived AEC2s, including those with genetic modifications, could facilitate translational studies of human disease13,53. In general, the bottom-up approach enabled by the ELT platform presents the opportunity to investigate the contributions of particular cell types to readouts of interest – such as AEC2 proliferation or differentiation state.
In summary, this protocol outlines a robust system for generating engineered lung tissues for co-culture studies of AEC2s, fibroblasts, and endothelial cells within acellular ECM lung slice scaffolds. ELTs represent a novel 3D culture strategy for primary AEC2s, which to date have typically relied upon less-physiologic gel-type matrices to maintain a well-differentiated phenotype6,11,12. The current platform builds upon previous work in the repopulation of decellularized lung slices24,25,26,27,28,29, but offers several advantages: 1) a tissue culture cassette system to facilitate ELT handling during decellularization, seeding, and culture; 2) a customized seeding bath to precisely seed a known number of cells on each slice scaffold; and 3) a tri-culture reseeding strategy that enables alveolar tissue repopulation with epithelial, mesenchymal, and endothelial cells. Thus, ELTs represent an important step forward toward creating reproducible in vitro models that capture the cellular and substrate complexity of the native alveolus and the AEC2 stem cell niche.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank Lorenzo Sewanan and Jorge Nunez for their work developing the tissue culture cassette design used in this protocol, the Kaminski lab for use of their vibratome, Maurizio Chioccioli and Jessica Nouws for assistance with lung slicing, Allie LaRocco for assistance with initial pilot experiments, and Hong Qian for careful reading of the protocol. This work was supported by NIH grants F30HL143880 (K.L.L.), the Medical Scientist Training Program Training Grant T32GM136651 (K.L.L.), and U01HL145567 (L.E.N.); and by an unrestricted research gift from Humacyte Inc. (L.E.N.).
Materials
| 3D Printer: Form 2 | Formlabs | ||
| 3-Isobutyl-1-methylxanthine (IBMX) | Sigma | I5879 | |
| 8-Bromo cAMP | Sigma | B7880 | |
| Agarose, UltraPure LMP | Invitrogen | 15517-014 | |
| Amphotericin B | Sigma | A2942 | |
| Barbed reducer fitting, 3/8 inch x 1/4 inch | McMaster-Carr | 5121K271 | |
| Benzonase nuclease | Sigma | E1014 | |
| Bovine serum albumin (BSA) Fraction V | Gemini | 700-104P | For AEC2 growth medium |
| Bovine serum albumin (BSA), standard grade | Gemini | 700-100P | For benzonase buffer |
| Check valve, polypropylene, 1/8 inch hose barb | Cole-Parmer | SK-98553-10 | |
| CHIR99021 | PeproTech | 2520691 | |
| Clear Resin, 1 L | Formlabs | RS-F2-GPCL-04 | |
| Cyanoacrylate glue, such as Krazy Maximum Bond Permanent Glue | Any hardware, craft, or drug store | KG483 or similar | |
| Dexamethasone | Sigma | D4902 | |
| DMEM (low glucose) | Gibco | 11885-084 | |
| DMEM (high glucose) | Gibco | 11965-092 | |
| DNA assay (Quant-iT PicoGreen dsDNA Assay Kit) | Invitrogen | P7589 | |
| EDTA, 0.5 M, pH 8.0 | AmericanBio | AB00502-01000 | |
| EdU kit (Click-iT EdU Cell Proliferation Kit for Imaging, Alexa Fluor 647) | Invitrogen | C10340 | Used according to manufacturer's directions |
| Elbow fitting, 3/8 inch | McMaster-Carr | 5121K907 | |
| F12 | Gibco | 11765-054 | |
| Fetal bovine serum (FBS), characterized | Hyclone | SH30071.03 | |
| Gentamicin sulfate | Gemini | 400-100P | Reconstituted in diH2O for a stock solution at 50 mg/mL |
| Hair clippers | Wahl | MiniArco | |
| Hank's balanced salt solution (HBSS), Phenol Red Free | Gibco | 14175-095 | |
| Heparin sodium injection, USP, 1000 U/mL | Sagent | NDC: 25021-400-30 | For intraperitoneal and intracardiac injection |
| Heparin sodium salt | Sigma | H4784 | For pulmonary artery perfusion; prepare stock solution at 100 U/mL in PBS |
| HEPES Buffer | Corning | 25-060-Cl | |
| Inline tee fitting, 3/8 inch x 1/8 inch | McMaster-Carr | 5121K851 | |
| Inoculating loop, disposable | Fisherbrand | 22-363-600 | |
| Insulin from bovine pancreas | Sigma | I6634 | |
| Ketamine injection, 100 mg/mL | Covetrus (Butler Animal Health) | 010177 | |
| KGF, recombinant human | PeproTech | 100-19 | |
| Laser cutter, VLS 3.50 30 watt | Universal Laser Systems | ||
| L-glutamine | Gibco | 25030-081 | |
| Luer-lock, female, 3/32 inch | Cole-Parmer | 45508-02 | |
| Luer-lock, male, 1/8 inch | Cole-Parmer | 30800-24 | |
| Luer-lock, male, 1/4 inch | McMaster-Carr | 51525K146 | |
| MCDB-131 Complete without serum | VEC Technologies | MCDB-131 WOFBS | |
| Magnesium chloride (MgCl2), 1 M | AmericanBio | AB09006-00100 | |
| NaCl | American Bioananalytical | AB01915 | |
| Phosphate buffered saline (PBS), without Ca2+ and Mg2+, 10X | Sigma | D1408 | Reconstitute to 1X with diH2O |
| Phosphate buffered saline (PBS), with Ca2+ and Mg2+ | Gibco | 21300-058 | |
| PDMS – SYLGARD 184 Silicone Elastomer Kit | Dow Corning Corporation | 4019862 | |
| Penicillin/Streptomycin (10,000 U/mL penicillin/10,000 μg/mL streptomycin) | Gibco | 15140-122 | |
| Petri dish, 150 mm | Falcon | 351058 | |
| Plastic film (parafilm) | Bemis | PM-996 | |
| Pharmed BPT tubing, LS 16 | Masterflex | 06508-16 | |
| Pharmed BPT tubing, LS 17 | Masterflex | 06508-17 | |
| Platinum-cured silicone tubing, LS 14 | Masterflex | 96420-14 | |
| Platinum-cured silicone tubing, LS 16 | Masterflex | 96420-16 | |
| Platinum-cured silicone tubing, LS 36 | Masterflex | 96410-36 | |
| Poloxamer 407 (Pluronic F-127) | Sigma | P2443 | |
| Povidone/iodine prep pads, 10% | Dynarex Corporation | 1108 | |
| PTFE sheet, 0.060 inch (1/16 inch) thick | ePlastics | PTFENAT0.060X12X12 | For tissue culture cassette tabs |
| PTFE sheet, 0.093 inch (3/32 inch) thick | ePlastics | PTFENAT0.093X12X12 | For tissue culture cassette frames and clips |
| Peristaltic pump drive: Masterflex L/S Variable-Speed Digital Drive | Cole-Parmer | ZM-07528-30 | |
| Peristaltic pump head: Masterflex L/S Easy-Load II Pump Head | Cole-Parmer | EW-77202-60 | |
| Rat, Sprague Dawley | Charles River | Strain Code: 400 | |
| Razor blade | Any hardware or craft store | Personna 94-120-71 or similar | |
| Retinoic acid | Sigma | R2625 | |
| Rotary blades, 28 mm | Omnigrid | 2046 | |
| Rotary cutter, 28 mm | Olfa | Model 9551 | |
| Sodium deoxycholate (SDC) | Sigma | D6750 | |
| Sodium nitrorusside (SNP) | Sigma | 71778 | |
| Stopcock, 4-way | Edwards | 594WSC | |
| Suture, 4-0 monofilament polypropylene | Covidien | VP-557-X | |
| Syringe, 10 mL | BD | 302995 | |
| Syringe, 50 mL | BD | 309653 | |
| Tissue culture dish, 35 mm | Falcon | 353001 | |
| Tissue culture dish, 100 mm | Corning | 430167 | |
| Tissue culture plate, 6-well | Falcon | 353046 | |
| Tissue culture plate 12-well | Falcon | 353043 | |
| Transferrin human | Sigma | T8158 | |
| Tris, 1 M solution, pH 8.0 | AmericanBio | AB14043-01000 | |
| Triton X-100 | American Bioanalytical | AB02025-00500 | |
| Vibratome, Compresstome VF-300-0Z | Precisionary Instruments LLC | ||
| Xylazine, 100 mg/mL | Henry Schein | NDC: 11695-4022-1 | |
| Y-connector, 1/16 inch barbed | Cole-Parmer | 30614-43 |
References
- Burri, P. H. Morphology and respiratory function of the alveolar unit. International Archives of Allergy and Applied Immunology. 76, 2-12 (1985).
- Barkauskas, C. E., Noble, P. W. Cellular Mechanisms of Tissue Fibrosis. 7. New insights into the cellular mechanisms of pulmonary fibrosis. American Journal of Physiology. Cell Physiology. 306 (11), 987-996 (2014).
- Taylor, M. S., et al. A Conserved Distal Lung Regenerative Pathway in Acute Lung Injury. American Journal of Pathology. 188 (5), 1149-1160 (2018).
- Carsana, L., et al. Pulmonary post-mortem findings in a series of COVID-19 cases from northern Italy: a two-centre descriptive study. Lancet Infectious Diseases. 20 (10), 1135-1140 (2020).
- Basil, M. C., et al. The Cellular and Physiological Basis for Lung Repair and Regeneration: Past, Present, and Future. Cell Stem Cell. 26 (4), 482-502 (2020).
- Barkauskas, C. E., et al. Type 2 alveolar cells are stem cells in adult lung. Journal of Clinical Investigation. 123 (7), 3025-3036 (2013).
- Desai, T. J., Brownfield, D. G., Krasnow, M. A. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature. 507 (7491), 190-194 (2014).
- Evans, M. J., Cabral, L. J., Stephens, R. J., Freeman, G. Renewal of alveolar epithelium in the rat following exposure to NO2. The American Journal of Pathology. 70 (2), 1-24 (1973).
- Beers, M. F., Moodley, Y. When is an alveolar type 2 cell an alveolar type 2 cell? A conundrum for lung stem cell biology and regenerative medicine. American Journal of Respiratory Cell and Molecular Biology. 57 (1), 18-27 (2017).
- Borok, Z., et al. Keratinocyte growth factor modulates alveolar epithelial cell phenotype in vitro: expression of aquaporin 5. American Journal of Respiratory Cell and Molecular Biology. 18 (4), 554-561 (1998).
- Salahudeen, A. A., et al. Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature. 588 (7839), 670-675 (2020).
- Sucre, J. M. S., et al. Successful establishment of primary Type 2 alveolar epithelium with 3D organotypic co-culture. American Journal of Respiratory Cell and Molecular Biology. 59 (2), 158-166 (2018).
- Jacob, A., et al. Differentiation of human pluripotent stem cells into functional lung alveolar epithelial cells. Stem Cell. 21 (4), 472-488 (2017).
- Yamamoto, Y., et al. Long-term expansion of alveolar stem cells derived from human iPS cells in organoids. Nature Methods. 14 (11), 1097-1106 (2017).
- Chen, Y. W., et al. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nature Cell Biology. 19 (5), 542-549 (2017).
- Korogi, Y., et al. In vitro disease modeling of hermansky-pudlak syndrome Type 2 using human induced pluripotent stem cell-derived alveolar organoids. Stem Cell Reports. 12 (3), 431-440 (2019).
- Strikoudis, A., et al. Modeling of fibrotic lung disease using 3D organoids derived from human pluripotent stem cells. Cell Reports. 27 (12), 3709-3723 (2019).
- Bissell, M. J., Hall, H. G., Parry, G. How does the extracellular matrix direct gene expression. Journal of Theoretical Biology. 99 (1), 31-68 (1982).
- Chapman, H. A. Epithelial responses to lung injury: Role of the extracellular matrix. Proceedings of the American Thoracic Society. 9 (3), 89-95 (2012).
- Guilak, F., et al. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell. 5 (1), 17-26 (2009).
- Zhou, Y., et al. Extracellular matrix in lung development, homeostasis and disease. Matrix Biology. 73, 77-104 (2018).
- Calle, E. A., et al. Targeted proteomics effectively quantifies differences between native lung and detergent-decellularized lung extracellular matrices. Acta Biomaterialia. 46, 91-100 (2016).
- Crapo, P. M., Gilbert, T. W., Badylak, S. F. An overview of tissue and whole organ decellularization processes. Biomaterials. 32 (12), 3233-3243 (2011).
- Bonvillain, R. W., et al. A nonhuman primate model of lung regeneration: Detergent-mediated decellularization and initial in vitro recellularization with mesenchymal stem cells. Tissue Engineering Part A. 18 (23-24), 2437-2452 (2012).
- O’Neill, J. D., et al. Decellularization of human and porcine lung tissues for pulmonary tissue engineering. The Annals of Thoracic Surgery. 96 (3), 1055 (2013).
- Wagner, D. E., et al. Three-dimensional scaffolds of acellular human and porcine lungs for high throughput studies of lung disease and regeneration. Biomaterials. 35 (9), 2664-2679 (2014).
- Burgstaller, G., et al. Distinct niches within the extracellular matrix dictate fibroblast function in (cell free) 3D lung tissue cultures. American Journal of Physiology-Lung Cellular and Molecular Physiology. 314 (5), 708-723 (2018).
- Sun, H., et al. Fibroblast engraftment in the decellularized mouse lung occurs via a β1-integrin-dependent, FAK-dependent pathway that is mediated by ERK and opposed by AKT. American Journal of Physiology-Lung Cellular and Molecular Physiology. 306 (6), 463-475 (2014).
- Booth, A. J., et al. Acellular normal and fibrotic human lung matrices as a culture system for in vitro investigation. American Journal of Respiratory and Critical Care Medicine. 186 (9), 866-876 (2012).
- Schwan, J., et al. Anisotropic engineered heart tissue made from laser-cut decellularized myocardium. Scientific Reports. 6, 32068 (2016).
- Petersen, T. H., et al. Tissue-engineered lungs for in vivo implantation. Science. 329 (5991), 538-541 (2010).
- Gerckens, M., et al. Generation of Human 3D Lung Tissue Cultures (3D-LTCs) for Disease Modeling. Journal of Visualized Experiments. (144), (2019).
- Lyons-Cohen, M. R., Thomas, S. Y., Cook, D. N., Nakano, H. Precision-cut mouse lung slices to visualize live pulmonary dendritic cells. Journal of Visualized Experiments. (122), e55465 (2017).
- Neuhaus, V., et al. Assessment of the cytotoxic and immunomodulatory effects of substances in human precision-cut lung slices. Journal of Visualized Experiments. (135), e57042 (2018).
- You, Y., Richer, E. J., Huang, T., Brody, S. L. Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. American Journal of Physiology-Lung Cellular and Molecular Physiology. 283 (6), 1315-1321 (2002).
- Vaccaro, C., Brody, J. S. Ultrastructure of developing alveoli. I. The role of the interstitial fibroblast. The Anatomical Record. 192 (4), 467-479 (1978).
- Calle, E. A., et al. Fate of distal lung epithelium cultured in a decellularized lung extracellular matrix. Tissue Engineering Part A. 21 (11-12), 1916-1928 (2015).
- Dobbs, L. G., Gonzalez, R., Williams, M. C. An improved method for isolating type II cells in high yield and purity. American Review of Respiratory Disease. 134 (1), 141-145 (1986).
- Dobbs, L. G. Isolation and culture of alveolar type II cells. American Journal of Physiology. 258, 134-147 (1990).
- Gonzalez, R. F., Dobbs, L. G. Isolation and culture of alveolar epithelial Type I and Type II cells from rat lungs. Methods in Molecular Biology. 945, 145-159 (2013).
- Dobbs, L. G., Pian, M. S., Maglio, M., Dumars, S., Allen, L. Maintenance of the differentiated type II cell phenotype by culture with an apical air surface. The American Journal of Physiology. 273 (2), 347-354 (1997).
- Bruce, M. C., Honaker, C. E. Transcriptional regulation of tropoelastin expression in rat lung fibroblasts: changes with age and hyperoxia. American Journal of Physiology. 274 (6), 940-950 (1998).
- Berk, J. L., Franzblau, C., Goldstein, R. H. Recombinant interleukin-1 beta inhibits elastin formation by a neonatal rat lung fibroblast subtype. Journal of Biological Chemistry. 266 (5), 3192-3197 (1991).
- Schultz, C. J., Torres, E., Londos, C., Torday, J. S. Role of adipocyte differentiation-related protein in surfactant phospholipid synthesis by type II cells. American Journal of Physiology-Lung Cellular and Molecular Physiology. 283 (2), 288-296 (2002).
- Maksvytis, H. J., et al. In vitro characteristics of the lipid-filled interstitial cell associated with postnatal lung growth: evidence for fibroblast heterogeneity. Journal of Cellular Physiology. 118, 113-123 (1984).
- Hopkinson, A., et al. Optimization of amniotic membrane (AM) denuding for tissue engineering. Tissue Engineering Part C: Methods. 14 (4), 371-381 (2008).
- Fernandez-Perez, J., Ahearne, M. The impact of decellularization methods on extracellular matrix derived hydrogels. Scientific Reports. 9 (1), 14933 (2019).
- Ott, H. C., et al. Regeneration and orthotopic transplantation of a bioartificial lung. Nature medicine. 16 (8), 927-933 (2010).
- Le, A. V., et al. Efficient and Functional Endothelial Repopulation of Whole Lung Organ Scaffolds. ACS Biomaterials Science & Engineering. 3 (9), 2000-2010 (2017).
- Ren, X., et al. Engineering pulmonary vasculature in decellularized rat and human lungs. Nature Biotechnology. 33 (10), 1097-1102 (2015).
- Wagner, D. E., et al. Comparative decellularization and recellularization of normal versus emphysematous human lungs. Biomaterials. 35 (10), 3281-3297 (2014).
- Balestrini, J. L., et al. Comparative biology of decellularized lung matrix: Implications of species mismatch in regenerative medicine. Biomaterials. 102, 220-230 (2016).
- Alysandratos, K. D., et al. Patient-specific iPSCs carrying an SFTPC mutation reveal the intrinsic alveolar epithelial dysfunction at the inception of interstitial lung disease. Cell Reports. 36 (9), 109636 (2021).

