Quantification of Global Histone Post Translational Modifications Using Intranuclear Flow Cytometry in Isolated Mouse Brain Microglia
Summary
This work describes a protocol for the quantification of global histone modifications using intranuclear flow cytometry in isolated brain microglia. The work also contains the microglia isolation protocol that was used for data collection.
Abstract
Gene expression control occurs partially by modifications in chromatin structure, including the addition and removal of posttranslational modifications to histone tails. Histone post-translational modifications (HPTMs) can either facilitate gene expression or repression. For example, acetylation of histone tail lysine residues neutralizes the positive charge and reduces interactions between the tail and negatively charged DNA. The decrease in histone tail-DNA interactions results in increased accessibility of the underlying DNA, allowing for increased transcription factor access. The acetylation mark also serves as a recognition site for bromodomain-containing transcriptional activators, together resulting in enhanced gene expression. Histone marks can be dynamically regulated during cell differentiation and in response to different cellular environments and stimuli. While next-generation sequencing approaches have begun to characterize genomic locations for individual histone modifications, only one modification can be examined concurrently. Given that there are hundreds of different HPTMs, we have developed a high throughput, quantitative measure of global HPTMs that can be used to screen histone modifications prior to conducting more extensive genome sequencing approaches. This protocol describes a flow cytometry-based method to detect global HPTMs and can be conducted using cells in culture or isolated cells from in vivo tissues. We present example data from isolated mouse brain microglia to demonstrate the sensitivity of the assay to detect global shifts in HPTMs in response to a bacteria-derived immune stimulus (lipopolysaccharide). This protocol allows for the rapid and quantitative assessment of HPTMs and can be applied to any transcriptional or epigenetic regulator that can be detected by an antibody.
Introduction
Epigenetics is the study of the mechanisms that regulate gene expression without altering the underlying DNA sequence. Epigenetic regulation of gene expression is dynamic within cells and can allow for rapid and coordinated responses to various environmental stimuli. The dynamic regulation occurs in part due to changes in the chromatin structure at the level of the nucleosome, which is comprised of histone proteins (H2A, H2B, H3, H4) assembled into an octamer core tightly wound by DNA1. The interactions between the histone proteins and the DNA can control the accessibility of DNA to transcription machinery, which can ultimately control gene expression and other aspects of chromatin biology2. Histone proteins have unstructured tails which feature positively charged residues that form electrostatic interactions with the negatively charged DNA backbone. These interactions result in tight packing of the DNA and reduced DNA accessibility. Covalent modifications to the histone tails, termed histone post-translational modifications (HPTMs), can regulate these interactions3,4. Some of the most well-characterized HPTMs include histone tail acetylation and methylation, which can change the affinity of electrostatic interactions between the histone tails and DNA, resulting in differential accessibility to the underlying DNA and recruitment of transcription factors which recognize these HPTMs at specific sites. HPTMs are regulated by three important classes of enzymes termed readers- which recognize, writers- which deposit, and erasers- which remove HPTMs. Thus, the recruitment or dissolution of reader, writer, or eraser enzymes can ultimately change the landscape of HPTMs and govern the structure and function of chromatin, making their regulation and readout essential for understanding cellular biology and function3,4.
The cells in the central nervous system (CNS) are epigenetically flexible as they change their transcriptome to adapt to environmental stimuli. Accumulating evidence suggests that changes in the epigenome, such as DNA methylation, non-coding RNAs, and HPTMs, play an essential role in memory formation and synaptic function5. Disrupting HPTM dynamics through manipulation of the relevant readers, writers, or erasers can block or enhance associative learning and long-term potentiation6,7,8. Microglia, the resident immune cell of the CNS, rapidly regulate their transcriptome in response to immune stimulation through dynamic changes in their epigenome9,10,11. This high level of adaptation to their local brain environment makes them difficult to examine in an isolated context as studies have shown that the epigenome and transcriptome of microglia becomes altered after only a few hours in culture media after removal from the brain environment11. In addition, as microglia only make up 10% of the brain's cells, measures examining changes at the whole tissue level lack sensitivity and specificity12,13. As a result, microglia need to be rapidly isolated to examine the epigenetic changes such as HPTM levels, ex vivo.
The methods commonly used to examine HPTMs include chromatin-immunoprecipitation sequencing (ChIP-seq) and cleavage under targets and tagmentation sequencing (CUT&Tag-seq)4. While these techniques are highly specific to an individual HPTM and can inform the presence of HPTMs within a specific genomic context, they can only examine one of the many possible HPTMs within a single experiment11,14 Therefore, before proceeding with such experiments, which require a significant time and money investment, it is highly valuable to narrow down the list of potentially interesting HPTMs for further investigation by first examining changes in global levels of HPTMs. The two main approaches for examining global HPTM levels are immunohistochemistry and western blot analysis, but both approaches are only semi-quantitative, low-throughput, and require large numbers of tissue sections or isolated cells15,16. Thus, we aimed to develop a highly sensitive, quantitative method that could be used to examine the global HPTM levels rapidly and at the single cell level.
The protocol presented enables detection of global HPTM levels rapidly using intranuclear flow cytometry. Previous studies in cancer cells have justified the importance of examining global levels from a clinical perspective17,18. It is also common for studies to use global levels as a screening method prior to assessing genomic location of specific HPTMs of interest19,20. For microglia, assessing global levels following isolation is challenging due to the low cell yield; Pan et al present global HPTM levels from isolated microglia, in which microglia from three animals was pooled to enable protein level detection by western blot19. Using our protocol, we are able to detect global changes with much lower cell inputs, enabling screening of multiple marks per animal and eliminating the need to pool samples.
Here, we describe a protocol to detect HPTM levels rapidly via quantitative intranuclear flow cytometry in isolated microglia. While we focus specifically on HPTM quantification for the sake of brevity, this protocol can be used in the same way to quantify global levels of reader, writer, and eraser enzymes. The protocol is delivered in two parts: first, the isolation method for microglia and, second, the flow cytometry-based method for determining HPTM levels. The isolation method yields cells that can be used for both RNA isolation and HPTM level assessment which allows for the evaluation of gene expression and HPTM levels from the same sample. In addition, the method for HPTM assessment can be used on other cell types as indicated in the protocol.
Protocol
All animal care protocols were approved by the University of British Columbia's Animal Care Committee in accordance with the Canadian Council on Animal Care guidelines.
1. Brain digestion for microglia isolation
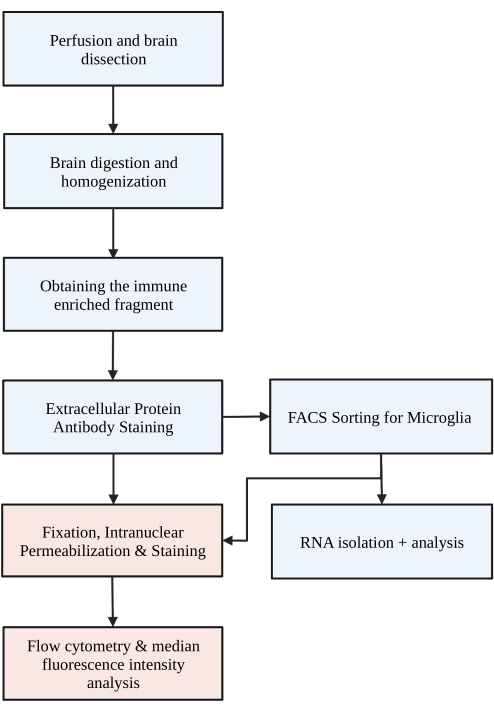
Figure 1: Simple flow chart of the protocol. The mice are first transcardially perfused with HBSS, and the brain is dissected. The brain is then dissociated through chemical digestion and mechanical disruption to result in a single cell homogenate. The immune enriched fraction is collected via discontinuous density gradient, after which the cells are stained for P2RY12. Stained cells are either 1) sorted via fluorescent activated cell sorting (FACS) to lead to RNA analysis or downstream protein analysis and/or 2) fixed, permeabilized, and stained for intranuclear proteins. The protein level is quantified by median fluorescence intensity in the channel of interest determined by flow cytometry. Boxes colored in blue are part of protocol step 1) Brain digestion for microglia isolation. Boxes colored in red are part of protocol step 2) Intranuclear flow staining for protein expression analysis. Created with BioRender.com. Please click here to view a larger version of this figure.
- Preparation of reagents
NOTE: If planning an extraction to collect both RNA and cells for HPTM analysis, refer to section 1.7.1 for modifications to include transcription and translation inhibitors. However, this is not required if just assessing protein signal as the cells are largely quiescent when kept on ice.- Fluorescence activated cell sorting (FACS) buffer (20 mL per sample): Dissolve bovine serum albumin (BSA) in 1x Hanks balanced salt solution (HBSS) to create a 2% BSA solution. Dissolve EDTA to a final concentration of 1 mM in the 2% BSA solution. Filter sterilize using a 0.2 µm filter and store at 4 °C for up to 1 week prior to use.
- Digestion buffer (1 mL per sample): Reconstitute a vial of papain in HBSS to a final concentration of 20 U/mL in 1 mM L-cysteine with 0.5 mM EDTA. Activate at 37 °C for a minimum of 10 min or until ready to digest tissue. Just prior to use, add DNase I to the activated papain solution to a final concentration of 200 U/mL. Prepare this on the day of experiment and do not store.
- Isotonic density gradient solution (5.5 mL per sample): Add 10x HBSS to cold density gradient medium to a final concentration of 1x HBSS, resulting in a final density of 1.117 g/mL. Vortex to mix for at least 30 s prior to use. Place on ice until use.
- 37% density gradient solution (4 mL per sample): Add isotonic density gradient to 1x HBSS to make a final concentration of 37% with a final density of 1.043 g/mL. Add 20 µL of phenol red for each mL of 37% density gradient to make a pink solution for visualization during layering. Vortex for at least 30 s prior to use. Place on ice until use.
- 70% density gradient solution (2 mL per sample): Add isotonic density gradient to 1x HBSS to make a final concentration of 70% with a final density of 1.082 g/mL. Add 5 µL of trypan blue for each mL of 70% density medium to make a blue solution for visualization during layering. Vortex for at least 30 s prior to use. Place on ice until use.
- Perfusion and brain dissection
NOTE: The perfusion protocol is similar to Posel et al. which features video depiction of mouse thoracotomy, transcardial perfusion, and brain removal21. Here, we use adult C57BL/6J male and female mice (10-15 weeks old, 20-30 g), but this protocol can be used to perform a thoracotomy for any mouse. All animal procedures must be approved by the institutional ethics committee prior to conducting experiments.- Mouse anesthesia: Anaesthetize mice with 4% Isoflurane in 100% oxygen until past the plane of surgical anesthesia, which can be confirmed with a toe pinch, or a lack of reflex upon firmly pinching the mouse's foot. Place the mouse on its back and firmly pin its four paws down into the surgical dissection board placed tilted in a plastic tray, ensuring the nose is secured into the isofluorane nose cone. After transfer, ensure the animal is still past the surgical plane of anesthesia before proceeding.
- Mouse thoracotamy: Grab and lift the abdominal skin using forceps and make a shallow incision through the skin and abdominal wall to expose the xyphoid without damaging the descending aorta or any underlying organs.
- Grip the xyphoid with forceps and make lateral incisions beneath the ribcage to expose the diaphragm and liver. Make careful shallow cuts through the diaphragm along the length of the rib cage using fine scissors and through the ribcage using tissue scissors and pin the sternum to the surgical station near the head of the mouse to expose the heart and lungs for transcardial perfusion.
- Transcardial perfusion: Prepare a peristaltic perfusion pump and attach a 26.5G needle onto one end of the tubing. Prime the tubing for the procedure by inserting one end of the tubing into a vial of cold 1x HBSS and switching on the pump to completely fill the tubing with 1x HBSS.
- While holding the heart with blunt forceps, insert the tip of a 26.5G needle with the attached perfusion tubing into the left ventricle of the heart and make a small incision in the right atrium. Turn on the perfusion pump to carefully perfuse the mouse at a rate of ~2-4 mL/min with at least 15-20 mL of cold 1x HBSS.
NOTE: A complete perfusion is often indicated when the liver begins to clear blood and becomes the same color as the heart. - Brain removal: Decapitate the mouse using tissue dissecting scissors and make a midline incision in the scalp from neck to nose. Peel the skin flaps to the sides to expose the skull and remove excess tissue and bones at the caudal end of the skull with dissecting scissors.
- Carefully slide one blade of the scissors under the skull into the foramen magnum with the sharp side facing the bone and carefully cut up the midline toward the nose. Make lateral cuts at both the base of the skull and near the nose using dissecting scissors. Using fine forceps, life the skull from the midline to the exterior to crack off the skull pieces and expose the brain. Gently lift the brain with a spatula and place on dissection blot paper.
- Brain dissection: Place the brain on a piece of dissection blot paper wetted with 1x HBSS on top of a closed Petri dish filled with ice. Remove the cerebellum and bisect the brain hemispheres using a clean razor blade.
- Remove the brainstem, striatum, and white matter from each hemisphere, while keeping the hippocampus and overlaying cortex intact. Transfer the hemispheres containing isolated cortex and hippocampal tissue into a 15 mL tube with 5 mL of cold 1x HBSS and keep on ice.
NOTE: It is important to perform the dissections as quickly as possible so that the tissue remains cold with no more than 2 min between decapitation and final placing of dissected tissue in 1x HBSS on ice. If isolating microglia from multiple animals, brains can be stored on ice in 1x HBSS for ~1 h before proceeding with processing the entire cohort of animals for digestion etc.
- Brain digestion and homogenization
- Mechanical and chemical dissociation: Place the brain tissue from each mouse and 1 mL of digestion buffer into individual Petri dishes on ice. Using a clean scalpel blade, thoroughly chop the brain into small pieces (<1 mm).
- Cut the tip off a plastic transfer pipette and carefully transfer each of the minced brains into separate wells within a 24-well plate on ice. Cover the plate with transparent flexible film and incubate on ice for 30 min.
NOTE: When chopped correctly, the brain tissue resembles well-minced garlic. - Dounce homogenization: Transfer digested brain solution from each well into individual 7 mL glass dounce homogenizers on ice each filled with 5 mL of cold FACS buffer. Dounce each brain gently with the loose pestle (A), approximately 30-40 times, until a single cell suspension is obtained. After douncing with the A pestle, gently dounce with the tight pestle (B) 3-4 times to ensure a single cell suspension.
NOTE: Do not push the pestle more than ¾ of the way down to avoid crushing the tissue at the bottom of the homogenizer. The final solution should be opaque and milky.
NOTE: If digesting multiple brains in a single experiment, time the transfer of the brain digest into FACS buffer so that each sample is only in the digest buffer for 30 min. Over-digestion can result in cleavage of surface proteins, reducing downstream antibody binding and signal.
- Obtaining immune enriched fragment
- Establishing density gradient: Transfer the homogenate from each brain into separate 15 mL polypropylene tubes and add 2.125 mL of isotonic density gradient and top to 8.5 mL with FACS buffer for each to obtain a final concentration of 25% density gradient. Gently invert the 15 ml tubes 20x to mix thoroughly.
- Using a narrow-graduated transfer pipette, gently underlay 4 mL of 37% density gradient to each tube, being very careful to establish clean layers. Switch transfer pipettes and gently underlay 2 mL of 70% density gradient (Figure 2A). Transfer to a centrifuge cooled to 4 °C and spin at 500 x g for 20 min with the braking ramp set to zero.
- Collecting immune enriched fragment: Using clean transfer pipettes, gently aspirate the myelin from the top of the volume in the 15 mL tube using a clean transfer pipette and discard. Carefully collect the top fragment of the density gradient into a clean 15 mL polypropylene tube using a transfer pipette.
- Carefully collect the immune enriched fragment (1.5 mL above and 1.5 mL below where the 70% and 37% density gradient layers meet) into a new 15 mL polypropylene tube (Figure 2B). Add 10 mL of FACS buffer to the immune-enriched sample to dilute out the density gradient medium and invert the tube gently 20x to mix thoroughly.
NOTE: As cells tend to stick to the sides of the tube, ensure to gather all cells in the sample during the collection steps by slowly circling the pipette along the sides of the tube while collecting the liquid. - Pellet the cells in the immune-enriched sample by centrifuging the 15 mL tubes in a 4 °C centrifuge at 500 x g for 10 min with the downhill ramp brake set to zero. Immediately upon the end of the spin, remove the supernatant carefully, leaving approximately 300 µL of liquid in the 15 mL tube, being careful not to disturb the pellet (which may not be visible).
- Collect the supernatant in another 15 mL tube to ensure that the cells were pelleted in the spin (discard this fraction once verification with the cell counts of the resuspended pellet is done). After resuspending the cell pellet in the 300 µL volume using a P1000 pipette, count cells with a hemacytometer to estimate the total cell yield.
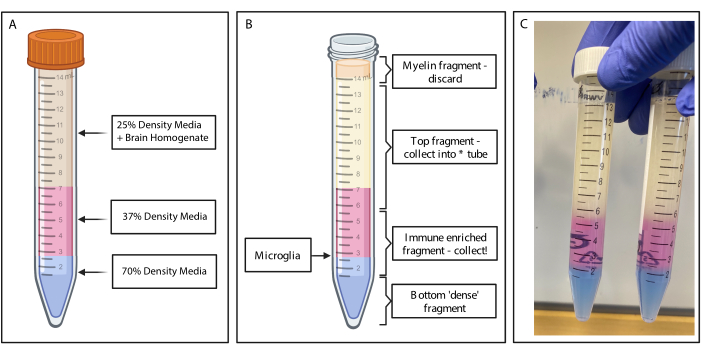
Figure 2: Obtaining the immune enriched fragment by discontinuous density gradient. (A) The brain homogenate is made to 25% density medium, underlay 4 mL of 37% density medium colored pink via phenol red and 2 mL of 70% density medium colored blue via trypan blue. (B) Following centrifugation, the fractions have separated. Microglia rests at the interface of 37% and 70% density media fragments. The myelin fragment is at the top of the 15 mL tube and will be discarded. The top fragment is collected as back up in case the spin fails, and no cells are recovered. If that occurs, the gradient can be repeated using this fraction. The immune enriched fraction is collected downstream. The bottom fraction containing any red blood cells remains in the tube and is discarded. (C) Example figure depicting complete layers. Created with BioRender.com. Please click here to view a larger version of this figure.
- Extracellular antibody staining
- Blocking: Transfer cells to a round bottom 96 well plate on ice and centrifuge at 500 x g with brake to pellet the cells. Quickly remove the supernatant in the sink by flicking the plate to dispose of the supernatant, leaving the cell pellet intact at the bottom of the well.
- Resuspend the cells in 50 µL of FACS buffer with anti-mouse CD16/32 FC-Receptor blocking reagent using a P200 pipette (final concentration 10 µg/mL, dilution factor 1:50) to prevent non-specific binding of antibodies to monocytes or other FcR bearing cells. Incubate for 10 min on ice.
- Antibody staining: Prepare the appropriate volume of a 2x master mix containing P2RY12- Allophycocyanin (APC; dilution factor 1:50, concentration 4 µg/mL for a final well concentration of 1:100, concentration 2 µg/mL) and violet 525 live dead stain (dilution factor 1:50 for a final well concentration of 1:100). Add 50 µL of the staining master mix to the cell suspension (obtained after blocking in section 1.5.1) and incubate the plate for 30 min in the dark on ice.
NOTE: For this protocol, we present staining the cells with P2RY12. Firstly, P2RY12 is a homeostatic marker for microglia that can be downregulated in certain disease contexts. For example, 5XFAD Alzheimer's model mice have downregulated P2RY12 levels which might make them difficult to identify22. Alternative stains that can be used for the isolation include Tmem119, Cd11b and CD4523. Secondly, the conjugate fluorochrome APC can be adjusted to suit the desired panel of antibodies. However, choosing a bright fluorochrome, such as APC or PE, will help to ensure the positive and negative populations are easily distinguishable24. - After staining, add 200 µL of FACS buffer directly to each well to wash the cells. Spin at 500 x g at 4 °C to remove supernatant by flicking. Resuspend cells in 200 µL of FACS buffer with a P200 pipette, spin at 500 x g at 4 °C, and flick plate to remove buffer from wells.
- Preparing flow controls: Prior to staining, separate necessary volumes of cells from each sample after blocking in step 1.5.1 for the required flow controls.
NOTE: Flow controls are required for each experiment to establish the gates. The flow controls can be taken from an additional animal or from a fraction of each of the experimental wells. When splitting cells ensure to assign enough cells per control as 10,000-30,000 cells per control is required to establish gates with high confidence.- There are three relevant flow controls: no stain, live dead, and P2RY12 isotype control. For the no stain control, do not add any antibody. In the P2RY12 isotype control, treat cells with viability dye (1:100) and an isotype control antibody conjugated to APC (1:100).
- To prepare the live dead control, aliquot cells into a separate well and move half of the cell volume into a 500 µL tube. Place the 500 µL tube into the -80 °C freezer for 5 min, followed placing in 37 °C incubator for 5 min to kill the cells. Return the aliquot of dead cells into the live dead control well and stain with an amine binding viability dye on violet 525 (dilution factor 1:100) to mark dead cells.
NOTE: The protocol is written for plate staining with a flick method for supernatant removal. However, this requires the supernatant to be removed immediately following completion of the spin and the flick needs to be done with enough force to quickly remove the supernatant without disturbing the pellet. Alternatively, 1.5 mL RNAse/DNase free tubes can be used for the staining, with the following modifications: Transfer cells into 1.5 mL microcentrifuge tube and pellet at 800 x g for 5 min at 4 °C. Aspirate supernatant with pipettes. Tip: For speed, a 5 mL transfer pipette with a P200 tip can quickly and accurately aspirate the supernatant. When aspirating, check for the pellet. If pellet is not visible, leave 50 µL of supernatant and adjust calculations accordingly. When washing out antibodies, add additional FACS to increase the dilution of antibodies (1000 µL instead of 200 µL) to account for incomplete removal of supernatant. Depending on the cytometer, use the 1.5 mL tubes for sorting, reducing the amount of supplies required.
- FACS sorting for microglia
- Preparation: Resuspend each well in 200 µL of FACS buffer with a P200 pipette and transfer into labelled flow sort tubes and add FACS buffer to a total of 500 µL for a concentration of approximately 5 x 105 events per mL. Store on ice in the dark until analysis. Prepare post sort tubes by adding 100 µL of FACS buffer as a cushion for cells in 1.5 mL RNAse free tubes.
- Cytometer settings: Sort cells on a flow cytometry cell sorter set up with the 100 µm nozzle. Sort cells using 18-20 psi.
- Gating: On the cytometer, gate for cell size using side-scatter (SSC) area versus forward scatter (FSC) height using the no stain control to help distinguish debris, put SSC-A on a log axis to visualize a cell population and gate closely to select for cells (gate S1; Figure 3). To remove any doublets, plot FSC-H vs FSC-W and gate closely around the cell population removing any debris and doublets (gate S2). Using the P2RY12 isotype control, examine the cells in the APC channel and set the gate for autofluorescence to determine P2RY12+ cells. Using the no stain and live dead controls, gate for the cells that are not fluorescent on violet 525 nm as live cells.
- Sorting: Plot violet 525 nm vs APC and determine the population that is P2RY12+ and live by FMOs (MG). Sort those cells into the labelled post sort tube (Figure 3). The final sort percentage is approximately 50% of the total events with the majority of event total loss being debris removed in gate S1 (~70% of events are cells; Table 1).
- RNA isolation and analysis
- Transcription and translation inhibitors: If planning RNA extraction, to eliminate the risk of isolation associated transcriptomic signatures, include inhibitors of translation and transcription in the buffer steps. Prepare the inhibitor cocktail as described by Marsh et al. including actinomycin D, anisomycin and triptolide25.
- Inhibitor preparation: Reconstitute inhibitor stocks and store as follows: Reconstitute actinomycin D in dimethylsulfoxide (DMSO) to 5 mg/mL and store at -20 °C. Reconstitute triptolide in DMSO to 10 mM and store at -20 °C, protected from light. Reconstitute Anisomycin in DMSO to 10 mg/mL and stored at 4 °C, protected from light. Store all inhibitor stocks for no longer than 1 month after being reconstituted.
- Buffer modifications: Add inhibitors in four different buffers in the protocol as follows: When performing the transcardial perfusion, prepare HBSS with actinomycin D (5 µg/mL, 1:1000 from stock) and triptolide (10 µM, 1:1000 from stock). Following perfusion, transport brains to the lab in HBSS containing actinomycin D (5 µg/mL, 1:1000 from stock), triptolide (10 µM, 1:1000 from stock) and anisomycin (27.1 µg/mL, 1:368.5 from stock). Prepare FACS buffer with actinomycin D (5 µg/mL, 1:1000 from stock), triptolide (10 µM, 1:1000 from stock) and anisomycin (27.1 µg/mL, 1:368.5 from stock). Prepare digestion buffer with actinomycin D (5 µg/mL, 1:1000 from stock), triptolide (10 µM, 1:1000 from stock) and anisomycin (27.1 µg/mL, 1:368.5 from stock).Prepare post sort wash buffer, with HBSS containing actinomycin D (5 µg/mL, 1:1000 from stock), triptolide (10 µM, 1:1000 from stock) and anisomycin (27.1 µg/mL, 1:368.5 from stock).
NOTE: When adding the inhibitors, ensure to add them immediately prior to use and protect any prepared buffers from light while in use. Avoid freeze-thaws of stock solutions.
- Post-sort washes: Because the cells have been sorted into 1.5 mL RNase free tubes in FACS buffer, which will interfere with RNA isolation, it is necessary to wash the cells. Spin the cells at 1000 x g at 4 °C for 5 min and remove supernatant, leaving approximately 50 µL of liquid.
- Add 200 µL of 1x HBSS containing actinomycin D (5 µg/mL, 1:1000 from stock), triptolide (10 µM, 1:1000 from stock) and anisomycin (27.1 µg/mL, 1:368.5 from stock) and mix thoroughly. Repeat the spin and remove supernatant leaving 50 µL of liquid (wash 1). Add 200 µL of post sort wash buffer, mix thoroughly and repeat the spin and remove supernatant leaving 25 µL of liquid (wash 2).
- RNA extraction: For RNA isolation from microglial cells, use a low-input RNA isolation kit for high and consistent RNA yields and RIN scores above 9 (see below and Table of Materials for product recommendations). To the cell pellet, add 350 µL of the lysis buffer from recommended kit + β-mercaptoethanol (1:100) and mix well.
NOTE: If necessary, the protocol can be suspended at this point. Samples can be stored in the lysis buffer in the -80 °C until RNA extraction. If extracting RNA after storage, thaw the lysate on ice and proceed with the kit-specific instructions for the isolation. - Transfer lysate into column-based cell shredder (see Table of Materials for product recommendations) and centrifuge at max speed at 4 °C for 2 min. Elute in a minimum of 14 µL of RNase-free water and determine the concentration as appropriate. RNA can be used for any downstream application after this point.
- Transcription and translation inhibitors: If planning RNA extraction, to eliminate the risk of isolation associated transcriptomic signatures, include inhibitors of translation and transcription in the buffer steps. Prepare the inhibitor cocktail as described by Marsh et al. including actinomycin D, anisomycin and triptolide25.
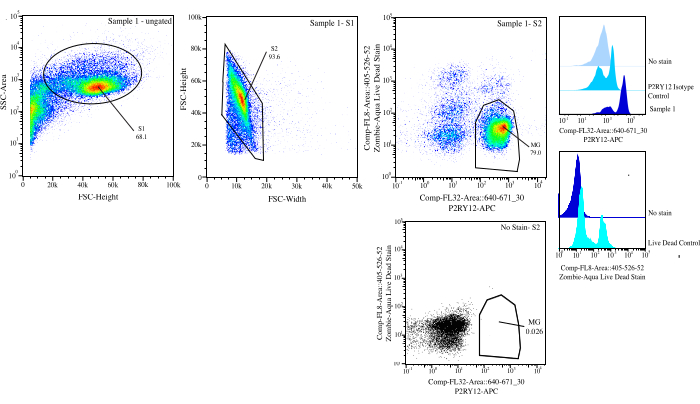
Figure 3: Gating strategy for flow sort. Events are gated for cell size on SSC-A vs FSC-H (S1). Then, cells are gated to be singlets on FSC-H vs FSC-W (S2). Singlet cells are sorted as live if negative on Comp-FL8-A::405-526-52 (violet 525 live dead stain) and as P2RY12+ if positive on Comp-FL32-A::640-671_30 (P2RY12-APC) using the P2RY12-isotype control. Cells are labelled as MG and sorted if both live and P2RY12+. Please click here to view a larger version of this figure.
| GATED POPULATION | Frequency of Parent | Frequency of Total | Count |
| S1 | 68.10% | 68.10% | 162186 |
| S2>S1 | 93.59% | 63.70% | 151707 |
| P2Ry12+ (670+) > S2 > S1 | 83.05% | 52.90% | 125986 |
| Live (525-) > S2 > S1 | 92.78% | 59.10% | 140752 |
| MG (P2RY12+ Live) >S2>S1 | 78.96% | 50.30% | 119794 |
Table 1: Example sample lineage table with gating percentages and expected event numbers.
2. Intranuclear flow staining for protein expression analysis
NOTE: Other cell types can be started at this point, this protocol is tested with cultured cells including HEK293 cells, BV2 microglia-like cells, and human IPSC-derived microglia.
- Fixation and staining of cells
NOTE: For the following protocol, use an intracellular staining kit that is optimized for nuclear staining. See Table of Materials for product recommendations.- Aliquot extracellularly stained cells from section 1.5.2 into 96 well plate (5 x 104– 1 x 106 cells). Spin cells for 5 min at 500 x g at 4 °C and flick to remove FACS buffer.
NOTE: To obtain data with high confidence of median levels, a minimum of 10,000 cells per well should be used. While there is no recommended maximum, it is best to keep the number of cells consistent throughout the experiment to ensure there is no significant effect of different coefficient of variations (CV). - Fixation and permeabilization: Add 200 µL of 1x fix concentrate and gently mix with P200 pipette to resuspend cells. Incubate in the dark for 45-60 min. Centrifuge plate for 5 min at 500 x g at room temperature (RT) and flick to discard supernatant.
NOTE: If necessary, the protocol can be suspended at this point. After discarding supernatant, re-suspend cells in long-term storage buffer for immune cells (see Table of Materials for product recommendations). Samples can be stored at 4 °C for 12-18 h, protected from light and covered in transparent film to protect buffer evaporation. - Add 200 µL of 1x permeabilization buffer to each well and pipette with a P200 to mix. Centrifuge plate for 5 min at 500 x g at RT and flick to discard supernatant. Repeat permeabilization buffer wash a total of 3x.
- Preparing flow controls: Split volume of cells from each sample for the required flow controls (10,000-30,000 cells per control well is sufficient).
- To prepare the no stain control, fix the no stain cells from the sort or aliquot unstained cells into a separate well that will not receive any antibody.
- To prepare the fluorescence minus one (FMO) control, aliquot cells for each of the antibodies on the panel except the one in that channel.
- For the relevant channels, include the isotype control antibody in the FMO for gating. For example, in a panel containing P2RY12-APC and H3K27Ac-AlexaFluor568 – there should be two FMOS: (1) the APC-FMO which contains only H3K27Ac-AlexaFluor568 and the P2RY12 isotype control antibody and (2) the 568-FMO which contains only P2RY12-APC and the isotype control primary and 568 secondary.
NOTE: This protocol is presented to test a single HPTM, however panels can be established that contain many HPTMs conjugated to different fluorophores.
- Primary antibody staining: Add 50 µL of 1x permeabilization buffer with the appropriate concentration of primary antibody to each well. Incubate for 30 min at RT in the dark. Wash 2x with 200 µL of 1x permeabilization buffer.
NOTE: The concentration of antibodies used for each HPTM is included in the Table of Materials. The concentration is determined by testing different concentrations of the antibodies on cultured cells treated with a stimulant that would cause a dramatic increase, e.g., an HDAC inhibitor for acetylation marks and ensuring both untreated and treated cells were well within the range of detection (above the isotype control and below the maximum detection range of the cytometer). The optimal antibody concentration for HPTMs should have an average median fluorescent intensity in the fluorophore channel between 5 x 104 and 1 x 105. - Secondary antibody staining: Block with 200 µL of 1x permeabilization buffer with 2% normal donkey serum (NDS) for 10 min at RT. Spin for 5 min at 500 x g at RT and flick to remove supernatant.
- Add 50 µL of 1x permeabilization buffer with 2% NDS and the appropriate concentration of secondary antibody and incubate for 30 min at RT in the dark. Add 200 µL of 1x permeabilization buffer to wells to dilute, centrifuge the plate for 5 min at 500 x g at RT, and flick to discard supernatant. Wash cells 2x with 200 µL of 1x permeabilization buffer.
NOTE: If necessary, suspend the protocol at this point. Resuspend cells in 200 µL of long-term storage buffer for immune cells with P200 pipette (see Table of Materials for recommendations) and store at 4 °C for 12-24 h protected from light. - Preparing for flow cytometry: Centrifuge plate for 5 min at 500 x g at RT and flick to discard supernatant. Resuspend cells in 200 µL of FACS buffer using a P200 pipette for flow cytometry. Seal with transparent film for transport to the cytometer.
- Aliquot extracellularly stained cells from section 1.5.2 into 96 well plate (5 x 104– 1 x 106 cells). Spin cells for 5 min at 500 x g at 4 °C and flick to remove FACS buffer.
- Flow cytometry
- To analyze the proposed antibody panel, ensure that the cytometer is equipped with at least four lasers including violet (405 nm), blue (488 nm), yellow (561 nm), and red (633 nm). The cytometer needs filters to detect FITC (blue-525 nm), KRO (violet-525 nm) PE (yellow-585 nm), and APC (red-660 nm). Add additional antibodies depending on the cytometer chosen.
- Calibration and standardization: At the start of each experiment, run rainbow fluorescent beads and adjust the photomultiplier tube (PMT) voltage until the bead peaks are comparable to the target values run for previous experiments. This method of standardization allows for accommodation of equipment drift over time.
- Compensation: After the PMT voltage and gain has been set for the experiment, use antibody captured compensation beads to establish the compensation matrix for the panel of antibodies. This calculation will ensure that the fluorophores are not contributing to the changes of signal in other channels. This is increasingly necessary when multiplexing multiple antibodies.
- Size gating: In a dot plot, plot SSC-A on log vs FSC-H on linear. Gate out debris and select for cell size using the S1 gate. Select for singlet cells in a dot plot of FSC-W vs FSC-H and gate as S2. (Figure 4).
- Establishing fluorophore gates: Using the relevant FMO for each fluorophore channel, establish the gates to determine what is a positive signal in each channel using single parameter histograms (Figure 4).
- Measuring the samples: Carefully record the samples using the established gating strategy. Identify the microglia using P2RY12+ signal, determine the expression of the protein in the respective channels for the microglia only.
- Flow cytometry data analysis
- Establishing analysis gates: Using the above steps for the cytometer on the analysis software user interface, use the same gates used for recording for analysis.
- Obtain MFI values using flow cytometry analysis software (see Table of Materials for recommendations): Recapitulate the cytometer gating strategy for flow analysis. Using the add statistics function, select median for the population of interest (e.g., 568+) on the compensated channel height. Using the table editor, export the median fluorescent intensity (MFI) values for the respective channels into a spreadsheet to proceed with statistical analysis (Table 2).
NOTE: Supplementary File S1 includes example data from lipopolysaccharide (LPS) and phosphate buffered saline (PBS) injected mice and an example analysis file with the gating strategy and MFI values. - Analyzing MFI values to protein fold change: After obtaining the MFI values, calculate the fold change of the MFI relative to the control or untreated population (Equation 1). The MFI fold change is reflective of the fold change in protein levels. Using the fold change values, assess the change in expression and calculate the statistical significance using a t-test or ANOVA.
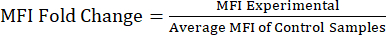 Equation 1
Equation 1
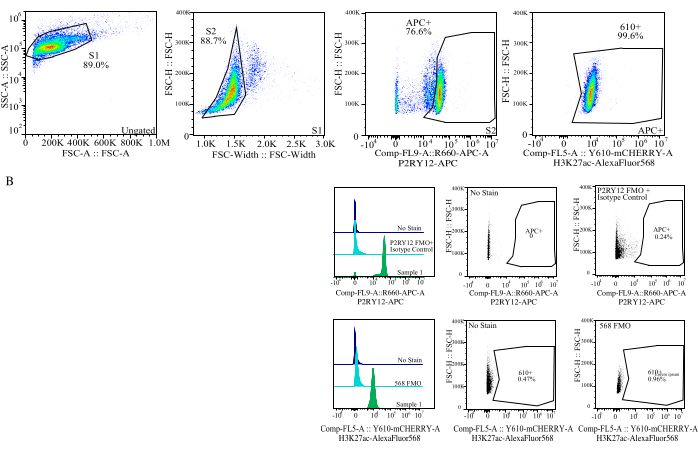
Figure 4: Gating strategy for protein MFI assessment. Events are gated first for cell size on SSC-A vs FSC-H (S1). The cells are then gated for singlets on FSC-H vs FSC-W (S2). Singlet cells are then identified as microglia by P2RY12-APC signal (APC+) with the gate established based on fluorescence in an APC-FMO control which contains an isotype control antibody. Cells are then gated for H3K27Ac-AlexaFluor568 signal on Comp-FL5-A::Y610-mCherry. The fluorescent intensity of the 610+ cells is determined as a proxy for protein expression. Please click here to view a larger version of this figure.
Representative Results
Adult mice were transcardially perfused and sacrificed for microglia isolation. Microglia were isolated on ice and stained with P2RY12-APC and violet 525 live dead antibodies. Cells that were determined to be positive for P2RY12 and negative for violet 525 live dead stain were sorted as live microglia. The average yield of microglia from a dissected mouse brain was 1.28 x 105 ± 0.05 (mean ± standard error of mean (SEM), N=100). There is no difference in the yield of microglia from female (1.25 x 105 ± 0.09 [mean ± SEM, N=46]) and male (1.32 x 105 ± 0.07 [mean ± SEM, N=54]) mice (t(98)=0.6365, p=0.526). When isolating from specific brain regions, the average yield of microglia from mouse cortices is 8.3 x 104 ± 0.08 (mean ± SEM, N=15) and from mouse hippocampus is 4.1 x 104 ± 0.02 (mean ± SEM, N=16). As expected, there is a significant difference in the yield of microglia from each brain region (F(2, 128)=25.25, P<0.0001). Following microglia isolation, RNA was extracted from the isolated cells using a low-input RNA isolation kit. Consistently the RNA integrity score (RIN) was above 9.0 (9.62 ± 0.05) and the average yield of RNA per cell was 0.25 ± 0.01 pg (mean ± SEM, N=32; Supplementary File S2).
Adult mice were intraperitoneally injected with 1 mg/kg lipopolysaccharide (LPS) 24 h prior to sacrifice. The mice were transcardially perfused with HBSS, and microglia isolated from the whole brain according to the described protocol (Figure 5A). For each stain, 20,000-30,000 cells were allocated to each panel of antibodies. The global levels of histone 3 lysine 27 acetylation (H3K27Ac) were assessed in isolated microglia via flow cytometry. For male and female mice, LPS treatment induced increase in H3K27Ac when the MFI is normalized within sex (t(6)=9.676, p<0.0001; Figure 5B). When examining the histograms for the stained cells, the populations remain normally distributed with similar variation; however, the cells have shifted to increased fluorescence resulting in the increase in MFI (Figure 5C). When examining H3K9Ac in the same treatment, there is a similar increase in H3K9Ac (t(6)=7.299, p=0.0003; Figure 5D,E) however the fold change of LPS relative to PBS of H3K9Ac signal is less than H3K27Ac signal.
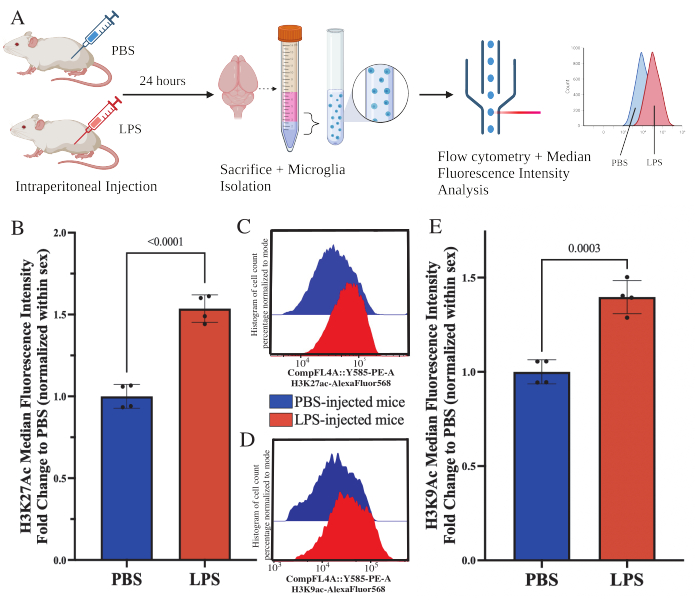
Figure 5: Global changes in histone acetylation in isolated microglia. (A) Mice are injected intraperitoneally with phosphate buffered saline (PBS) or 1 mg/kg lipopolysaccharide (LPS) 24 h prior to sacrifice. The microglia are collected from the immune enriched fraction and fixed for flow cytometry and global histone post translational modification assessment. The median fluorescent intensity is assessed as a proxy for protein expression. Created with BioRender.com. (B) Global levels of H3K27Ac increased in response to LPS treatment. Fold change to PBS normalized within experiment and sex. Unpaired two tailed t-test, t(6)=9.676, p<0.0001. Bar graph depicts the mean ± SEM. N=8 animals; 2 per condition in 2 independent experiments. (C) Example histograms depicting shift of H3K27Ac fluorescent intensity. Modal depicts histograms from PBS-injected versus LPS-injected mice. (D) Example histograms depicting shift of H3K9Ac fluorescent intensity. Modal depicts histograms from PBS-injected versus LPS-injected mice. (E) Global levels of H3K9Ac increased in response to LPS treatment. Fold change to PBS normalized within experiment and sex. Unpaired two tailed t-test, t(6)=7.299, p=0.0003. Bar graph depicts the mean ± SEM. N=8 animals; 2 per condition in 2 independent experiments. Please click here to view a larger version of this figure.
To confirm that the method described was comparable to other previously used methods for global histone modification quantification, we aimed to use immunoblot as a comparative tool. However, the yield from the isolated microglia is simply too low to enable reasonable assessment. Therefore, we used cultured BV2 cells to compare the intracellular flow cytometry method to a Western blot (WB). BV2 cells were grown in complete media (DMEMF12, 10% FBS, 1x penicillin/streptamycin, and 1x L-glutamine) at 37 °C, 5% CO2. Cells were passaged with 0.25% trypsin-EDTA and plated at a density of 250,000 cells/well and treated in reduced serum media (DMEM F12, 2% FBS, 1x penicillin/streptamycin, and 1x L-glutamine) and allowed to recover for 12 h at 37 °C, 5% CO2. Cells were treated with 25 ng/mL LPS for 24 h prior to fixation as described above or lysis with a WB lysis buffer. Signal of H3K27Ac was performed by both methods with GAPDH used as a loading control for WB. Analysis of the normalized fluorescent intensity compared to the PBS control was determined for each group (Figure 6A). When examining the change in normalized H3K27Ac signal by WB, there was a 1.527-fold increase in the LPS treated condition relative to the H2O control which was determined to be significant by unpaired t-test (t=3.024, df=5; p=0.0293). When examining the change using flow cytometry, there was a 1.482-fold increase in the LPS treated condition which was determined to be significant (t=7.843, df=10; p<0.0001). Using a 2-way ANOVA to compare the methods, there was determined to be a significant effect of the treatment (F(1,15)=45.21,p<0.0001), but not the method (F(1,15)=0.05545, p=0.8697) or interaction (F(1,15)=0.02785, p=0.8697). In addition, we verify here that there is no change in histone H3 levels by both Western blot and flow cytometry as 2-way ANOVA revealed no significant effect of the LPS treatment (F(1,7)=0.02170,p=0.8870), the method (F(1,7)=0.01191, p=0.9162) or the interaction (F(1,7=0.01191, p=0.9162; Figure 6B). Example blots and histogram shifts for this data are shown as well (Figure 6C,D).
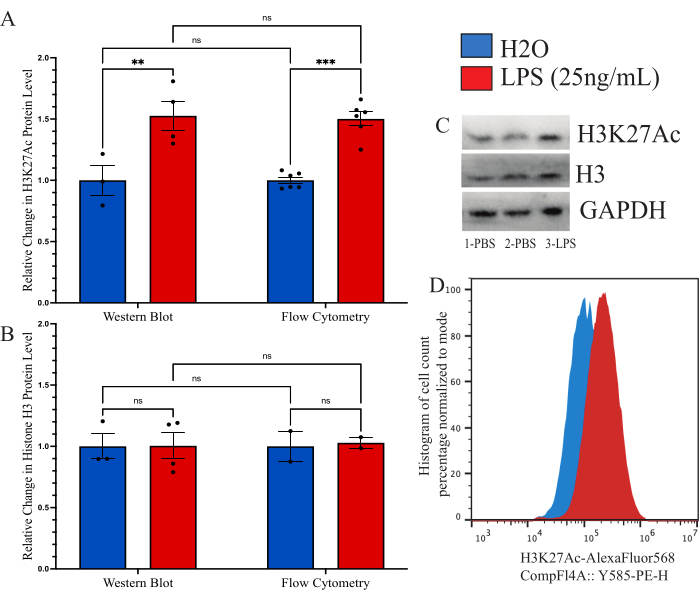
Figure 6: Method comparison for quantification of global histone modification change between flow cytometry and western blot. (A) BV2 cells are treated with 25 ng/mL lipopolysaccharide (LPS) or H2O for 24 h prior to analysis. The fluorescent intensity of H3K27Ac is depicted as fold change to the vehicle control, phosphate buffered saline (PBS), for both flow cytometry and western blot. 2-way ANOVA revealed significant effect of the LPS treatment (F(1,15)=45.21, p<0.0001), but not the method (F(1,15)=0.05545, p=0.8697) or interaction (F(1,15)=0.02785, p=0.8697). Tukey's correction for multiple hypothesis testing was applied for the residuals. * presents 0.0332, ** presents 0.0021. (B) The fluorescent intensity for histone H3 is depicted as fold change to PBS for both flow cytometry and western blot. 2-way ANOVA revealed no significant effect of the LPS treatment (F(1,7)=0.02170, p=0.8870) or the method (F(1,7)=0.01191, p=0.9162) or the interaction (F(1,7=0.01191, p=0.9162). (C) Example blots and (D) flow cytometry shifts are portrayed. Histogram size is normalized to percent based on number of cells present at the mode fluorescent intensity. Bar graph depicts the mean SEM. n=2 independent experiments, 2 per condition per experiment. Please click here to view a larger version of this figure.
All together, these results show that this technique can be used to quantitatively assess the global HPTM levels in isolated microglia. In addition, the method was shown to be comparable to previous techniques but requiring much lower cell inputs. In addition, while not shown, with proper compensation, the present technique can be used with multiple antibodies on the same panel assessing different HPTMs.
Supplementary file S1: Example analysis files. This file contains a wsp analysis file and 7 fcs files including the no stain, P2RY12FMO, 568FMO, two PBS treated animals and two LPS treated animals stained with H3K27Ac. The purpose of this file is to demonstrate the analysis and gating on an experiment that could depict what a successful experiment looked like. Please click here to download this File.
Supplementary File S2: Isolation data. The file included contains the relevant data post microglia sort which contains the microglia and RNA yield from the described protocol. Please click here to download this File.
| GATED POPULATION | Frequency of Parent | Frequency of Total | Count |
| S1 | 89.00% | 89.00% | 25672 |
| S2>S1 | 88.73% | 78.97% | 22779 |
| APC+ > S2 > S1 | 76.61% | 60.50% | 17452 |
| 610+ > APC+ > S2 > S1 | 99.56% | 60.24% | 17376 |
Table 2: Example sample lineage chart depicts percentage and event numbers required for accurate protein detection.
Discussion
The protocol presented enables quantitative assessment of global HPTM levels through flow cytometry. While this protocol presents a novel method, previous studies have done quantitative assessment of proteins using a similar approach26. Previous methods used to assess global levels of HPTMs include immunohistochemistry and western blot16,17,19,20. The flow cytometry-based method presented is an easily quantifiable method, whereas western blot and immunohistochemistry are semi-quantitative and have lower throughput. Western blot relies on cell lysis and thus requires both protein normalization and a loading control protein that is assumed to be unchanged by the experimental condition27. Immunohistochemistry is semiquantitative and very low throughput as it is difficult to quantitatively assess the amount of protein without examining on a single cell level16. Similarly, for the isolated microglia, there is a benefit to using the flow cytometry method due to the limited yield as western blot requires much larger protein input19. The low cell number requirements allow for multiple staining panels to be run from the same animal.
However, as with any other method, there are limitations to this technique including antibody cost and availability, as not all antibodies work well in a flow cytometry setting. In addition, compared to immunoblot, the concentration of antibody required is much higher. While multiplexing allows for multiple antibodies to be used on the same panel of cells, cells cannot be stripped of the antibody after analysis, thus limiting the cell usage to one per antibody species. This is different from immunoblot in which the same blot can be used repeatedly. However, depending on the availability of antibodies and the number of detection channels on a cytometer, it would be possible to examine up to a dozen marks simultaneously.
The current method captures only global levels of protein expression and not the specific genomic location, and changes in global levels may not reflect changes at individual genomic loci. Similarly, a lack of change in global levels may not mean that no genomic loci are undergoing changes, simply that the global changes sum to no differences between treatments. As such, this technique is meant to be used as a screen to identify HPTMs of interest for genomic analysis. In addition, this method does not allow for comparison across different protein marks except for when assessed as a fold change to control. Therefore, this is limited compared to a standard curve-based method such as ELISA for protein determination.
The protocol presented offers a strategy for isolating live brain microglia. This protocol relies on P2RY12 protein expression for microglia isolation. However, P2RY12 is a homeostatic marker in microglia and can be downregulated in disease models, such as 5XFAD22. Therefore, when using a disease model animal be sure to choose other marker proteins such as TMEM119, CD11b, or CD45 to aid in isolation of microglia23. Similarly, we present this protocol as isolation from the hippocampus and/or the cortex. This protocol would work to isolate microglia from other brain regions including white matter regions, however, multiple animals may be required to obtain enough microglia depending on the size of the regions of interest.
The protocol presented can robustly isolate live, brain microglia, but there are several steps, described below, in the isolation stage that can decrease cell yield if performed incorrectly.
Perfusions for this protocol result in a higher percentage of microglia in the immune enriched fragment which will reduce the amount of time at the sorter. However, perfusion is not required, and other methods of euthanasia can be used if required.
During microglia isolation, myelin should be completely removed. Flow cytometers rely on cells being able to travel through narrow tubing at a rapid pace. Due to its viscosity and tendency to clump, myelin causes problems with cytometers, often causing clogs which can both damage the equipment and destroy the sample, reducing yield drastically. Be cautious to remove all of the myelin during collection of immune-enriched fragments to avoid having issues downstream.
Plate staining versus tube staining: In this protocol, we described two options for staining cells in either 1.5 mL tubes or a 96 well plate. The use case for each depends on the experiment; however generally tube staining is lower risk for impacting yield than plate staining as the flick risks loss of cells if done incorrectly. Plate staining is much faster as aspirating the supernatant for each tube is time consuming. Prior to fixation (for sorting, etc.), use tube staining to maximize yield and reduce risk of loss. However, for HPTM analysis, once cells are fixed for intranuclear staining, the pellet is more stable, and there is reduced risk of loss with flicking.
Establishing the discontinuous density gradient: When establishing the layering, setting up the layers properly is essential to obtain the immune enriched fraction. If the layers are disturbed or mixed and appear cloudy, the cells will not sort to their desired location, and there will be difficulty in obtaining the immune-enriched cell fraction. If this occurs, spin with the density medium to remove myelin and then collect the entire remaining fractions, dilute with 3 mL of FACS buffer to 1 mL of density medium and mix well (this will require multiple tubes). Spin at 500 x g for 10 min with the brake on 0. Discard the supernatant, leaving only ~300 µL solution. Collect the entire sample and stain. This will yield in reduced sort percentages and a higher amount of time spent at the cytometer, but the yield can still be comparable.
When using the isolation method, it is beneficial to be able to collect cells for RNA and for HPTM evaluation from the same mouse brain. In this situation, after sorting the live microglia, cells can be divided to allocate a portion to RNA evaluation (minimum input number of cells to obtain a decent RNA yield is 75,000 cells) and a portion for further flow cytometry analysis (minimum 10,000 cells per well for good determination of MFI). In this case, flow cytometer sorting is required. However, when only planning to use the cells for HPTM analysis, sorting is not required, and the immune fraction can be stained with the P2RY12 antibody and HPTM antibody. Gating on the cytometer can then be set for P2RY12+ microglia, as would be done for flow sorting, to analyze only HPTM signal within microglia. Eliminating the sorting allows for the protocol to be faster and more cost effective. In addition, if evaluating HPTMs from cultured cells, starting at the staining protocol is sufficient and no cell marker antibodies are required as demonstrated in Figure 6. The HPTM evaluation protocol can be used for many cell types including cultured, primary, and IPSC derived cells.
Finally, while we have presented only two potential uses of microglia downstream of isolation, there are many others including epigenetic techniques such as ChIP, CUT&Tag and CUT&RUN. In the case of genomic epigenetic techniques, where characterizing changes at specific loci is of interest, choose specific inhibitors for writers and erasers of chromatin marks11 tailored to the experiments to ensure that any microglial epigenetic modifications profiled are not technical artifacts from any steps in the isolation procedure such as enzymatic digestion. When assessing changes in global levels of epigenetic marks, such as by using quantitative flow cytometry, any procedure-induced changes are not expected to be so large that they are detected at the global level.
Overall, the discussed methods provide a novel, single cell method for quantifying global levels of histone modifications and other epigenetic changes by flow cytometry. We demonstrated that this method is sufficiently sensitive to detect global changes in the enhancer marker H3K27ac in microglia in response to LPS in vivo. This is consistent with previous ChIP-sequencing of H3K27ac following LPS stimulation showing dramatic remodeling of enhancers responsive to LPS28. Applications of this method will allow for examination of global epigenetic changes across different brain cell types in development and disease.
Disclosures
The authors have nothing to disclose.
Acknowledgements
Thanks to Yanyang Bai for helping with the immunoblot in Figure 5. This work was supported by the Canadian Institutes for Health Research [CRC-RS 950-232402 to AC]; Natural Sciences and Engineering Research Council of Canada [RGPIN-2019-04450, DGECR-2019-00069 to AC]; Scottish Rite Charitable Foundation [21103 to AC] and the Brain Canada Foundation [AWD-023132 to AC]; University of British Columbia Aboriginal Graduate Fellowship (6481 to MT); British Columbia Graduate Scholarship (6768 to MT); Canadian Open Neuroscience Platform Student Scholar Award (10901 to JK); University of British Columbia Four Year Doctoral Fellowship (6569 to JK). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Materials
| 0.5M EDTA | Invitrogen | AM9260G | |
| 15 mL Falcon Centrifuge Tubes, Polypropylene, Sterile | Falcon | 352196 | |
| 24-well Clear Not Treated Plates | Costar | 3738 | |
| 2-Mercaptoethanol | Gibco | 21985023 | |
| 96 Well Clear Polystyrene Microplate, clear round bottom, non treated surface | Corning | 3788 | |
| Acetyl Histone 3 K9 (C5B11) | Cell Signalling Technology | 9649S | Dilution: 1:100 |
| Acetyl Histone H4 K8 (2594) | Cell Signalling Technology | 2594S | Dilution: 1:100 |
| Acetyl-Histone H3 K27 (D5E4) | Cell Signalling Technology | 8173S | Dilution: 1:100 |
| Acetyl-Histone H3 Lys27 (MA523516) | Invitrogen | MA523516 | Dilution: 1:100 |
| Actinomycin D | New England Biolabs | 15021S | |
| Anisomycin | New England Biolabs | 2222S | |
| Anti-Histone H3 (tri methyl K4) | Abcam | ab213224 | Dilution: 1:100 |
| Anti-Lactyl-Histone H4 (Lys 12) Rabbit mAb | PTM Biolabs | PTM-1411RM | Dilution: 1:250 |
| Anti-L-Lactyllysine Rabbit pAb | PRM Biolabs | PTM-1401RM | Dilution: 1:250 |
| Apc anti-P2RY12 Antibody, Clone: S16007D | BioLegend | 848006 | |
| BSA | Tocris | 5217 | |
| Cyto-Last Buffer | BioLegend | 422501 | |
| dimethylsulfoxide, sterile | Cell Signalling Technology | 12611S | |
| DNAse I | STEMCELL Technologies | 07900 | |
| Donkey Anti Mouse AlexaFluor488 | Jackson ImmunoResearch | 715-546-150 | Dilution: 1:500 |
| Donkey Anti Rabbit AlexaFluor488 | ABclonal | AS035 | Dilution: 1:500 |
| Donkey Anti Rabbit AlexaFluor568 | Invitrogen | A10042 | Dilution: 1:500 |
| Donkey Anti Rabbit Brilliant Violet 421 | BioLegend | 406410 | Dilution: 1:500 |
| Fisherbrand Disposable Graduated Transfer Pipettes | Fisherbrand | 13-711-9AM | |
| Fisherbrand Disposable PES Filter Unit, 250mL | Fisherbrand | FB12566502 | |
| H3K18ac Polyclonal Antibody | Invitrogen | 720095 | Dilution: 1:100 |
| HBSS (10X), no calcium, no magnesium, no phenol red | Gibco | 14185052 | |
| HBSS, no calcium, no magnesium, no phenol red | Gibco | 14175103 | |
| Histone 3 Trimethyl K27 (ab6002) | Abcam | ab6002 | Dilution: 1:100 |
| KONTES Dounce Tissue Grinders 125mm 7mL | VWR | 885300-0007 | |
| Lactyl-Histone H3 (Lys 18) Rabbit mAb | PTM BIolabs | PTM-1406RM | Dilution: 1:250 |
| Lipopolysacharide | Sigma-Aldrich | L5418 | |
| Normal Donkey Serum | Jackson ImmunoResearch | 017-000-121 | |
| OneComp eBeads Compensation Beads | Invitrogen | 01-1111-41 | |
| PDS Kit, Papain Vial – Worthington Biochemical | Cedarlane | LK003178 | |
| Percoll | Sigma-Aldrich | GE17-0891-02 | |
| Phenol Red | VWR | RC57004 | |
| QIAshredder | Qiagen | 79656 | |
| Rainbow Fluorescent Particles, 1 peak (3.0-3.4 uM – Mid Range Intensity | BioLegend | 422905 | |
| RNase-free Microfuge Tubes, 1.5 mL | Invitrogen | AM12400 | |
| Rneasy Plus Micro Kit | Qiagen | 74034 | |
| Round Bottom Polypropylene Tubes with Caps, 5 mL | Corning | 352063 | |
| Triptolide | New England Biolabs | 97539 | |
| True Nuclear Transcription Factor Buffer Set | BioLegend | 424401 | |
| TruStain FcX PLUS (anti-mouse CD16/32) Antibody | BioLegend | 156604 | |
| Trypan Blue | VWR | 97063-702 | |
| Zombie Aqua Fixable Viability Kit | BioLegend | 423102 |
References
- Miller, J. L., Grant, P. A. The Role of DNA Methylation and Histone Modifications in Transcriptional Regulation in Humans. Epigenetics: Development and Disease. 61, 289-317 (2013).
- Kouzarides, T. Chromatin Modifications and Their Function. Cell. 128 (4), 693-705 (2007).
- Bannister, A. J., Kouzarides, T. Regulation of chromatin by histone modifications. Cell Research. 21 (3), 381-395 (2011).
- Barski, A., et al. High-resolution profiling of histone methylations in the human genome. Cell. 129 (4), 823-837 (2007).
- Vogel Ciernia, A., LaSalle, J. The landscape of DNA methylation amid a perfect storm of autism aetiologies. Nature Reviews. Neuroscience. 17 (7), 411-423 (2016).
- Keiser, A. A., et al. Systemic HDAC3 inhibition ameliorates impairments in synaptic plasticity caused by simulated galactic cosmic radiation exposure in male mice. Neurobiology of Learning and Memory. 178, 107367 (2021).
- McQuown, S. C., et al. HDAC3 is a critical negative regulator of long-term memory formation. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 31 (2), 764-774 (2011).
- Barrett, R. M., et al. Hippocampal Focal Knockout of CBP Affects Specific Histone Modifications, Long-Term Potentiation, and Long-Term Memory. Neuropsychopharmacology. 36 (8), 1545-1556 (2011).
- Datta, M., et al. Histone Deacetylases 1 and 2 Regulate Microglia Function during Development, Homeostasis, and Neurodegeneration in a Context-Dependent Manner. Immunity. 48 (3), 514.e6-529.e6 (2018).
- Belhocine, S., et al. Context-dependent transcriptional regulation of microglial proliferation. Glia. 70 (3), 572-589 (2022).
- Gosselin, D., et al. An environment-dependent transcriptional network specifies human microglia identity. Science (New York, N.Y.). 356 (6344), eaal3222 (2017).
- Kettenmann, H., Hanisch, U. -. K., Noda, M., Verkhratsky, A. Physiology of Microglia. Physiological Reviews. 91 (2), 461-553 (2011).
- Sullivan, O., Ciernia, A. V. Work hard, play hard: how sexually differentiated microglia work to shape social play and reproductive behavior. Frontiers in Behavioral Neuroscience. 16, 989011 (2022).
- Das, P. M., Ramachandran, K., vanWert, J., Singal, R. Chromatin immunoprecipitation assay. BioTechniques. 37 (6), 961-969 (2004).
- Mahmood, T., Yang, P. C. Western blot: technique, theory, and trouble shooting. North American Journal of Medical Sciences. 4 (9), 429-434 (2012).
- Crowe, A., Yue, W. Semi-quantitative Determination of Protein Expression Using Immunohistochemistry Staining and Analysis: An Integrated Protocol. BIO-PROTOCOL. 9 (24), (2019).
- Seligson, D. B., et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 435 (7046), 1262-1266 (2005).
- Liu, B., et al. Global Histone Modification Patterns as Prognostic Markers to Classify Glioma Patients. Cancer Epidemiology, Biomarkers & Prevention. 19 (11), 2888-2896 (2010).
- Pan, R. Y., et al. Positive feedback regulation of microglial glucose metabolism by histone H4 lysine 12 lactylation in Alzheimer’s disease. Cell Metabolism. 34 (4), 634.e6-648.e6 (2022).
- Zhang, D., et al. Metabolic regulation of gene expression by histone lactylation. Nature. 574 (7779), 575-580 (2019).
- Pösel, C., Möller, K., Boltze, J., Wagner, D. C., Weise, G. Isolation and Flow Cytometric Analysis of Immune Cells from the Ischemic Mouse Brain. Journal of Visualized Experiments. (108), 53658 (2016).
- Oblak, A. L., et al. Comprehensive Evaluation of the 5XFAD Mouse Model for Preclinical Testing Applications: A MODEL-AD Study. Frontiers in Aging Neuroscience. 13, 713726 (2021).
- Bohlen, C. J., Bennett, F. C., Bennett, M. L. Isolation and Culture of Microglia. Current Protocols in Immunology. 125 (1), e70 (2019).
- McKinnon, K. M. Multiparameter Conventional Flow Cytometry. Flow Cytometry Protocols. 1678, 139-150 (2018).
- Marsh, S. E., et al. Dissection of artifactual and confounding glial signatures by single-cell sequencing of mouse and human brain. Nature Neuroscience. 25 (3), 306-316 (2022).
- Wang, L., Gaigalas, A. K., Marti, G., Abbasi, F., Hoffman, R. A. Toward quantitative fluorescence measurements with multicolor flow cytometry. Cytometry Part A. 73A (4), 279-288 (2008).
- Rumbaugh, G., Miller, C. A. Epigenetic changes in the brain: measuring global histone modifications. Methods in Molecular Biology (Clifton, N.J). 670, 263-274 (2011).
- Xavier, A. M., et al. Systematic delineation of signaling and epigenomic mechanisms underlying microglia inflammatory activity in acute and chronic brain pathologies. BioRvix. , (2022).

