Standardized In vitro Assays to Visualize and Quantify Interactions between Human Neutrophils and Staphylococcus aureus Biofilms
Summary
The present protocol describes the study of neutrophil-biofilm interactions. Staphylococcus aureus biofilms are established in vitro and incubated with peripheral blood-derived human neutrophils. The oxidative burst response from neutrophils is quantified, and the neutrophil localization within the biofilm is determined by microscopy.
Abstract
Neutrophils are the first line of defense deployed by the immune system during microbial infection. In vivo, neutrophils are recruited to the site of infection where they use processes such as phagocytosis, production of reactive oxygen and nitrogen species (ROS, RNS, respectively), NETosis (neutrophil extracellular trap), and degranulation to kill microbes and resolve the infection. Interactions between neutrophils and planktonic microbes have been extensively studied. There have been emerging interests in studying infections caused by biofilms in recent years. Biofilms exhibit properties, including tolerance to killing by neutrophils, distinct from their planktonic-grown counterparts. With the successful establishment of both in vitro and in vivo biofilm models, interactions between these microbial communities with different immune cells can now be investigated. Here, techniques that use a combination of traditional biofilm models and well-established neutrophil activity assays are tailored specifically to study neutrophil and biofilm interactions. Wide-field fluorescence microscopy is used to monitor the localization of neutrophils in biofilms. These biofilms are grown in static conditions, followed by the addition of neutrophils derived from human peripheral blood. The samples are stained with appropriate dyes prior to visualization under the microscope. Additionally, the production of ROS, which is one of the many neutrophil responses against pathogens, is quantified in the presence of a biofilm. The addition of immune cells to this established system will expand the understanding of host-pathogen interactions while ensuring the use of standardized and optimized conditions to measure these processes accurately.
Introduction
A biofilm is a community of surface-associated microbes or non-attached aggregates encased in an extracellular polymeric substance (EPS)1,2. These communities protect the encased microorganisms from environmental stressors, including tolerance to antimicrobial agents and the immune system3. Several pathogenic microbial species form biofilms that have been associated with chronic infections4. The development of biofilms is an intricate process involving attachment to surfaces, EPS production, cell proliferation, biofilm structuring, and cell detachment5. Once cells disperse to form a biofilm, they remain planktonic or translocate to a new substratum and re-initiate biofilm development6.
Staphylococcus aureus, an opportunistic pathogen, follows a general scheme of biofilm development, including attachment, proliferation, maturation, and dispersal7. The attachment process in S. aureus biofilms is dictated by hydrophobic interactions, teichoic acids, and microbial surface components recognizing adhesive matrix molecules (MSCRAMMs)8,9. As the proliferation of S. aureus begins, EPS, which primarily consists of polysaccharides, proteins, extracellular DNA, and teichoic acids, is produced5. As EPS components are produced, various exoenzymes and small molecules are also produced, contributing to the biofilm 3-dimensional structure and aiding in detachment5. S. aureus takes advantage of this highly coordinated lifestyle to establish various chronic infections, including infections due to the indwelling of medical devices10.
Methicillin-resistant S. aureus (MRSA) is one of the leading causes of infections related to indwelling medical devices, such as central venous and urinary catheters, prosthetic joints, pacemakers, mechanical heart valves, and intrauterine devices11. During such infections, neutrophils are the first host immune cells recruited to the infection site to combat pathogens via multiple strategies12. These include phagocytosis, degranulation, reactive oxygen and nitrogen species (ROS/RNS) production, or release of neutrophil extracellular traps (NETs) to eliminate pathogens13.
Generation of ROS upon phagocytosis of microbes is one of the key antimicrobial responses exhibited by neutrophils14. Phagocytosis is enhanced if microbes are coated in opsonins, particularly immunoglobulins and complement components found in serum15. The opsonized microbes are then recognized by cell surface receptors on neutrophils and engulfed, forming a compartment called the phagosome15. Neutrophils generate and release ROS in the phagosome via the membrane-associated NADPH-oxidase16. This multi-component enzyme complex generates superoxide anions by transferring electrons to molecular oxygen16. Additionally, neutrophils also generate RNS through the expression of inducible nitric oxide synthase (iNOS)17. These high superoxide and nitric oxide radicals within the phagosome have broad antimicrobial activities. They can interact with metal centers in enzymes and damage nucleic acids, proteins, and cell membranes of the pathogen18,19,20,21. Numerous microbes adopt a biofilm lifestyle and employ different strategies to evade killing by ROS22,23. Thus, standardized assays that couple biofilms with neutrophils to quantify ROS are beneficial for consistent results.
While assays, such as quantifying neutrophil ROS production, provide information about the responses of neutrophils to biofilms, the ability to visualize the interactions of neutrophils within a biofilm can also serve as a powerful tool. The use of fluorescent dyes for microscopy often requires optimization to obtain high-quality images that can be used for microscopy imaging analysis. The flexibility to optimize some conditions is limited as neutrophils can undergo cell death post-isolation. Furthermore, biofilms are typically washed to remove the planktonic population from the experimental set-up before the addition of neutrophils. While washing, variability between replicate biofilms may arise due to loss of partial biomass if biofilms are loosely adhered to the surface.
Broadly, current methods in the field to analyze interactions between neutrophils and biofilms mainly include microscopy, flow cytometry, and colony-forming units (CFU) enumeration24,25,26,27. Microscopy involves the use of dyes that either directly stain the neutrophils and biofilms, or target various neutrophil responses against microbes such as NET formation, degranulation, and cell death25,28. A subset of these responses, such as neutrophil cell death and degranulation, can also be analyzed via flow cytometry, but requires neutrophils to be preferentially unassociated with large aggregates of microbes in a biofilm28,29. Flow cytometry can also quantify some biofilm parameters, such as cell viability27. These processes, however, require disruption of the biofilm biomass and would not be useful to visualize other important interactions such as the spatial distribution of neutrophils and their components within a biofilm27,29,30.
The present protocol focuses on adapting some of the traditionally used methods to study neutrophil-biofilm interactions on biofilms that have been optimized to provide minimal variability during handling. This protocol thus provides standardized methods to grow and quantify biofilms, isolate primary human neutrophils from peripheral blood, quantify ROS production, and visualize biofilm-neutrophil interactions via microscopy. This protocol can be adapted to different systems to understand biofilm-neutrophil interactions while considering the heterogeneity among donor pools.
Protocol
All procedures were approved by the Ohio State University Institutional Review Board (IRB) (2014H0154). Informed written consent was obtained from all the donors for collecting peripheral blood to isolate primary human neutrophils. Staphylococcus aureus (USA300 LAC)31 was used as the model organism for performing the experiments. The experiments were performed with proper personal protective equipment (PPE) due to potential exposure to a bloodborne pathogen.
1. Preparation of in vitro biofilm
- Obtain isolated colonies of S. aureus from a cryopreserved stock31 using a streak-plate technique32,33 on a nutrient-rich agar plate, such as Tryptic Soy Agar (see Table of Materials).
- Coat individual wells of a 96-well plate with 100 µL of 0.001% (v/v) poly-L-Lysine (PLL) diluted in sterile H2O and incubate at room temperature for 30 min. Aseptically, aspirate the PLL solution using a vacuum-assisted aspiration trap. Allow the wells to dry overnight at room temperature.
NOTE: All aspiration steps in the protocol are performed using a vacuum-assisted aspiration trap unless otherwise stated. - Prepare an overnight culture by inoculating a colony of S. aureus in minimal essential media alpha (MEMα) supplemented with 2% glucose and incubate at 37 °C, shaking at 200 rpm for 16-18 h.
- Dilute the overnight culture by transferring 50 µL to 5 mL of fresh MEMα supplemented with 2% glucose and incubate at 37 °C, shaking at 200 rpm, until mid-logarithmic phase, generally between optical density 600 (OD600nm) of 0.5-0.8. Use MEMα to normalize mid-logarithmic culture to an OD600nm of 0.1.
- Transfer 150 µL of normalized culture to each well of the PLL treated 96-well plate. Incubate statically for 18-20 h in a humidified chamber at 37 °C.
NOTE: The biofilms can also be grown in other formats such as µ-channel slides (see Table of Materials). - Aspirate the supernatant to remove the planktonic cells. Gently wash remaining biomass with 150 µL of Hanks' Balanced Salt Solution (HBSS) to remove the unattached cells. Add HBSS dropwise to avoid disrupting the biofilm.
NOTE: While aspirating the supernatant and HBSS during washes, leave just enough liquid (supernatant or HBSS) in the wells containing biofilm such that the biofilm is still immersed. This prevents the disruption of the biofilm structure when HBSS is added dropwise to wash the biofilm. - Repeat step 1.6 at least two more times to remove all the planktonic cells. At this point, biofilms are ready for immediate downstream experiments.
NOTE: If the biofilms are not used for neutrophil experiments, HBSS can be substituted with phosphate-buffered saline (PBS). HBSS is preferred over PBS as HBSS contains components, including glucose, that provide optimum conditions for neutrophil activation34.
2. Quantification of biofilm biomass
- Prepare a stock of 0.1% (w/v) Crystal Violet (CV) solution (see Table of Materials) by dissolving in 20% (v/v) ethanol and 80% (v/v) H2O. Ensure that the CV is completely dissolved in ethanol prior to adding H2O. Filter-sterilize the solution.
- Add 150 µL of 0.1% CV solution to the washed biofilm and incubate for 20 min at room temperature. Use at least three empty wells as media-only controls.
- Aspirate the 0.1% CV solution from the biofilms and wash the stained biofilms with 200 µL of 1x PBS. Repeat this process for a total of three washes to remove any excess CV from the wells.
- Add 150 µL of 33% (v/v) glacial acetic acid diluted with H2O. Incubate at room temperature on a rocker at 50 rpm for 30 min to allow the CV bound to the biomass to dissolve completely.
CAUTION: Perform this step in a laminar flow hood with appropriate PPE as glacial acetic acid is a corrosive chemical. - In the meantime, set up the microplate reader (see Table of Materials) to read the CV stain values. Following glacial acetic acid treatment, read the plate at 595 nm wavelength.
NOTE: The wavelength used to measure the OD of CV can range from 500-600 nm35.
3. Neutrophil isolation
NOTE: Neutrophils were isolated following a previously published method with minor changes36. This isolation protocol combines density gradient centrifugation first, followed by 3% dextran sedimentation. This section only covers the overall neutrophil isolation protocol, focusing on the changes made to the published protocol. Furthermore, the protocol outlined below is one of the many methods that can isolate neutrophils, and can be substituted as needed. Other methods for isolating neutrophils include the use of cell separation media or magnetic antibody cell separation37.
- Draw blood from an adult donor via venipuncture, as per the protocol outlined in the institutional IRB. Prior to the blood draw, ensure that the syringe has sufficient preservative-free heparin, such that the final concentration of heparin is 20 U/mL.
- Dilute the heparinized blood with 3/4 the volume of endotoxin-free 0.9% NaCl (see Table of Materials) in H2O at room temperature.
- For every 20 mL of the diluted blood sample, aliquot 14 mL of a commercially available density gradient medium (see Table of Materials) in a fresh 50 mL conical tube. Carefully layer the diluted blood sample on top of the density gradient medium.
- Centrifuge the layered blood sample at 400 x g for 40 min at room temperature. Ensure that the centrifuge has a slow break to avoid disturbing the layer once the centrifugation is completed.
NOTE: The blood sample will have five layers containing a mixture of saline and plasma, a mononuclear cell layer, density gradient medium, neutrophils, and erythrocytes. - Using a serological pipette, aspirate all the layers above the neutrophils and erythrocyte pellet, followed by a gentle resuspension of the pellet in cold endotoxin-free 0.9% NaCl in H2O. For each pellet generated from a 20 mL blood sample, resuspend the pellet back to 20 mL volume total. Add 1:1 volume of 3% dextran (see Table of Materials). Incubate the tube upright for 18-20 min on ice.
NOTE: Ensure that the 3% dextran is made with endotoxin-free 0.9% NaCl in H2O. - Remove 20 mL of the upper layer that contains neutrophils and some erythrocytes onto a new 50 mL conical tube and centrifuge it at 355 x g for 10 min at 4 °C. Pour off the supernatant leaving behind a red pellet.
- Gently resuspend the pellet in 10 mL of cold, sterile H2O for 30 s to lyse the remaining erythrocytes. Immediately add 10 mL of cold endotoxin-free 0.9% saline to the mixture to restore tonicity. Centrifuge the solution at 233 x g for 3 min at 4 °C.
- Pour off the supernatant and resuspend the pellet containing 95%-97% neutrophils in 1 mL cold HBSS per 20 mL of the blood sample.
- Transfer 10 µL of the resuspended neutrophils in 90 µL of 0.4% trypan blue exclusion dye and count the cells using a hemocytometer (see Table of Materials).
NOTE: Non-viable cells are stained blue as trypan blue exclusion dye is impermeable in viable cells. This protocol provides >99% cell viability37,38. - Add additional HBSS such that the final concentration of neutrophils is 4 x 106 cells/mL.
NOTE: For instances with <99% cell viability, the final concentration of 4 x 106 cells/mL can still be achieved; however, the total volume of solution containing 4 x 106 cells/mL obtained will decrease. The final concentration of neutrophils can be adjusted according to the user's experimental needs. The neutrophils were resuspended at a 4 x 106 cells/mL final concentration for all the experiments described below. To account for donor-to-donor variability, it is strongly recommended that all experiments involved with neutrophils be performed with at least three different donors.
4. Measurement of ROS produced by neutrophils
- Add 100 µL of 20% normal human serum (diluted in HBSS) dropwise to the washed biofilm (step 1.6) and incubate at 37 °C under static condition for 30 min to opsonize the biofilm.
- Aspirate the 20% serum solution and wash the biofilms dropwise with 150 µL of HBSS once. Aspirate the HBSS, leaving behind wells with opsonized biofilms.
NOTE: For interpretation of the experiment, a minimum of four groups is recommended: (A) Neutrophils + Biofilm, (B) Neutrophils + PMA (positive control, see Table of Materials), (C) Neutrophils only, and (D) Biofilm only. - Add luminol (see Table of Materials) to the neutrophils resuspended in HBSS at a concentration of 4 x 106 cells/mL such that the final luminol concentration is 50 µM. This solution is ready to use for groups (A) and (C). Add 4 x 105 neutrophils mixed with luminol to the wells with opsonized biofilms.
- In a separate tube, prepare 50 µM luminol solution in HBSS without any neutrophils and add it to the well containing biofilm (group D).
- Aliquot 350 µL of neutrophils mixed with luminol and add phorbol 12-myristate 13-acetate (PMA) at a final concentration of 500 ng/mL to the mixture. For group (B), add 4 x 105 neutrophils from this mixture into wells without biofilm. This serves as a positive control.
NOTE: The concentration of PMA indicated in this step is relatively high to ensure robust burst response as PMA stimulated neutrophils is a positive control. PMA can be used at a lower concentration to activate neutrophils, depending on the experiment. - Centrifuge the plate at 270 x g for 30 s at 4 °C.
- Ensure the plate reader is set to 37 °C along with the setting for luminescence and kinetic read for 60 min with 3 min intervals. Place the plate in the plate reader to measure ROS production by neutrophils for 60 min.
NOTE: For this assay, biofilms were grown in white plates used for luminescence assays. PMA is a known agonist for the oxidative burst response39. When performing studies involving PMA, ensure that PMA is added at the final step while the solution containing neutrophils is cold since PMA immediately initiates the burst response.
5. Imaging biofilm-neutrophil interactions
- Set up a biofilm using steps 1.2-1.6. To facilitate biofilm imaging, employ a fluorescent strain of S. aureus, such as USA300 expressing green fluorescent protein (GFP)40,41, to increase the ease of microscopy imaging.
NOTE: A 6 µ-channel slide (see Table of Materials) was used instead of a 96-well plate to demonstrate the in vitro biofilm model (step 1). - Incubate 4 x 106 cells/mL of neutrophils with 100 µM of Blue CMAC (7-amino-4-chloromethylcoumarin) Dye (BCD, see Table of Materials) for 30 min in a rocker at 37 °C and 5% CO2. Ensure the samples are incubated in the dark and limit exposure to light for the remaining steps.
- To wash excess BCD, centrifuge neutrophils at 270 x g for 5 min and aspirate the supernatant. Resuspend the neutrophils in fresh HBSS. At this point, add ethidium homodimer-1 (see Table of Materials) to the BCD-stained neutrophils at a final concentration of 4 µM to monitor neutrophil and bacterial death.
- Add 150 µL of neutrophils to the S. aureus biofilm that has been grown in µ-slides, such that the neutrophil to bacteria ratio is 1:30 (neutrophil: bacteria). Incubate the µ-slides in a humidified chamber for 30 min. The number of bacterial cells is based on the cell counts obtained from plating an 18 h biofilm.
- Image the neutrophil-biofilm interaction using fluorescent channels corresponding to the excitation and emission wavelengths of the fluorescent dyes/proteins.
NOTE: For the present study, BCD is 353/466 nm, ethidium homodimer-1 is 528/617 nm, and GFP is 395/509 nm. Limit the exposure of the sample to the laser or the light to prevent photobleaching of the samples. - Analyze the images using microscopy image analysis software or programs such as FIJI/ImageJ, COMSTAT2, BiofilmQ, and BAIT, among many more42,43,44,45.
NOTE: When working with stains, it is important to consider the specificity of the dyes in use. Some stains work on prokaryotic and eukaryotic cells, while others work only on one. If neutrophils and biofilms are separately stained using dyes that can stain both cell types, ensure to wash off any remaining dye before combining neutrophils and biofilms to prevent cross-staining.
Representative Results
The media used to grow bacterial biofilms influence the survival of neutrophils. Different media were tested to reduce the effect of media alone on the viability of neutrophils for studying neutrophil-biofilm interactions (Figure 1). Bacterial growth media such as Tryptic Soy Broth minimizes the viability of neutrophils, such that ~60% of neutrophils are alive after a 30 min incubation period at 37 °C with 5% CO2. Mammalian cell culture media, such as MEMα, does not affect the viability of neutrophils and supports the growth of S. aureus biofilms. In fact, minimal media promotes robust growth of biofilms in other bacteria46,47.
To assess the effect of media on biofilm growth and variability in biofilm biomass quantification after washing the biomass to eliminate planktonic cells, an 18 h S. aureus biofilm was grown in a 96-well plate, with wells treated or untreated with poly-L-Lysine. A nutrient-rich (Tryptic Soy Broth (TSB)) and minimal (MEMα) media were used as-is or supplemented with 2% glucose. The biofilm biomass stained with CV revealed that S. aureus biofilm grown in MEMα supplemented with 2% glucose produced the most robust biofilm among all tested media (Figure 2A). Furthermore, biofilms grown in PLL pretreated wells containing MEMα + 2% glucose showed less variability than biofilms in PLL-untreated wells containing MEMα + 2% glucose. These biofilms showed less variability in quantification via CV assay35 and the CFU/mL when plated after precisely handling biofilms for biomass quantification. These biofilms contained, on average, 1 x 108 CFU/mL, as demonstrated by plating the biofilms in 3 separate days (Figure 2B). This number is useful in determining the number of neutrophils to add to the biofilms for neutrophil functionality assays.
To measure ROS production by neutrophils in response to biofilms, S. aureus biofilms were grown statically for 18-20 h in a 96-well plate. Biofilms were then opsonized, and neutrophils were added. ROS production was then measured for 60 min (Figure 3A). The area under the curve is calculated from the kinetic curve to quantify total ROS production by neutrophils. Neutrophils treated with an agonist, such as PMA, used as a control, show an increased ROS production. In the absence of biofilms, neutrophils treated with PMA showed robust ROS production. In the presence of S. aureus biofilm, the overall ROS production by neutrophils treated with PMA decreased. In the absence of PMA, neutrophils solely rely on their interaction with the biofilm, which further reduces the amount of ROS produced (Figure 3B).
To visualize the neutrophil-biofilm interactions using fluorescence microscopy, a GFP-expressing strain of S. aureus, Blue CMAC dye, and ethidium homodimer-1, that stains the cytoplasm of live cells and DNA of dead cells, respectively, were used. S. aureus biofilm was grown for 18 h in a 6 µ-channel slide. Blue CMAC dye-labeled neutrophils were added along with ethidium homodimer-1 to the washed biofilms and incubated for 30 min at 37 °C with 5% CO2 prior to imaging. Wide-field fluorescent microscopy revealed that many neutrophils were localized to the surface of S. aureus biofilms, while a few are within the biofilm (Figure 4A). The interaction between S. aureus cells within neutrophils was also apparent (Figure 4C). Most of the S. aureus cells interacting with neutrophils (cyan) were dead (magenta), while a few remained alive (yellow) as determined by live-dead staining (Figure 4C). For comparison, GFP-expressing S. aureus biofilms were stained with ethidium homodimer-1, which revealed a fraction of the dead S. aureus population within the biofilm (Figure 4B). Non-viable neutrophils that were positive for ethidium homodimer-1 were quantified using analysis software (see Table of Materials) after incubation with S. aureus biofilms. Approximately 48% of neutrophils were already dead within 30 min of incubation with S. aureus biofilm. During optimization of the microscopy protocol, the effect of washing the biofilm and neutrophils after 30 min of incubation to remove non-adhered neutrophils was also assessed, revealing around 33% of dead neutrophils still attached to the biofilm (Figure 4D).
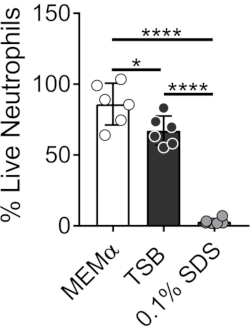
Figure 1: LIVE-DEAD assay compares neutrophil survival between bacterial and mammalian growth media. Neutrophils were isolated and incubated in HBSS, MEMα, TSB, or 0.1% SDS for 30 min. LIVE-DEAD staining was performed using Calcein AM (live) and ethidium homodimer-1 (dead). Percent of live neutrophils was determined, where HBSS-incubated neutrophils were treated as 100% live neutrophils. Results represent an average of two independent experiments performed in triplicate, with neutrophils obtained from two different donors. Data are presented as mean ± SD (*p < 0.05, ****p < 0.0001. One-way ANOVA). Please click here to view a larger version of this figure.
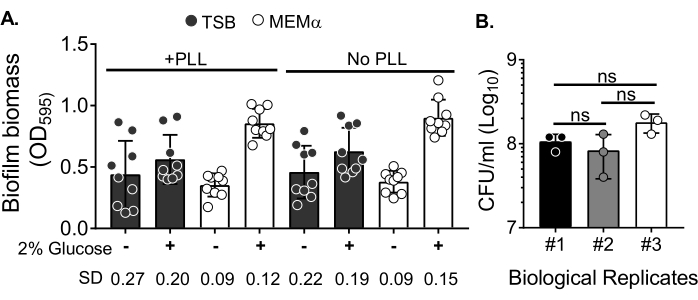
Figure 2: Quantification of biofilm biomass in different conditions and bacterial viability count of biofilms grown in the optimized conditions. (A) S. aureus was seeded in a 96-well plate either coated or uncoated with poly-L-Lysine (PLL). Biofilms were grown in TSB, MEMα, or either of the media supplemented with 2% glucose under static conditions for 18 h. Crystal violet (CV) staining was performed to stain biofilm biomass. The eluted CV stain was diluted at 1:10 and read in a microplate reader. Results represent an average of three independent experiments performed in triplicate. Data are presented as mean ± SD. The SD for each group is shown at the bottom to demonstrate different biofilm growth conditions variability. (B) Bacterial CFU counts were obtained from biofilms grown in an optimized medium (MEMα + 2% glucose). The 18 h static biofilms were subjected to the same number of washes followed by a 10 min sonication to loosen the biofilm biomass and passed through a 22G needle to disrupt the aggregates prior to plating. Results represent three replicates performed in triplicate. Data are presented as mean ± SD (ns = not significant. One-way ANOVA). Please click here to view a larger version of this figure.
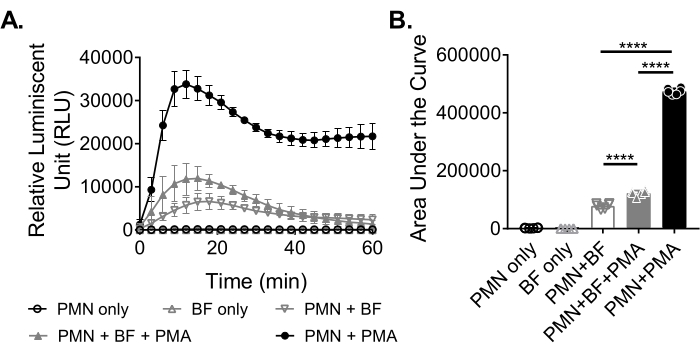
Figure 3: Quantification of ROS production by neutrophils via chemiluminescence assay. (A) Neutrophils (PMN) were incubated with HBSS-washed S. aureus biofilms (BF) either in the presence (closed gray triangle) or absence (open gray inverted triangle) of PMA to measure ROS production by neutrophils. Luminol was used to detect ROS every 3 min for 60 min in a microplate reader. While neutrophils treated with PMA in the absence of a biofilm (closed black circle) served as a positive control, neutrophil only (open black circle) and biofilm only (open gray triangle) groups served as negative controls. Data represent an average of two independent experiments performed in triplicate with neutrophils obtained from two different donors. Data are presented as mean ± SD. (B) The area under the curve from (A) was calculated to quantify the total ROS generated by the neutrophils. The data are represented as mean ± SD. (***p < 0.0001. One-way ANOVA). Please click here to view a larger version of this figure.
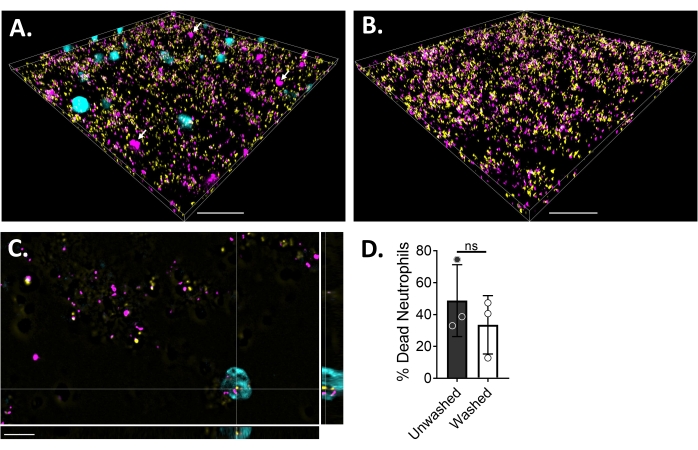
Figure 4: Visualization of the interaction between S. aureus biofilm and neutrophils using wide-field fluorescence microscopy. Blue CMAC dye-labeled neutrophils (cyan) were supplemented with ethidium homodimer-1 (magenta; dead) prior to incubating with an 18 h S. aureus biofilm (yellow). Biofilm-neutrophil interactions were imaged using wide-field fluorescent microscopy and images processed using an image analysis software. Experiments were performed with three different donors. Representative images are presented as (A) 3D view of S. aureus biofilm with live (cyan) and dead (magenta; a few indicated with white arrows) neutrophils, (B) 3D view of an S. aureus biofilm in the absence of neutrophils with either live S. aureus expressing GFP (yellow) or dead S. aureus stained with ethidium homodimer-1 (magenta), (C) an orthogonal view of S. aureus and neutrophil interaction as depicted by the xy, yz, and xz planes, and (D) quantification of neutrophil viability in the presence of S. aureus biofilm after 30 min either immediately (unwashed) or after three rounds of washes with HBSS to remove non-adhered neutrophils (washed). Neutrophil cell death is presented as mean ± SD (Student's t-test). Scale bar indicates 50 µm in (A) and (B) and 10 µm in (C). Please click here to view a larger version of this figure.
Discussion
There have been numerous efforts to grow robust and reproducible S. aureus biofilms for downstream experiments in vitro48,49,50. A standardized protocol is outlined that takes advantage of the cationic nature of PLL, as well as supplementing the media with glucose for the growth of robust in vitro S. aureus biofilms. The addition of PLL allows for better attachment of the negatively charged bacterial cell to the positively charged PLL coated surfaces. It is important to note that PLL at a 10 µg/mL concentration has antimicrobial activity against Pseudomonas aeruginosa, Escherichia coli, and S. aureus when incubated for 24 h51. The same concentration is used to coat surfaces; however, excess PLL is aspirated, making the concentration of PLL lower than 10 µg/mL when seeding for biofilm growth.
It is important to note that PLL has worked only in specific growth media such as MEMα with 2% glucose, where it was observed that S. aureus produced robust biofilms with minimal variability (Figure 2A). PLL concentration to be used in conjunction with other media types would require further optimization, such as using an increased concentration of PLL to coat the wells. Additionally, these conditions have been optimized for a monospecies S. aureus biofilm. While chronic wound biofilms are often polymicrobial, standardizing assays to study monospecies biofilm and its interactions with neutrophils and other immune cells is key in understanding their contribution to pathogenesis52. These standardized protocols can be optimized further to sustain and study polymicrobial biofilms and their interactions with neutrophils.
It was also observed that rich bacterial culture media, such as TSB, led to a loss of neutrophil viability (Figure 1). Therefore, growth conditions of S. aureus biofilms in MEMα, used for mammalian cell cultures, were optimized. For studies involving neutrophils, this media supports neutrophil viability and promotes S. aureus growth. While it was observed that media affects the viability of neutrophils, it is also important to consider that neutrophils isolated from peripheral human blood undergo apoptosis ex vivo with approximately 70% apoptotic neutrophils by 20 h53. This necessitates proper handling, such as storing the neutrophils on ice when preparing for experiments, using endotoxin-free reagents, and preventing activation of neutrophils by avoiding vortexing of samples with neutrophils.
The assessment of oxidative burst in neutrophils is routinely performed to determine the killing effect of neutrophils on the pathogen14,54,55. These studies are frequently performed with planktonic bacteria where neutrophils are added, and the oxidative burst response is quantified using luminol-amplified chemiluminescence that detects superoxide anions produced by neutrophils. The present protocol is modified by replacing planktonic bacteria with statically grown 18 h S. aureus biofilm. As such, neutrophils can be directly added to the biofilm to assess their activation. On the other hand, bacteria in biofilms produce enzymes, such as catalase and superoxide dismutase to detoxify ROS23,56. Staphylococcus epidermidis biofilms produce higher catalase than its planktonic counterpart under stress57. The total chemiluminescence of PMA-stimulated neutrophils in an S. aureus biofilm is significantly lower than the PMA-stimulated neutrophils where biofilm is absent (Figure 2). This may be due to the activity of these detoxifying enzymes. Furthermore, S. aureus biofilms produce several pore-forming toxins called leukocidins that kill neutrophils58. The reduced burst response is also likely due to the reduced viability of neutrophils in the presence of S. aureus biofilm. While this study uses luminol that detects the total ROS produced both inside and outside of the cells, other reagents, such as CM-H2DCFDA (5-(and-6)-chloromethyl-2'7'-dichlorodihydrofluorescein diacetate) or isoluminol, need to be considered if the goal of the work is to study intracellular or extracellular ROS production14,53,54 specifically.
The ability to visualize neutrophil-biofilm interactions via microscopy can be informative about the behavior of neutrophils and biofilms in the presence of each other. The excitation and emission spectra of the fluorescent dyes and proteins represent a snapshot of the interaction between an 18 h S. aureus biofilm and neutrophils after a 30 min incubation. To effectively capture signals from stained cells, it is important to limit exposure of the samples to light sources while setting up the samples for microscopy. While imaging, rapid photobleaching of the samples was avoided by lowering the intensity of the light source when adjusting all the parameters such as Z-stack height and exposure time for different channels.
These simple practices allowed for proper microscopy imaging where it was observed that few neutrophils are localized within the biofilm (Figure 4A). This may be due to spaces present within the biofilm as 18 h S. aureus biofilm grown in MEMα with 2% glucose does not uniformly cover the surface (Figure 4B). However, other studies' use of rich media has shown a uniform lawn of S. aureus biofilm growth and leukocytes penetrating through the biofilm30,58. Furthermore, it is also observed that there was neutrophil cell death after 30 min of incubation with S. aureus biofilms due to S. aureus biofilm-produced leukocidins that lyse neutrophils58 (Figure 4A,D). Addition of a wash step to remove non-adhered neutrophils after incubating them with biofilm for 30 min removed ~15% of dead neutrophils from the system compared to the unwashed group, in which microscopy was performed immediately after 30 min of incubation (Figure 4D). Neutrophils interacting with S. aureus were also observed (Figure 4C). Further experiments are required to assess whether S. aureus is engulfed by neutrophils or attached to the cell surface of neutrophils54. Imaging neutrophils and biofilms is the first step to evaluate several neutrophil functionalities downstream, such as phagocytosis and NETosis54,59. The effect of neutrophils on biofilms can also be assessed by quantifying the biofilm biomass, structural changes of the biofilm, and biofilm viability, among many others, using image analysis tools listed in step 5.6. Lastly, donor-to-donor variability exists in neutrophils; thus, it is recommended that at least three different donors be used for studies involving neutrophils.
Overall, standardized in vitro assays were combined to assess interactions between neutrophils and biofilms. Though these assays utilize S. aureus, the protocols described can easily be adapted to study other pathogens. While there are various in vivo models to study host-pathogen interactions, they can be expensive and labor-intensive, especially if the conditions are not optimized. Working with standardized in vitro assays allows one to optimize experimental conditions and confirm observations prior to moving to an in vivo system. Finally, various animal infection models have been used to study biofilm-neutrophil interactions in vivo. However, it is important to consider immunological differences between humans and animal models60,61,62,63. This necessitates using neutrophils derived from humans to study these complex host-pathogen interactions.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was funded by the National Institute of Allergy and Infectious Diseases (R01AI077628) to DJW and an American Heart Association Career Development Award (19CDA34630005) to ESG. We thank Dr. Paul Stoodley for providing us with USA 300 LAC GFP strain. Furthermore, we acknowledge resources from the Campus Microscopy and Imaging Facility (CMIF) and the OSU Comprehensive Cancer Center (OSUCCC) Microscopy Shared Resource (MSR), The Ohio State University. We also thank Amelia Staats, Peter Burback, and Lisa Coleman from the Stoodley lab for performing blood draws.
Materials
| 0.9% sodium chloride irrigation, USP | Baxter | 2F7124 | Endotoxin-free; Used for isolation of neutrophils |
| 150 mL rapid-flow filter unit | Thermo Scientific | 565-0020 | |
| 200 proof ethanol | VWR | 89125-188 | |
| 3 mL syringe | BD | 309657 | Used for blood draw |
| 50 mL conical centrifuge tubes | Thermo Scientific | 339652 | |
| 60 mL syringe | BD | 309653 | Used for blood draw |
| Agar | Fisher Bioreagents | BP1423-2 | |
| Alcohol swab | BD | Used for blood draw | |
| Band-aids | Used for blood draw | ||
| BD Bacto Tryptic Soy Broth | BD | DF0370-07-5 | Combine with 1.5% agar to make Tryptic Soy Agar |
| Cell counter | Bal Saupply | 202C | |
| CellTracker blue CMCH | Invitrogen | C2111 | Blue CMAC Dye (BCD) |
| Clear bottom 96-well flat bottom polystyrene plates | Costar | 3370 | |
| Cotton gauze | Fisherbrand | 13-761-52 | Used for blood draw |
| Crystal violet | Acros Organic | 40583-0250 | |
| Culture tubes | Fisherbrand | 14-961-27 | Borosilicate Glass 13 x 100 mm |
| D-(+)-glucose | Sigma | G-8270 | |
| Dextran from Leuconostoc spp. | Sigma | 31392-250G | Used for isolation of neutrophils |
| Dulbecco's phosphate buffered saline (DPBS) 1x | Gibco | 14190-144 | |
| Ethidium homodimer-1 | Invitrogen | L3224 B | |
| Ficoll-Paque plus | Cytiva | 17144003 | Used for isolation of neutrophils (density gradient medium) |
| Hanks' balanced salt solution (HBSS) 1x | Corning cellgro | 21-022-CV | without calcium, magnesium, and phenol red |
| Hemacytometer | Bright Line | ||
| Heparin | Novaplus | NDC 63323-540-57 | 1000 USP units/mL, Used for blood draw |
| IMARIS 9.8 | Oxford Instruments | Microscopy image analysis software | |
| Luminol | Sigma | A8511-5G | |
| Minimal essential media (MEM) Alpha 1x | Gibco | 41061-029 | |
| Needle (23 G1) | BD | 305145 | Used for blood draw |
| Nikon Eclipse Ti2 | Nikon | ||
| NIS-Elements | Nikon | Quantification of dead neutrophils | |
| Normal human serum | Complement Technology | NHS | |
| Petri Dish (100 x 15 mm) | VWR | 25384-342 | |
| Phorbol 12-myristate 13-acetate | |||
| Poly-L-lysine solution | Sigma | P4707-50ML | |
| Sodium chloride | Fisher Bioreagents | BP358-10 | Used for neutrophil isolation |
| SoftMax Pro Software | Molecular Devices | Microplate reader software used for data acquisition | |
| SpectraMax i3x | Molecular Devices | Microplate reader | |
| Sterile water for irrigation, USP | Baxter | 2F7114 | Endotoxin-free; Used for neutrophil isolation |
| Surflo winged infusion set | Terumo | SC*19BLK | 19 G x 3/4", used for blood draw |
| Trypan blue stain (0.4%) | Gibco | 15250-061 | |
| Turnicate | Used for blood draw | ||
| UltraPure distilled water | Invitrogen | 10977015 | |
| White opaque 96-well plates | Falcon | 353296 | Tissue culture treated and flat bottom plate |
| μ-Slide VI 0.4 | Ibidi | 80601 | μ-channel slide |
References
- Donlan, R. M. Biofilms: microbial life on surfaces. Emerging Infectious Diseases. 8 (9), 881-890 (2002).
- Alhede, M., et al. Phenotypes of non-attached Pseudomonas aeruginosa aggregates resemble surface attached biofilm. PLoS One. 6 (11), 27943 (2011).
- Hall-Stoodley, L., Costerton, J. W., Stoodley, P. Bacterial biofilms: from the natural environment to infectious diseases. Nature Reviews: Microbiology. 2 (2), 95-108 (2004).
- Donlan, R. M., Costerton, J. W. Biofilms: survival mechanisms of clinically relevant microorganisms. Clinical Microbiology Reviews. 15 (2), 167-193 (2002).
- Schilcher, K., Horswill, A. R. Staphylococcal biofilm development: structure, regulation, and treatment strategies. Microbiology and Molecular Biology Reviews. 84 (3), 0002 (2020).
- Kaplan, J. B. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. Journal of Dental Research. 89 (3), 205-218 (2010).
- Otto, M. Staphylococcal Biofilms. Microbiology Spectrum. 6 (4), 10 (2018).
- Moormeier, D. E., Bayles, K. W. Staphylococcus aureus biofilm: a complex developmental organism. Molecular Microbiology. 104 (3), 365-376 (2017).
- Gross, M., Cramton, S. E., Gotz, F., Peschel, A. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infection and Immunity. 69 (5), 3423-3426 (2001).
- Zheng, Y., He, L., Asiamah, T. K., Otto, M. Colonization of medical devices by staphylococci. Environmental Microbiology. 20 (9), 3141-3153 (2018).
- Donlan, R. M. Biofilms and device-associated infections. Emerging Infectious Diseases. 7 (2), 277-281 (2001).
- Kolaczkowska, E., Kubes, P. Neutrophil recruitment and function in health and inflammation. Nature Reviews Immunology. 13 (3), 159-175 (2013).
- Amulic, B., Cazalet, C., Hayes, G. L., Metzler, K. D., Zychlinsky, A. Neutrophil function: from mechanisms to disease. Annual Review of Immunology. 30, 459-489 (2012).
- Chen, Y., Junger, W. G. Measurement of oxidative burst in neutrophils. Methods in Molecular Biology. 844, 115-124 (2012).
- van Kessel, K. P., Bestebroer, J., van Strijp, J. A. Neutrophil-mediated phagocytosis of Staphylococcus aureus. Frontiers in Immunology. 5, 467 (2014).
- Nguyen, G. T., Green, E. R., Mecsas, J. Neutrophils to the ROScue: Mechanisms of NADPH oxidase activation and bacterial resistance. Frontiers in Cellular and Infection Microbiology. 7, 373 (2017).
- Saini, R., Singh, S. Inducible nitric oxide synthase: An asset to neutrophils. Journal of Leukocyte Biology. 105 (1), 49-61 (2019).
- Imlay, J. A. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nature Reviews Microbiology. 11 (7), 443-454 (2013).
- Segal, A. W. The function of the NADPH oxidase of phagocytes and its relationship to other NOXs in plants, invertebrates, and mammals. The International Journal of Biochemistry and Cell Biology. 40 (4), 604-618 (2008).
- Fang, F. C. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nature Reviews Microbiology. 2 (10), 820-832 (2004).
- Bogdan, C. Nitric oxide and the immune response. Nature Immunology. 2 (10), 907-916 (2001).
- Chua, S. L., et al. Reactive oxygen species drive evolution of pro-biofilm variants in pathogens by modulating cyclic-di-GMP levels. Open Biology. 6 (11), 160162 (2016).
- El Haj, C., Lichtenberg, M., Nielsen, K. L., Bjarnsholt, T., Jensen, P. O. Catalase protects biofilm of Staphylococcus aureus against daptomycin activity. Antibiotics. 10 (5), 511 (2021).
- Ghimire, N., et al. Direct microscopic observation of human neutrophil-Staphylococcus aureus interaction in vitro suggests a potential mechanism for initiation of biofilm infection on an implanted medical device. Infection and Immunity. 87 (12), 00745 (2019).
- Bhattacharya, M., et al. Leukocidins and the nuclease nuc prevent neutrophil-mediated killing of Staphylococcus aureus biofilms. Infection and Immunity. 88 (10), 00372 (2020).
- Bogachev, M. I., et al. Fast and simple tool for the quantification of biofilm-embedded cells sub-populations from fluorescent microscopic images. PLoS One. 13 (5), 0193267 (2018).
- Kerstens, M., et al. A flow cytometric approach to quantify biofilms. Folia Microbiologica. 60 (4), 335-342 (2015).
- Meyle, E., et al. Destruction of bacterial biofilms by polymorphonuclear neutrophils: relative contribution of phagocytosis, DNA release, and degranulation. The International Journal of Artificial Organs. 33 (9), 608-620 (2010).
- Oveisi, M., et al. Novel assay to characterize neutrophil responses to oral biofilms. Infection and Immunity. 87 (2), 00790 (2019).
- Leid, J. G., Shirtliff, M. E., Costerton, J. W., Stoodley, P. Human leukocytes adhere to, penetrate, and respond to Staphylococcus aureus biofilms. Infection and Immunity. 70 (11), 6339-6345 (2002).
- Fey, P. D., et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio. 4 (1), 00537 (2013).
- Cody, W. L., et al. Skim milk enhances the preservation of thawed -80 degrees C bacterial stocks. Journal of Microbiological Methods. 75 (1), 135-138 (2008).
- Sanders, E. R. Aseptic laboratory techniques: plating methods. Journal of Visualized Experiments. 63, 3064 (2012).
- Freitas, M., Porto, G., Lima, J. L., Fernandes, E. Optimization of experimental settings for the analysis of human neutrophils oxidative burst in vitro. Talanta. 78 (4-5), 1476-1483 (2009).
- Merritt, J. H., Kadouri, D. E., O’Toole, G. A. Growing and analyzing static biofilms. Current Protocols in Microbiology. , (2005).
- Nauseef, W. M. Isolation of human neutrophils from venous blood. Methods in Molecular Biology. 412, 15-20 (2007).
- Zhou, L., et al. Impact of human granulocyte and monocyte isolation procedures on functional studies. Clinical and Vaccine Immunology. 19 (7), 1065-1074 (2012).
- Quach, A., Ferrante, A. The application of dextran sedimentation as an initial step in neutrophil purification promotes their stimulation, due to the presence of monocytes. Journal of Immunology Research. 2017, 1254792 (2017).
- Karlsson, A., Nixon, J. B., McPhail, L. C. Phorbol myristate acetate induces neutrophil NADPH-oxidase activity by two separate signal transduction pathways: dependent or independent of phosphatidylinositol 3-kinase. Journal of Leukocyte Biology. 67 (3), 396-404 (2000).
- Staats, A., et al. Rapid aggregation of Staphylococcus aureus in synovial fluid is influenced by synovial fluid concentration, viscosity, and fluid dynamics, with evidence of polymer bridging. mBio. , 0023622 (2022).
- Chiu, I. M., et al. Bacteria activate sensory neurons that modulate pain and inflammation. Nature. 501 (7465), 52-57 (2013).
- Hartig, S. M. Basic image analysis and manipulation in ImageJ. Current Protocols in Molecular Biology. 102 (1), 14-15 (2013).
- Heydorn, A., et al. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 146 (10), 2395-2407 (2000).
- Hartmann, R., et al. Quantitative image analysis of microbial communities with BiofilmQ. Nature Microbiology. 6 (2), 151-156 (2021).
- Luo, T. L., et al. Introducing BAIT (Biofilm Architecture Inference Tool): a software program to evaluate the architecture of oral multi-species biofilms. Microbiology. 165 (5), 527-537 (2019).
- Naves, P., et al. Measurement of biofilm formation by clinical isolates of Escherichia coli is method-dependent. Journal of Applied Microbiology. 105 (2), 585-590 (2008).
- Eze, E. C., El Zowalaty, M. E. Combined effects of low incubation temperature, minimal growth medium, and low hydrodynamics optimize Acinetobacter baumannii biofilm formation. Infection and Drug Resistance. 12, 3523-3536 (2019).
- Harris, L. G., Tosatti, S., Wieland, M., Textor, M., Richards, R. G. Staphylococcus aureus adhesion to titanium oxide surfaces coated with non-functionalized and peptide-functionalized poly(L-lysine)-grafted-poly(ethylene glycol) copolymers. Biomaterials. 25 (18), 4135-4148 (2004).
- Miao, J., et al. Biofilm formation of Staphylococcus aureus under food heat processing conditions: first report on cml production within biofilm. Scientific Reports. 9 (1), 1312 (2019).
- Lade, H., et al. Biofilm formation by Staphylococcus aureus clinical isolates is differentially affected by glucose and sodium chloride supplemented culture media. Journal of Clinical Medicine. 8 (11), 1853 (2019).
- Guzel Kaya, G., et al. Antibacterial activity of linezolid against gram-negative bacteria: utilization of epsilon-Poly-l-Lysine capped silica xerogel as an activating carrier. Pharmaceutics. 12 (11), 1126 (2020).
- Clinton, A., Carter, T. Chronic wound biofilms: pathogenesis and potential therapies. Laboratory Medicine. 46 (4), 277-284 (2015).
- Scheel-Toellner, D., et al. Reactive oxygen species limit neutrophil life span by activating death receptor signaling. Blood. 104 (8), 2557-2564 (2004).
- Pestrak, M. J., et al. Pseudomonas aeruginosa rugose small-colony variants evade host clearance, are hyper-inflammatory, and persist in multiple host environments. PLoS Pathogens. 14 (2), 1006842 (2018).
- Guerra, F. E., et al. Staphylococcus aureus SaeR/S-regulated factors reduce human neutrophil reactive oxygen species production. Journal of Leukocyte Biology. 100 (5), 1005-1010 (2016).
- Suo, Y., Huang, Y., Liu, Y., Shi, C., Shi, X. The expression of superoxide dismutase (SOD) and a putative ABC transporter permease is inversely correlated during biofilm formation in Listeria monocytogenes 4b G. PLoS One. 7 (10), 48467 (2012).
- Olwal, C. O., Ang’ienda, P. O., Ochiel, D. O. Alternative sigma factor B (sigma(B)) and catalase enzyme contribute to Staphylococcus epidermidis biofilm’s tolerance against physico-chemical disinfection. Scientific Reports. 9 (1), 5355 (2019).
- Bhattacharya, M., et al. Staphylococcus aureus biofilms release leukocidins to elicit extracellular trap formation and evade neutrophil-mediated killing. Proceedings of the National Academy of Sciences of the United States of America. 115 (28), 7416-7421 (2018).
- Masuda, S., et al. NETosis markers: Quest for specific, objective, and quantitative markers. Clinica Chimica Acta. 459, 89-93 (2016).
- Dworsky, E. M., et al. Novel in vivo mouse model of implant related spine infection. Journal of Orthopaedic Research. 35 (1), 193-199 (2017).
- Pletzer, D., Mansour, S. C., Wuerth, K., Rahanjam, N., Hancock, R. E. New mouse model for chronic infections by gram-negative bacteria enabling the study of anti-infective efficacy and host-microbe interactions. mBio. 8 (1), 00140 (2017).
- Davis, M. M. A prescription for human immunology. Immunity. 29 (6), 835-838 (2008).
- Mestas, J., Hughes, C. C. Of mice and not men: differences between mouse and human immunology. The Journal of Immunology. 172 (5), 2731-2738 (2004).

