Detecting Wolbachia Strain wAlbB in Aedes albopictus Cell Lines
Summary
Four methods were used to detect intracellular Wolbachia, which complemented each other and improved the detection accuracy of Wolbachia infection of Aedes albopictus-derived Aa23 and Aa23-T cured of native Wolbachia infection using antibiotics.
Abstract
As a maternally harbored endosymbiont, Wolbachia infects large proportions of insect populations. Studies have recently reported the successful regulation of RNA virus transmission using Wolbachia-transfected mosquitoes. Key strategies to control viruses include the manipulation of host reproduction via cytoplasmic incompatibility and the inhibition of viral transcripts via immune priming and competition for host-derived resources. However, the underlying mechanisms of the responses of Wolbachia-transfected mosquitoes to viral infection are poorly understood. This paper presents a protocol for the in vitro identification of Wolbachia infection at the nucleic acid and protein levels in Aedes albopictus (Diptera: Culicidae) Aa23 cells to enhance the understanding of the interactions between Wolbachia and its insect vectors. Through the combined use of polymerase chain reaction (PCR), quantitative PCR, western blot, and immunological analytical methods, a standard morphologic protocol has been described for the detection of Wolbachia-infected cells that is more accurate than the use of a single method. This approach may also be applied to the detection of Wolbachia infection in other insect taxa.
Introduction
The Asian tiger mosquito Aedes albopictus (Skuse) (Diptera: Culicidae), which is a key vector of dengue virus (DENV) in Asia and other parts of the world1, is a natural host of two types of the intracellular bacteria, Wolbachia (wAlbA and wAlbB), which are distributed throughout the germ line and somatic tissue2,3. The Aa23 cell line derived from A. albopictus embryos consists of at least two morphological cell types, both of which support infection4 and may be cured of native Wolbachia infection using antibiotics (Aa23-T). Given that Aa23 retains only wAlbB, it is a useful model for the study of host-endosymbiont interactions4,5,6.
Wolbachia is maternally transmitted and infects an estimated 65% of insect species8,9 and 28% of mosquito species10. It infects a variety of tissues and forms an intimate symbiotic relationship with the host, usually inducing cytoplasmic incompatibility (CI)11 and population replacement by manipulating the host reproductive system12,13. These host responses have been observed in natural populations of Drosophila simulans14 and in A. aegypti in a laboratory cage and field trial15. An important nonreproductive manipulation elicited by Wolbachia is induced host resistance to a variety of pathogens, including DENV, Chikungunya virus (CHIKV), and West Nile virus (WNV)16,17, that may be mediated by an improved innate immune system of the symbiont18,19, competition between Wolbachia and viruses for essential host resources20, and manipulation of host viral defense pathways21.
This protocol has been developed for studying these underlying mechanisms of Wolbachia-induced host antiviral responses. It uses four methods of detection of intracellular Wolbachia infection of Aa23 cells. These methods provide a strong theoretical basis for studies of intracellular Wolbachia infection of other host species. The first method, PCR-a powerful technique allowing the enzymatic amplification of specific regions of DNA without utilizing conventional cloning procedures-was used to detect Wolbachia DNA and determine the presence/absence of Wolbachia infection22. The second method measures Wolbachia DNA copy density using quantitative PCR (qPCR) for reliable detection and measurement of products generated during each PCR cycle that is directly proportionate to the amount of template before PCR23. The third method detects the presence of intracellular Wolbachia proteins, using western blot-one of the most powerful tools for detecting specific proteins in complex mixtures by combining the high separation power of electrophoresis, the specificity of antibodies, and the sensitivity of chromogenic enzymatic reactions. The final method is an immunofluorescence assay (IFA) that combines immunology, biochemistry, and microscopy to detect the Wolbachia surface protein (wsp) through an antigen-antibody reaction to confirm the cellular uptake of Wolbachia and determine its cellular localization.
This paper describes the four methods listed above to verify the existence of Wolbachia in the cells, which can be used to detect whether the exogenous Wolbachia was successfully transfected and the Wolbachia in the cell was cleared. After determining whether Wolbachia is present in the cells or not, a variety of different analyses can be performed, including genomics, proteomics, or metabolomics. This protocol demonstrates the detection of Wolbachia through Aa23 cells but can also be used in other cells.
Protocol
1. Materials and reagents
- Use pyrogen-free solutions and media for cell culture (see the Table of Materials).
- Use ultrapure water to prepare all solutions.
- Be careful in selecting fetal bovine serum (FBS) for cell culture, following a lot-check process.
NOTE: As FBS lots are subject to regular change, it is impossible to state the relevant catalog and lot numbers in this protocol. - Select Aa23 cell strains derived from A. albopictus eggs, corresponding to Wolbachia-free Aa23-T.
2. Cell culture
- Prepare 25 cm2 cell culture flasks with 4 mL of Schneider's medium with 10% FBS.
- Incubate the flasks at 28 °C under a 5% CO2 atmosphere for 5 to 7 days7,24.
- Observe unstained Aa23 and Aa23-T cells in the flasks using light microscopy at 100x magnification (Figure 1).
3. DNA extraction
- Culture the cells for 5-7 days until 70-80% confluence per flask (step 2). Harvest the cells with manual pipetting from 1 mL of resuspended culture medium by centrifugation for 5 min at 100 × g in a microcentrifuge.
- Remove the supernatant by aspiration and store the resulting cell pellet at -20 °C.
- Resuspend the pellets in 0.40 mL of phosphate-buffered saline (PBS), place 50 µL of the solution in a microcentrifuge tube, add 5 µL of proteinase K (20 mg/mL), and incubate at 55 °C for 1 h. Finally, heat these tubes at 99 °C for 15 min to obtain the crude DNA and store it at -20 °C.
4. Detection of Wolbachia nucleic acid
- PCR analysis
- Perform PCR in a total reaction volume of 20 µL, containing 8 µL of ddH2O, 10 µL of Taq polymerase (see the Table of Materials), 1 µL of DNA template (from step 3), 0.5 µL of forward and reverse primers (10 µM): wsp81F (5' TGG TCC AAT AAG TGATGA AGAAAC 3') and wsp691R (5' AAA AAT TAA ACG CTA CTC CA 3')25, respectively.
- Use the following PCR conditions: initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 94 °C, annealing at 55 °C, and extension at 72 °C; then, a final extension at 72 °C for 7 min.
- Detect the DNA on a 1% agarose gel (1% w/v agarose in PBS) using a gel imager.
- qPCR analysis
- Analyze the Wolbachia genome copy for three biological replicates (n = 6) from the extracted DNA (step 3) using wsp primers 183 F (5' AAGGAACCGAAGTTCATG 3') and QBrev R (5' AGTTGAGAGTAAAGTCCC 3')22, normalized with ribosomal protein S6 (RPS6) forward and reverse primers (5' GAAGTTGAACGTATCGTTTC 3' and 5' GAGATGGTCAGCGGTGATTT 3', respectively)3.
- Generate a standard curve for wAlbB and RPS6.
- Extract DNA from PMD-18T (a special vector for efficient cloning of PCR products (TA cloning)) recombinant plasmid containing wsp and rps6 gene fragments (Supplemental File) and measure the plasmid DNA concentration (see the Table of Materials).
- Convert the plasmid DNA concentration to copy number concentration using Eq (1).
Plasmid copy number =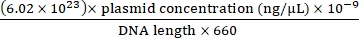 (1)
(1) - Dilute the plasmid copy number from 101 to 108 with ultrapure water, and set 3 replicates for each concentration gradient.
- Perform qPCR amplification using TB Green and a real-time PCR system (see the Table of Materials), and record the results of each reaction. Use the Ct value to develop a standard curve. Analyze the DNA in a total reaction volume of 20 µL, comprising 7 µL of ddH2O, 10.4 µL of TB Green (see the Table of Materials), 1 µL of DNA template (provided by Step 3), and 0.8 µL of forward and reverse primers (10 µM). Use the following reaction conditions: initial denaturation at 95 °C for 5 min, followed by 40 cycles of denaturation at 95 °C for 10 s, and annealing at 60 °C for 35 s.
- Calculate the WSP and RPS6 gene copy numbers in the cells according to the established standard curve and the Wolbachia relative density by the copy number of wsp/RPS6. Analyze the data using an independent samples t-test.
- Western blot analysis of Wolbachia protein
- Protein extraction
- Collect 2 × 107 Aa23 and Aa23-T cells from the six-well plate (from step 2) in a microcentrifuge tube, wash them three times with PBS, and centrifuge them at 100 × g for 5 min at 4 °C.
- Prepare 1 mL lysates with RIPA lysis buffer (50 mM Tris (pH 7.4), 150 mM NaCl, 1%Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 2 mM sodium pyrophosphate, 25 mM β-glycerophosphate, 1 mM EDTA, 1 mM Na3VO4, 0.5 µg/mL leupeptin) and PMSF (final concentration 1 mM).
- Add 200 µL of lysis buffer to the cell pellet and maintain for 30 min on ice. Centrifuge the lysates at 12,000 × g for 10 min at 4 °C and remove the supernatant.
- Assay the wsp concentration of the supernatant using a bicinchoninic acid (BCA) protein assay kit (see the Table of Materials) following the manufacturer's instructions.
- Load equal amounts of protein on a 15% SDS-PAGE gel [10% distilled water; 50% (30% Acr-Bis (29:1), 38% 1 M Tris, pH 8.8; 1% (10% SDS), 1% (10% ammonium persulfate), 0.04% TEMED].
- Transfer the protein to nitrocellulose membranes, block the membranes with 5% skimmed milk for 2 h26,27 at room temperature, and then wash the membranes three times with TBST (1 mL of Tween 20 in 1 L of TBS) (see the Table of Materials).
- Incubate the membranes overnight at 4 °C with 100 µL of anti-wsp polyclonal antibody (prepared in-house, see Supplemental File)), prepared by diluting the primary antibody with 5% skimmed milk (1:12,000).
- Wash the primary antibody three times with TBST.
- Incubate the membranes in 100 µL of horseradish peroxidase (HRP)-conjugated secondary antibody on a shaker for 1 h at room temperature.
- Wash the membranes three times with TBST, and mix 20 µL of solution A (HRP Substrate Luminol Reagent) and 20 µL of solution B (HRP Substrate Peroxide Reagent) in the chemiluminescence detection kit at a ratio of 1:1. Immerse the entire membrane into the luminescence solution at the same time, and observe the signals using a multifunctional imaging system.
- Protein extraction
- Immunofluorescence localization of Wolbachia protein
- Incubate 5 × 105 Aa23 and Aa23-T cells at 28 °C on a Laser confocal Petri dish with a thickness of 0.17 mm at the bottom, under a 5% CO2 atmosphere for 3 to 4 days7,24. Wait until the cell is 60%-70% confluence and wash the cells three times with PBS.
- Fix the cells for 20 min at 4 °C in 1 mL of 4% (w/v) paraformaldehyde in PBS.
- Wash the fixed cells using PBST (0.2% v/v Triton X-100 in PBS) for 5 min and wash the cells again twice with PBS.
- Incubate the slides for 1 h at 37 °C in 1 mL of 3% (w/v) bovine serum albumin in PBS.
- Incubate the slides overnight in 100 µL of mouse anti-wsp polyclonal antibody (prepared in-house) and mouse serum (1:1,000 dilution in PBS) in a wet box (it can maintain humidity and prevent moisture from evaporating) at 4 °C.
- Remove the slides from the wet box and return them to room temperature; wash them three times with PBS and remove the PBS.
NOTE: From step 4.4.7 onward, all operations must be protected from light. - Incubate the slides with 100 µL of an Alexa Fluor 488-conjugated goat anti-mouse antibody (1:2,000 dilution in PBS) at 37 °C for 30 min.
- Incubate the sample slides with 50 µL of 4',6-diamidino-2-phenylindole (DAPI diluted in PBS to 10 µg/mL, binds strongly to DNA) at 37 °C for 5 min and then wash them three times with PBS.
- Observe the immunostaining under a confocal microscope.
- Turn on the power and the laser.
- Turn on the normal and mercury lamps of the microscope.
- Start the system and run the operation software. Put a drop of cedar oil on the sample slides, place the sample slides on the microscope stage, and move the stage to the appropriate position.
- Click the mercury lamp observation button in the software, open the mercury lamp shutter, and observe the sample under an inverted microscope to focus.
- Use the manual control panel to select green and blue fluorescence to take fluorescence photos under 100x magnification.
- Turn off the mercury lamp observation button and start acquiring images.
- To ensure good image quality, prescan by scan size (512 pixels x 512 pixels). Adjust the scan size to get clear images (1,024 pixels x 1,024 pixels).
- Carry out noise reduction if the background noise is too high.
- Stop scanning and save the images.
- Turn off the mercury lamp and lasers.
- Wipe the objective with 75% alcohol and lower the objective to the lowest position.
- Turn off the shutter and microscope power.
- After the instrument has cooled, cover it with the dust cover.
Representative Results
Before Wolbachia was detected, Aa23 and Aa23-T cells were observed under a light microscope to determine any morphological differences between the two cell lines. Aa23 and Aa23-T cells have at least two cell morphologies but no obvious morphological difference between the two cell types (Figure 1). Here, Aa23 cells were used as a model system to detect Wolbachia infection using four methods. Positive amplification of the Wolbachiawsp gene using the diagnostic primers 81F/691R was possible in Aa23 cells (Lanes 1 and 2) but not in Aa23-T cells (Lanes 3 and 4) (Figure 2A). The analysis of Wolbachia density in the cell lines using qPCR showed a wsp/rps6 ratio of 2.4 in Aa23 cells but no Wolbachia in Aa23-T cells (Figure 2B). Western blot analysis of protein extracts showed a strong wsp signal for Aa23 and no signal for Aa23-T (Figure 3A), while the indirect immunofluorescence assay detected the wsp protein (Figure 3B) in Aa23 cells (green), while only DNA stained with DAPI (blue) was detected in Aa23-T cells.
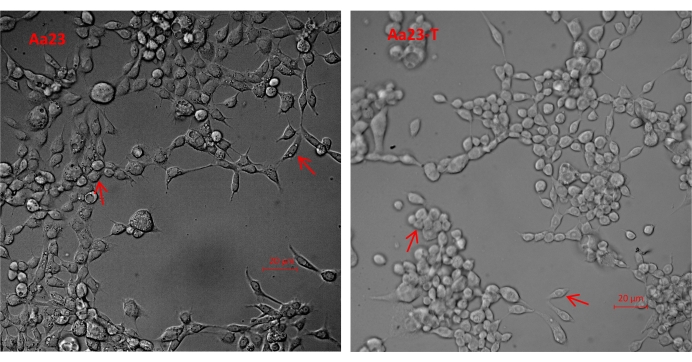
Figure 1: Light microscopy of unstained Aa23 and Aa23-T cells. Heterogeneous morphology indicates that more than one cell type is present in both cell lines, as indicated by the arrows. Scale bar = 20 µm. Please click here to view a larger version of this figure.
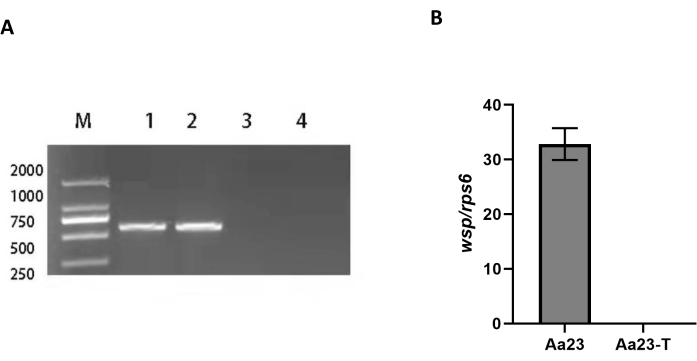
Figure 2: Detection of Wolbachia infection in cells at the nucleic acid level. (A) PCR analysis of Aa23 and Aa23-T cells resolved on a 1% agarose gel. Positive PCR amplification of the Wolbachiawsp gene using the diagnostic primers, 81F/691R, observed in Aa23 cells (Lanes 1 and 2) but not in Aa23-T cells (Lanes 3 and 4). (B) Wolbachia wsp copy density of Aa23 and Aa23-T cells. Density was greater in Aa23 cells and undetectable in Aa23-T cells. Copy number was normalized using Aedes albopictus rps6; data are mean ± SEM, n = 6. Please click here to view a larger version of this figure.
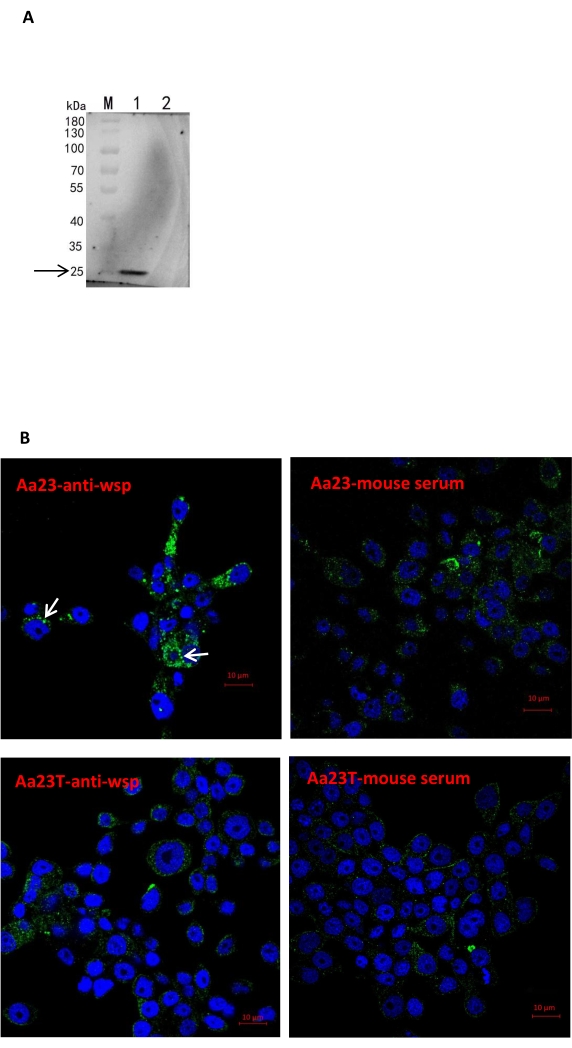
Figure 3: Detection of Wolbachia infection in cells at the protein level. (A) Immunoblotting with wsp antibody in Aa23 cells (Lane 1), with an obvious band near 25 kDa; Aa23-T (Lane 2), which was treated with tetracycline. (B) Immunofluorescence labeling of cells showing the localization of Wolbachia (green); cells were probed with polyclonal anti-wsp antibody (Wolbachia), followed by goat anti-mouse (green) Alexa Fluor 488-conjugated antibodies, and nuclei (blue) are stained with DAPI. Mouse serum was used as a negative control because the anti-wsp antibody was not purified, and the polyclonal antibody was mouse-derived. Aa23 cells (Aa23-anti-wsp) infected with Wolbachia are indicated with a white arrow. Scale bars = 10 µm. Abbreviation: DAPI = 4',6-diamidino-2-phenylindole; M = marker. Please click here to view a larger version of this figure.
Supplemental File: Plasmid generation and anti-wsp antibody preparation. Please click here to download this File.
Discussion
Detection of intracellular Wolbachia infection is essential for the study of Wolbachia-host interactions and the confirmation of successful transfection of cells with novel strains. In this protocol, four methods were used to successfully detect intracellular Wolbachia infection at the nucleic acid and protein levels. These four experimental methods corroborated and improved the detection accuracy of Wolbachia infection of cells.
PCR was used to detect the presence of Wolbachia infection of cells at the nucleic acid level. However, as it does not quantify levels of infection28,29, qPCR was used to quantify DNA copy density28,30. As the accuracy of PCR methods may be affected by low amplification efficiency and false positives, it is important to ensure good quality of DNA, especially for qPCR detection, where reagents and templates must be placed on ice to prevent denaturation, and operations should be carried out according to standard methods. Low amplification efficiency tends to be caused by reagent degradation in the reaction system, insufficient template amounts, and/or excessive length of extended primer fragments28. False positives are usually caused by contamination of reagents and templates, high levels of primer concentration, and/or dimer and hairpin primer structures. Therefore, it is important to ensure that reagents are of high quality, primers are optimized based on the literature, and appropriate software used for primer design.
Two methods described above detect intracellular Wolbachia infection at the nucleic acid level; the other two detect intracellular Wolbachia infection at the protein level. Western blotting was used to determine intracellular Wolbachia infection of Aa23 and Aa23-T cells at the protein level. Problems associated with this method are related to sample preparation, antibody incubation, and detection31. Critical steps in the western blotting detection process include using fresh and fully lysed samples and correct secondary antibodies, which must be validated and diluted according to the instructions at a dilution of 1:12,000. According to the experimental requirements, this method can also be combined with other immunization methods. Therefore, Immunofluorescence was used to localize intracellular infection by Wolbachia31 , because this is not directly detected using PCR, qPCR, or western blot analyses. There are some critical steps to note about immunofluorescence. First, ensure the optimal quality of cell samples and avoid contamination. Contamination of cells by black glue bug, which can also be stained, will seriously affect the interpretation of the results. Second, cell density in the laser confocal dishes must be ~60-70%. Finally, washing with PBS after sample incubation is critical to get clear fluorescence images.
Using relative and absolute quantitative and visual methods to detect intracellular Wolbachia infection at the nucleic acid and protein levels, this protocol is more comprehensive and accurate than the use of any single method. In addition, constructing a double standard curve (for wsp and RPS6) results in less stringent requirements for the PCR system and amplification efficiency and partial elimination of the experimental error in the final data processing, which greatly reduces the experimental work. However, there were some limitations to this protocol. First, the anti-wsp antibody is prepared in-house and has not yet been commercialized. Second, although there were no extra bands due to the unpurified polyclonal anti-wsp antibody in western blotting, indicating antibody purity, the negative control group using mouse serum also had a green fluorescence background, which was not conducive to the interpretation of the results. While the accuracy of the western blot and immunofluorescence methods may be improved by ensuring the use of purified antibodies or accurate fluorescent probes, further study and analysis of empirical data are required. In summary, a more comprehensive and accurate protocol is reported for the detection of Wolbachia infection in Aa23 cells using four methods used in previous studies of Wolbachia. The protocol described here is applicable to the detection of Wolbachia in other types of cells and tissues.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Dr. Xin-Ru Wang from the University of Minnesota for insightful suggestions and guidance. This work was supported by a grant from the National Natural Science Foundation of China (No.81760374).
Materials
| Microscope | Zeiss | SteREO Discovery V8 | |
| Petri dish | Fisher Scietific | FB0875713 | |
| Pipette | Pipetman | F167380 | P10 |
| inSituX platform | |||
| Analysis software | In-house developed | ||
| Cerium doped yttrium aluminum garnet | MSE Supplies | Ce:Y3Al5O12, YAG single crystal substrates | |
| Chip holder | In-house developed | ||
| Control software | In-house developed | ||
| Immersion oil | Cargille Laboratories | 16482 | Type A low viscosity 150 cSt |
| inSituX platform | In-house developed | ||
| IR light source | Thorlabs Incorporated | LED1085L | LED with a Glass Lens, 1085 nm, 5 mW, TO-18 |
| Outer ring | In-house developed | ||
| Pump lasers | Thorlabs Incorporated | LD785-SE400 | 785 nm, 400 mW, Ø9 mm, E Pin Code, Laser Diode |
| Raspberry Pi | Raspberry Pi Fundation | ||
| Retaining ring | Thorlabs Incorporated | SM1RR | SM1 retaining ring for Ø1" lens tubes and mounts |
| Seedless quartz crystal | University Wafers, Inc. | U01-W2-L-190514 | 25.4 mm diameter Z-cut 0.05 mm thickness double side polish 8 mm on -X |
| Shim | In-house developed | ||
| X-ray beam stop | In-house developed |
References
- Wiwatanaratanabutr, I., Kittayapong, P. I. Effects of crowding and temperature on Wolbachia infection density among life cycle stages of Aedes albopictus. Journal of Invertebrate Patholology. 102 (3), 220-224 (2009).
- Sinkins, S. P., Braig, H. R., O’Neill, S. L. Wolbachia superinfections and the expression of cytoplasmic incompatibility. Proceedings of Biologial Sciences. 261 (1362), 325-330 (1995).
- Dobson, S. L., et al. Wolbachia infections are distributed throughout insect somatic and germ line tissues. Insect Biochemistry and Molecular Biology. 29 (2), 153-160 (1999).
- O’Neill, S. L., et al. In vitro cultivation of Wolbachia pipientis in an Aedes albopictus cell line. Insect Molecular Biology. 6 (1), 33-39 (1997).
- Sinha, A., Li, Z., Sun, L., Carlow, C. K. S. Complete genome sequence of the Wolbachia wAlbB endosymbiont of Aedes albopictus. Genome Biology and Evoution. 11 (3), 706-720 (2019).
- Sinkins, S. P. Wolbachia and cytoplasmic incompatibility in mosquitoes. Insect Biochemistry and Molecular Biology. 34 (7), 723-729 (2004).
- Fallon, A. M. Cytological properties of an Aedes albopictus mosquito cell line infected with Wolbachia strain wAlbB. In Vitro Cellular Developmental Biology – Animals. 44 (5-6), 154-161 (2008).
- Hilgenboecker, K., Hammerstein, P., Schlattmann, P., Telschow, A., Werren, J. H. How many species are infected with Wolbachia?-A statistical analysis of current data. Microbiology Letters. 281 (2), 215-220 (2008).
- Werren, J. H., Baldo, L., Clark, M. E. Wolbachia: master manipulators of invertebrate biology. National Review of Microbiology. 6 (10), 741-751 (2008).
- Kittayapong, P., Baisley, K. J., Baimai, V., O’Neill, S. L. Distribution and diversity of Wolbachia infections in Southeast Asian mosquitoes (Diptera: Culicidae). Journal of Medical Entomology. 37 (3), 340-345 (2000).
- O’Neill, S. L., Hoffmann, A., Werren, J. . Influential passengers: inherited microorganisms and arthropod reproduction. , (1997).
- McGraw, E. A., O’Neill, S. L. Beyond insecticides: new thinking on an ancient problem. National Review of Microbiology. 11 (3), 181-193 (2013).
- Bourtzis, K., et al. Harnessing mosquito-Wolbachia symbiosis for vector and disease control. Acta Tropica. 132, 150-163 (2014).
- Turelli, M., Hoffmann, A. A. Rapid spread of an inherited incompatibility factor in California Drosophila. Nature. 353 (6343), 440-442 (1991).
- Hoffmann, A. A., et al. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature. 476 (7361), 454-457 (2011).
- Walker, T., et al. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature. 476 (7361), 450-453 (2011).
- Hughes, G. L., Koga, R., Xue, P., Fukatsu, T., Rasgon, J. L. Wolbachia infections are virulent and inhibit the human malaria parasite Plasmodium falciparum in Anopheles gambiae. PLoS Pathogens. 7 (5), 1002043 (2011).
- Bian, G., Xu, Y., Lu, P., Xie, Y., Xi, Z. The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS Pathogens. 6 (4), 1000833 (2010).
- Moreira, L. A., et al. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell. 139 (7), 1268-1278 (2009).
- Caragata, E. P., et al. Dietary cholesterol modulates pathogen blocking by Wolbachia. PLoS Pathogens. 9 (6), 1003459 (2013).
- Zhang, G., Hussain, M., O’Neill, S. L., Asgari, S. Wolbachia uses a host microRNA to regulate transcripts of a methyltransferase, contributing to dengue virus inhibition in Aedes aegypti. Proceedings of the National Academy of Sciences of the United States of America. 110 (25), 10276-10281 (2013).
- Tortosa, P., Courtiol, A., Moutailler, S., Failloux, A. B., Weill, M. Chikungunya-Wolbachia interplay in Aedes albopictus. Insect Molecular Biology. 16 (7), 677-684 (2008).
- Lu, P., Bian, G., Pan, X., Xi, Z. Wolbachia induces density-dependent inhibition to dengue virus in mosquito cells. PLoS Neglected Tropical Diseases. 6 (7), 1754 (2012).
- Ghosh, A., Jasperson, D., Cohnstaedt, L. W., Brelsfoard, C. L. Transfection of Culicoides sonorensis biting midge cell lines with Wolbachia pipientis. Parasite Vectors. 12 (1), 483 (2019).
- Zhou, W., Rousset, F., O’Neill, S. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. The Royal Society Publishing. Proceedings B. 265 (1395), 509-515 (1998).
- Park, M. S., Takeda, M. Cloning of PaAtg8 and roles of autophagy in adaptation to starvation with respect to the fat body and midgut of the Americana cockroach, Periplaneta americana. Cell Tissue Research. 356 (2), 405-416 (2014).
- Geng, S. C., Li, X. L., Fang, W. H. Porcine circovirus 3 capsid protein induces autophagy in HEK293T cells by inhibiting phosphorylation of the mammalian target of rapamycin. Journal of Zhejiang University Science B. 21 (7), 560-570 (2020).
- Taylor, S. C., Laperriere, G., Germain, H. Droplet digital PCR versus qPCR for gene expression analysis with low abundant targets: from variable nonsense to publication quality data. Scientific Reports. 7 (1), 2409 (2017).
- Kosea, H., Karr, T. L. Organization of Wolbachia pipientis in the Drosophila fertilized egg and embryo revealed by an anti-Wolbachia monoclonal antibody. Mechanisms of Development. 51 (2-3), 275-288 (1995).
- Ye, Y. H., et al. Wolbachia reduces the transmission potential of dengue-infected Aedes aegypti. PLoS Neglected Tropical Diseases. 9 (6), (2015).
- Jensenius, M., et al. Comparison of immunofluorescence, Western blotting, and cross-adsorption assays for diagnosis of African tick bite fever. Clinical and Diagnostic Laboratory Immunology. 11 (4), 786-788 (2004).

