An R-Based Landscape Validation of a Competing Risk Model
Summary
The present protocol describes codes in R for evaluating the discrimination and calibration abilities of a competing risk model, as well as codes for the internal and external validation of it.
Abstract
The Cox proportional hazard model is widely applied for survival analyses in clinical settings, but it is not able to cope with multiple survival outcomes. Different from the traditional Cox proportional hazard model, competing risk models consider the presence of competing events and their combination with a nomogram, a graphical calculating device, which is a useful tool for clinicians to conduct a precise prognostic prediction. In this study, we report a method for establishing the competing risk nomogram, that is, the evaluation of its discrimination (i.e., concordance index and area under the curve) and calibration (i.e., calibration curves) abilities, as well as the net benefit (i.e., decision curve analysis). In addition, internal validation using bootstrap resamples of the original dataset and external validation using an external dataset of the established competing risk nomogram were also performed to demonstrate its extrapolation ability. The competing risk nomogram should serve as a useful tool for clinicians to predict prognosis with the consideration of competing risks.
Introduction
In recent years, emerging prognostic factors have been identified with the development of precision medicine, and prognostic models combining molecular and clinicopathological factors are drawing increasing attention in clinical settings. However, non-graphical models, such as the Cox proportional hazard model, with results of coefficient values, are difficult for clinicians to understand1. In comparison, a nomogram is a visualization tool of regression models (including the Cox regression model, competing risk model, etc.), a two-dimensional diagram designed for the approximate graphical computation of a mathematical function2. It enables the valuation of different levels of variables in a clinical model and the calculation of risk scores (RS) to predict prognosis.
Model evaluation is essential in model construction, and two characteristics are generally accepted for evaluation: discrimination and calibration. In clinical models, discrimination refers to the ability of a model to separate individuals who develop events from those who do not, such as patients who die versus those who remain alive, and the concordance index (C-index) or the area under the receiver operating characteristic curve (AUC) are typically used to characterize it3,4. Calibration is a process of comparing the predicted probabilities of a model with the actual probabilities, and calibration curves have been widely used to represent it. In addition, model validation (internal and external validation) is an important step in model construction, and only validated models can be further extrapolated5.
The Cox proportional hazard model is a regression model used in medical research for investigating the associations between prognostic factors and survival status. However, the Cox proportional hazard model only considers two statuses of outcome [Y (0, 1)], while study subjects often face more than two statuses, and competing risks arise [Y (0, 1, 2)]1. Overall survival (OS), which is defined as the time from the date of origin (e.g., treatment) to the date of death due to any cause, is the most important endpoint in survival analysis. However, the OS fails to differentiate cancer-specific death from non-cancer-specific death (e.g., cardiovascular events and other unrelated causes), thus ignoring competing risks6. In these situations, the competing risk model is preferred for the prediction of survival status with the consideration of competing risks7. The methodology of constructing and validating Cox proportional hazard models is well-established, while there have been few reports regarding the validation of competing risk models.
In our previous study, a specific competing risk nomogram, a combination of a nomogram and competing risk model, and a risk score estimation based on a competing risk model were established8. This study aims to present different methods of evaluation and validation of the established competing risk nomogram, which should serve as a useful tool for clinicians to predict prognosis with the consideration of competing risks.
Protocol
The Surveillance, Epidemiology, and End Results (SEER) database is an open-access cancer database that only contains deidentified patient data (SEER ID: 12296-Nov2018). Therefore, this study was exempted from the approval of the review board of the Affiliated Jinhua Hospital, Zhejiang University School of Medicine.
1. Data preparation and R packages preparation
- Prepare and import the data.
> Dataset <- read.csv("…/Breast cancer Data.xlsx") #Import data.
NOTE: The data are uploaded in Supplementary File 1. - Install and load the R packages.
> packages <- c("rms","cmprsk","mstate","survival","riskRegression","
prodlim")
> req.pcg <- function(pcg){
new <- pcg[!(pcg %in% installed.packages()[, "Package"])]
if (length(new)) install.packages(new, dependencies = T)
sapply(pcg, require, ch = T)
}
> req.pcg(packages)
NOTE: Perform the following procedures based on R software (version 3.6.2) using the packages rms, cmprsk, mstate, survival, riskRegression, and prodlim (http://www.r-projectrg/).
2. Establish competing risk nomograms in two distinct methods
- Establish the competing risk nomogram in a direct method.
> mod_cph <- cph(Surv(Survivalmonths, status) ~ factor1+ factor2+…,
x=T, y=T, surv=T, data=Dataset)
> nom <- nomogram(mod_cph, fun=list(function(x) 1-surv_cph(36, x)…),
funlabel=c("3-year event1 Prob."…), lp=F) #Take the 36th month as an example.
> mod_crr <- crr(Survivalmonths, fstatus, failcode=1, cov1=cov)
> score <- log(log((1-real.3y),(1-cif.min36)))/(maxbeta/100)
> plot(nom) - Establish the competing risk nomogram in a weighted method.
> df.w <- crprep("Survivalmonths"," fstatus",
data=Dataset, trans=c(1,2), cens=0,
keep=c("factor1"," factor2"…))
> mod.w <- cph(Surv(Tstart, Tstop, status==1) ~ factor1+factor2+…,
data=df.w, weight=weight.cens, subset=failcode==1, surv=T)
> nom.w <- nomogram(mod.w…)
3. Discrimination ability of the competing risk nomogram
- C-index for discrimination
- Fit the matrix cov into the competing risk model mod_crr. and get a predicted matrix suv.
> suv <- predict.crr(mod_crr, cov) - Get the cumulative incidences in a certain month from suv and calculate the C-index with the function rcorr.cens.
> cif36 <- suv[which(suv[,1]==36),][-1]
> rcorr <- rcorr.cens(1-cif36,Surv(Dataset$Survivalmonths,Dataset$tumordeath))
> cindex <- rcorr[1]
- Fit the matrix cov into the competing risk model mod_crr. and get a predicted matrix suv.
- AUC for discrimination
- Score the predictive performance of the competing risk model using the function Score (riskRegression package).
> fgr.w <- FGR(Hist(Survivalmonths, fstatus) ~ factor1+ factor2+…, data=Dataset, cause=1)
> score <- Score(list("Fine-Gray" = fgr.w), - Extract the AUC from the "score".
> score$AUC
- Score the predictive performance of the competing risk model using the function Score (riskRegression package).
4. Calibration ability of competing risk models
- Calibration curves with a 95% confidence interval of the competing risk model
- Get a data frame with the cumulative incidences of each individual in a certain failure time.
> cif36 <- data.frame(cif36) #Take the 36th month as an example.
> colnames(cif36.36_o)<-c("36m") - Divide the cohort according to the estimated cumulative incidences into five subgroups and calculate the average predicted cumulative incidences of each subgroup.
> group36 <- cut(cif36$`36m`,
quantile(cif36$`36m`, seq(0, 1, 0.2)),
include.lowest = TRUE, labels = 1:5)
> mean36 <- as.vector(by(cif36 $`36m`, group36, mean)) - Calculate the observed cumulative incidences, that is, the actual cumulative incidences, using the function cuminc, and then get the observed cumulative incidences with a 95% confidence interval in a certain failure time.
> cum36 <- cuminc(Dataset$Survivalmonths,Dataset$fstatus,group36)
> obs36 <- timepoints(cum36,Dataset$Survivalmonths)$est[c(1:5),36]
> obs36var <- timepoints(cum36,Dataset$Survivalmonths)$var[c(1:5),36]
> df <- data.frame(mean36, obs36, obs36var) - Plot the calibration curve with the predicted cumulative incidences as the x-axis and the observed cumulative incidences as the y-axis using the function ggplot.
> ggplot(df)+ geom_point(aes(x=mean36,y=obs36),col="red")+
geom_point(aes(x=mean36,y=obs36),col="red",pch=4)+
geom_line(col="red",aes(x=mean36,y=obs36))+
geom_errorbar(col="red",aes(x=mean36,y=obs36+1.96
*sqrt(obs36var)),
ymin =obs36-1.96*sqrt(obs36var), ymax = obs36+1.96
*sqrt(obs36var))
geom_abline(lty=3,lwd=2,col=c(rgb(0,118,192,
maxColorValue=255)))
- Get a data frame with the cumulative incidences of each individual in a certain failure time.
- Calibration curve with risk scores of the competing risk model
- Valuate each level of all the variables and obtain the total RS.
> Dataset$factor1[Dataset$factor1==1] <- factor1.scale["Factor1_level1"]
> … #For example, Dataset$histology[Dataset$histology==1]<-histology.scale["Histology1"]
> Dataset$rs <- Dataset$factor1+Dataset$factor2+Dataset$factor3+…
NOTE: Obtain the total RS for each patient by summing the points of each variable. - Count the frequencies and calculate the observed cumulative incidences of the different total risk scores.
> rs.freq <- as.data.frame(table(Dataset$rs))
> obs.36 <- vector(mode="numeric", length=nrow(rs.freq))
> for (i in 1: nrow(rs.freq)) {
dataset <- subset(Dataset,Dataset$rs== rs.freq [i,1])
cif.dataset <- cuminc(dataset$Survivalmonths,dataset$death3)
cif36.dataset <- timepoints(cif.dataset,36)
obs.36[i] <- cif36.dataset$est[1]} - Set the range of the x-axis and calculate the predicted cumulative incidences of the total risk scores.
> RS <- range(nom$total.points)
> x.36 <- seq(min(RS),max(RS),0.01)
> pre.36 <- 1-(1-cif.min36)^exp(x.36*maxbeta/100) - Plot the calibration curve with risk scores.
> plot(x.36, pre.36, type='l'…)
> par(new=TRUE)
> plot(as.vector(rs.freq[,1]), obs.36… )
- Valuate each level of all the variables and obtain the total RS.
5. Decision curve analysis of competing risk models
- Source the stdca function to perform the decision curve analysis.
> source("stdca.R") - Extract the polynomial equations from the nomogram to calculate the survival probability.
> nomogramEx(nomo = nom)
> Dataset$predictors <- A * (Dataset$rs ^3) + B * (Dataset$rs ^2) + C * Dataset$rs + D #predictors are predicted probabilities of cancer-specific death calculated by the established nomogram - Perform the decision curve analysis.
> stdca(data = Dataset, outcome = "status", ttoutcome = "Survivalmonths", timepoint = 36,
predictors = "predictors", cmprsk = TRUE, smooth = FALSE, probability = FALSE)
NOTE: For evaluating an outcome in the presence of a competing risk, TRUE should be chosen for cmprsk.
6. Internal validation using the bootstrap method
- Get the average predicted cumulative incidences using the bootstrap method.
- Resample the original dataset (Dataset) with replace to generate the bootstrap dataset (Dataset_in). Establish a competing risk model (mod.in_crr) with the bootstrap dataset. Use the function predict.crr to predict mod.in_crr and loop b times to generate suvall.in.
B=b
suvall.in <- list()
for(j in 1:B){
Dataset_in <- Dataset[sample(c(1:nrow(Dataset)),nrow(Dataset),
replace = TRUE),]
attach(Dataset_ in)
cov. in <- model.matrix(~factor1+ factor2+…)[,-1]
mod. in _crr <- crr(Survivalmonths, fstatus, failcode=1, cov1=cov.in)
detach(Dataset. inner)
suv. in <- predict.crr(mod. in _crr, cov)
suvall.in[[j]] <- suv.in} - Get the average predicted cumulative incidences in a certain month.
cif36all. inner <- vector(mode="numeric", length=nrow(Dataset))
for (k in 1:B) {
cif36all. inner<- cif36all. inner+ suvall. inner[[k]][which(suvall. inner[[k]][,1]==36),][-1]
}
cif36.in <- cif36all.in/B
- Resample the original dataset (Dataset) with replace to generate the bootstrap dataset (Dataset_in). Establish a competing risk model (mod.in_crr) with the bootstrap dataset. Use the function predict.crr to predict mod.in_crr and loop b times to generate suvall.in.
- Calculate the C-index using internal cross-validation with the function rcorr.cens.
rcorr. inner <- rcorr.cens(1-cif36.in,Surv(Dataset$Survivalmonths,Dataset$tumordeath))
cindex. inner <- rcorr. inner[1] - Calibrate using the cross internal validation.
NOTE: The codes of the calibration curve of the competing risk model with internal validation are similar to the codes in section 4, while suv was replaced by suv.in.
7. External validation of the competing risk model
- Get the predicted cumulative incidences using external data. Get the predicted cumulative incidences with the matrix of external data variables (cov.ex).
suv.ex <- predict.crr(mod_crr,cov.ex)
cif36.ex <- suv.ex [which(suv.ex $time=="36"),][-1] - Calculate the C-index using external validation.
rcorr.ex <- rcorr.cens(1-cif36.ex,Surv(Dataset.ex$Survivalmonths,Dataset.ex$tumordeath))
cindex.ex <- rcorr.ex[1] - Calibrate using external validation.
NOTE: The codes of the calibration curve of the competing risk model with internal validation are similar to the codes in section 4, while suv is replaced by suv.ex.
Representative Results
In this study, data of patients with breast cancer were retrieved from the SEER database and served as example data. The SEER database provides data on cancer representing around 34.6% of the United States population, and permission to access the database was obtained (reference number 12296-Nov2018).
Two nomograms (Figure 1), both including histological type, differentiated grade, T stage, and N stage, were established using the direct method and the weighted method, respectively. The points of each level of variables and the probabilities corresponding to the total points were almost the same, while some slight differences were observed. Zhang et al. introduced a "weighted" approach to establish a competing risk nomogram, which first transformed the original data to weighted data (using function crprep), then constructed a Cox regression model with the weighted data (using function coxph), and finally established a competing risk nomogram with the Cox regression model (using function nomogram)9. In contrast, the "direct" approach in this study is totally different from the "weighted" one. In short, the parameters generated from the competing risk model (using function crr) replaced the parameters in the Cox regression model (using function coxph), which was finally used to establish the competing risk nomogram (using function nomogram). In the comparison of the nomograms established by the "weighted" method and the "direct" method, the two nomograms were similar in general, while some slight differences could be observed. The "direct" method in the study is more precise because it obtains the parameters for constructing the nomogram directly (with the formula "score=log(log((1-real.3y),(1-cif.min36)))/(maxbeta/100)" in section 2).
In rcorr.cens(X, Surv), X is a numeric predictor with cumulative incidences at any time point, and Surv is a survival object containing survival months and status. When Surv defines patients experiencing competing events as censored and then generates a life table, the evaluable ordered pair is identical. The C-index of the competing risk model without validation was 0.7978 (95% CI = 0.7650-0.8305), indicating this model had moderate discrimination ability. Bootstrap analysis was repeated 500 times, and the 500 results were then averaged to produce a single estimation for calculating the C-index. The C-index in the internal validation was 0.7978 (95% CI = 0.7651-0.8305), which was similar to the C-index in the original dataset. An external dataset was fitted into the competing risk model, and the C-index in the external validation was 0.5071 (95% CI = 0.4637-0.5505). The AUC of the competing risk model was calculated from the original dataset in the study. The 36th month AUC was 0.8461 (95% CI = 0.8232-0.8691), demonstrating the discrimination ability of the model.
As shown in Figure 2A, points on the calibration curve were close to the equivalence line, and the 95% CI of the observed frequency fell into the equivalence line in each group, indicating the accurate calibration ability of the model. Calibration curves using internal and external validation are shown in Figure 2B and Figure 2C, respectively, indicating that the constructed model had a good calibration ability in the internal validation but a poor one in the external validation.
As shown in Supplementary Figure 1, the points representing the observed cumulative incidences distributed around the line representing the predicted cumulative incidences, and no significant differences were observed between the observed and predicted incidences. The results of the decision curve analysis are shown in Figure 3, which presents the changes in net benefit with increasing threshold probability.
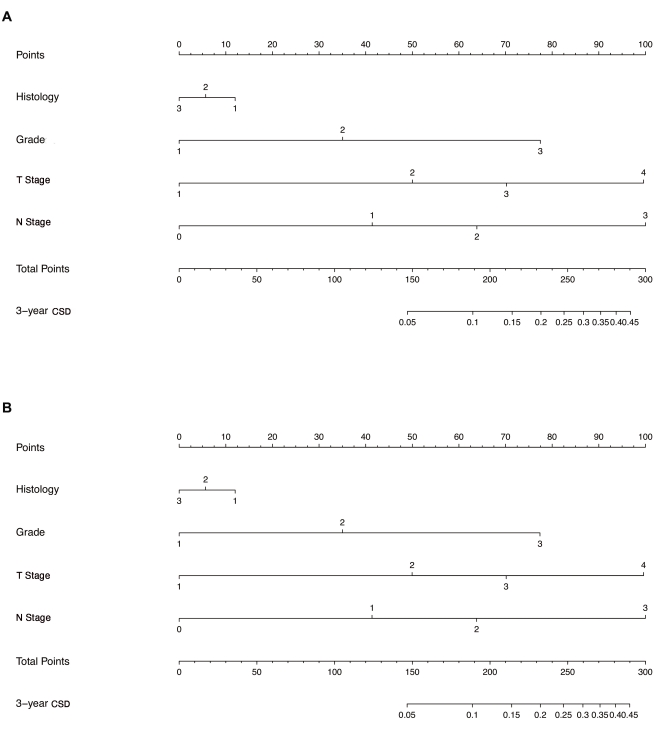
Figure 1: Establishment of the competing risk nomogram with two methods. (A) Nomogram established using the direct method. (B) Nomogram established using the weighted method. Histology: 1,invasive ductal carcinoma; 2, invasive lobular carcinoma; 3, invasive ductal carcinoma + invasive lobular carcinoma. Grade: 1, well-differentiated; 2, moderately differentiated; 3, poorly differentiated. T Stage: 1, T1 stage; 2, T2 stage; 3, T3 stage; 4, T4 stage. N Stage: 0, N0 stage; 1, N1 stage; 2, N2 stage; 3, N3 stage. Abbreviation: CSD = cancer-specific death. Please click here to view a larger version of this figure.
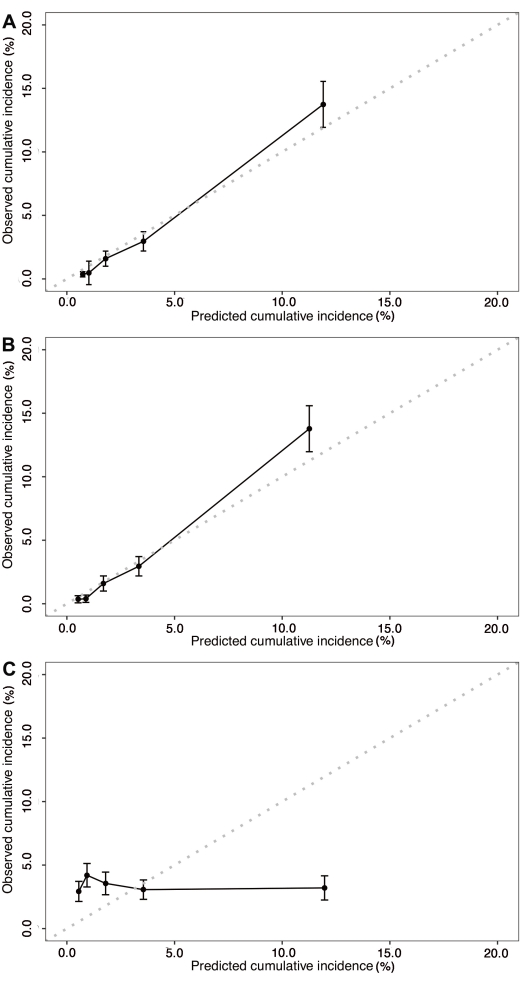
Figure 2: Calibration curves of the competing risk nomogram. (A) Calibration curve with a confidence interval of the established competing risk model. (B) Calibration curve of the competing risk model in the internal validation. (C) Calibration curve of the competing risk model in the external validation. Please click here to view a larger version of this figure.
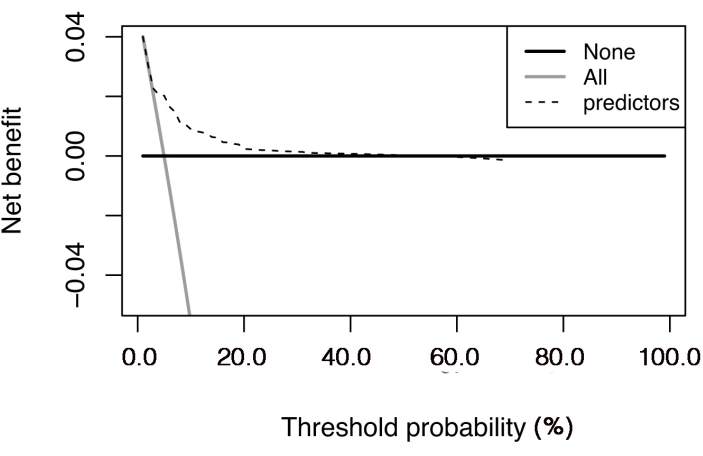
Figure 3: Decision curve analysis of the competing risk nomogram. The net benefit is plotted against the threshold probability. The "all" line shows the net benefit by considering all the patients that suffered cancer-specific death, and the "none" line is the net benefit by considering all the patients that did not suffer cancer-specific death. Please click here to view a larger version of this figure.
Supplementary File 1: Breast cancer data. Title definition: grademodify, differentiated grade; histology, histological type; stageT, tumor T stage; stageN, tumor N stage; Survivalmonths, the time from the date of treatment to the date of death due to any cause or censor; death, death (including cancer-specific death and non-cancer-specific death) or censor; death3, cancer-specific death, non-cancer-specific death, or censor. Please click here to download this File.
Supplementary Figure 1: Calibration curve with risk scores of the established competing risk model. The points representing the observed cumulative incidences were distributed around the line representing the predicted cumulative incidences. Please click here to download this File.
Discussion
This study compared competing risk nomograms established by two distinct methods and conducted evaluation and validation of the established nomograms. Specifically, this study provided a step-by-step tutorial for establishing the nomogram based on a direct method, as well as calculating the C-index and plotting the calibration curves.
The rms package in R software is widely used for the construction and evaluation of Cox proportional hazard models, but it is not applicable for competing risk models. For models with multiple outcomes, Zhang et al. reported a validation of competing risk models using the riskRegression package in R software, which calculates the AUC and Brier score to evaluate the discrimination ability and plots calibration curves to validate the calibration ability9,10.
However, the riskRegression package has some shortcomings, such as a failure to calculate the C-index. The C-index is the proportion of concordant evaluable ordered pairs in all the evaluable ordered pairs (C-Index = concordant evaluable ordered pairs/all evaluable ordered pairs)3,11. In traditional two-status outcome survival analysis, only pairs in which the two patients both suffer death (Di = 1, Dj = 1) or the patient who suffers death has shorter survival months than the patient who fails can become an evaluable ordered pair (Di = 1, Dj = 0, Ti < Tj) (i = patient i, j = patient j, T = time, D = status, 1 = death, 0 = failure). In the context of competing risks, patients who fail from the competing risk still remain in the risk set because it can only deduce that patients experiencing competing events have longer survival months than observed. Additionally, only pairs in which the two patients both suffer death (Di = 1, Dj = 1) or the patient who suffers death has shorter survival months than the patient who fails or experiences a competing event can become an evaluable ordered pair (Di = 1, Dj = 0|2, Ti < Tj) (i = patients i, j = patient j, T = time, D = status, 2 = competing event, 1 = death, 0 = failure). Therefore, this study used the function rcorr.cens to calculate the C-index.
In the internal validation of the competing risk model, this study applied the bootstrap method and demonstrated good performance of the constructed model12. A randomized split into training and test datasets may be problematic since the test dataset is likely to be particularly easy (or hard) to predict13. The K-fold method can also be used for cross-validation, while it has been applied less frequently in model validation5,14. This study also conducted an external validation of the competing risk model, but it did not perform well in the external context. This may attribute to the fact that our data used for the external validation were just a resample of the original data.
However, there are several limitations in this study. Firstly, our methodology is based on R software; therefore, the users need a certain programming knowledge, which may limit its target audience. In addition, there are hundreds of lines of code, and some codes need changes for different data; we hope to develop an "all-in-one" R package in future research that can be applied to all kinds of data. This study does not have other breast cancer data to conduct an external validation and had no choice but to resample from the original data, but the methods and the codes for external validation are the same. Most importantly, the linearity between covariates and outcome assumed in the study may not hold true in a real-world study, and interaction and non-linearity should be considered, for which ensemble modeling may be helpful15.
In conclusion, this study established the competing risk nomogram in a "direct" method and evaluated its discrimination and calibration abilities in original, internal, and external datasets. It is hoped that the competing risk nomogram will serve as a supplement to the riskRegression package in R and provide assistance in handling clinical competing risk events.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The study was supported by grants from the Medical Science & Technology Plan Project of Zhejiang Province (grant numbers 2013KYA212), the general program of Zhejiang Province Natural Science Foundation (grant number Y19H160126), and the key program of the Jinhua Municipal Science & Technology Bureau (grant number 2016-3-005, 2018-3-001d, and 2019-3-013).
Materials
| R software | None | Not Applicable | Version 3.6.2 or higher |
| Computer system | Microsoft | Windows 10 | Windows 10 or higher |
References
- Andersen, P. K., Gill, R. D. Cox’s regression model for counting processes: A large sample study. The Annals of Statistics. 10 (4), 1100-1120 (1982).
- Lubsen, J., Pool, J., vander Does, E. A practical device for the application of a diagnostic or prognostic function. Methods of Information in Medicine. 17 (2), 127-129 (1978).
- Harrell, F. E., Lee, K. L., Mark, D. B. Multivariable prognostic models: Issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Statistics In Medicine. 15 (4), 361-387 (1996).
- Hung, H., Chiang, C. -. T. Estimation methods for time-dependent AUC models with survival data. The Canadian Journal of Statistics / La Revue Canadienne de Statistique. 38 (1), 8-26 (2010).
- Moons, K. G. M., et al. Risk prediction models: I. Development, internal validation, and assessing the incremental value of a new (bio)marker. Heart. 98 (9), 683-690 (2012).
- Fu, J., et al. Real-world impact of non-breast cancer-specific death on overall survival in resectable breast cancer. Cancer. 123 (13), 2432-2443 (2017).
- Fine, J. P., Gray, R. J. A proportional hazards model for the subdistribution of a competing risk. Journal of the American Statistical Association. 94 (446), 496-509 (1999).
- Wu, L., et al. Establishing a competing risk regression nomogram model for survival data. Journal of Visualized Experiments. (164), e60684 (2020).
- Zhang, Z., Geskus, R. B., Kattan, M. W., Zhang, H., Liu, T. Nomogram for survival analysis in the presence of competing risks. Annals of Translational Medicine. 5 (20), 403 (2017).
- Zhang, Z. H., et al. Overview of model validation for survival regression model with competing risks using melanoma study data. Annals Of Translational Medicine. 6 (16), 325 (2018).
- Newson, R. Confidence intervals for rank statistics: Somers’ D and extensions. Stata Journal. 6 (3), 309-334 (2006).
- Davison, A. C., Hinkley, D. V., Schechtman, E. Efficient bootstrap simulation. Biometrika. 73 (3), 555-566 (1986).
- Roecker, E. B. Prediction error and its estimation for subset-selected models. Technometrics. 33 (4), 459-468 (1991).
- Steyerberg, E. W., Harrell, F. E. Prediction models need appropriate internal, internal-external, and external validation. Journal of Clinical Epidemiology. 69, 245-247 (2016).
- Zhang, Z., Chen, L., Xu, P., Hong, Y. Predictive analytics with ensemble modeling in laparoscopic surgery: A technical note. Laparoscopic, Endoscopic and Robotic Surgery. 5 (1), 25-34 (2022).

