Whole-Mount Fluorescence In Situ Hybridization to Study Spermatogenesis in the Anopheles Mosquito
Summary
Given their simple anatomy, Anopheles testes offer a good cytological model for studying spermatogenesis. This protocol describes whole-mount fluorescence in situ hybridization, a technique used to investigate this biological process, as well as the phenotype of transgenic strains harboring mutations in the genes involved in sperm production.
Abstract
Spermatogenesis is a complex biological process during which diploid cells undergo successive mitotic and meiotic division followed by large structural changes to form haploid spermatozoa. Besides the biological aspect, studying spermatogenesis is of paramount importance for understanding and developing genetic technologies such as gene drive and synthetic sex ratio distorters, which, by altering Mendelian inheritance and the sperm sex ratio, respectively, could be used to control pest insect populations. These technologies have proven to be very promising in lab settings and could potentially be used to control wild populations of Anopheles mosquitoes, which are vectors of malaria. Due to the simplicity of the testis anatomy and their medical importance, Anopheles gambiae, a major malaria vector in sub-Saharan Africa, represents a good cytological model for studying spermatogenesis. This protocol describes how whole-mount fluorescence in situ hybridization (WFISH) can be used to study the dramatic changes in cell nuclear structure through spermatogenesis using fluorescent probes that specifically stain the X and Y chromosomes. FISH usually requires the disruption of the reproductive organs to expose mitotic or meiotic chromosomes and allow the staining of specific genomic regions with fluorescent probes. WFISH enables the preservation of the native cytological structure of the testis, coupled with a good level of signal detection from fluorescent probes targeting repetitive DNA sequences. This allows researchers to follow changes in the chromosomal behavior of cells undergoing meiosis along the structure of the organ, where each phase of the process can clearly be distinguished. This technique could be particularly useful for studying chromosome meiotic pairing and investigating the cytological phenotypes associated with, for example, synthetic sex ratio distorters, hybrid male sterility, and the knock-out of genes involved in spermatogenesis.
Introduction
Malaria imposes an enormous burden on the health and well-being of the global human population. In 2021, the World Health Organization (WHO) estimated that malaria caused 619,000 deaths, of which 96% occurred in Sub-Saharan Africa1. The disease is transmitted by mosquitoes belonging to the Anopheles genus, and in Sub-Saharan Africa, three species, namely Anopheles gambiae (An. gambiae), Anopheles coluzzi (An, coluzzi) and Anopheles arabiensis (An. Arabiensis) have a disproportionately large role in malaria transmission, accounting for 95% of malaria cases globally. Control programs relying on traditional methods such as insecticides and antimalarial drugs have saved millions of lives; however, in recent years, the rising resistance to these control methods has challenged their efficacy1,2. In addition, restrictions imposed by the COVID-19 pandemic have affected the availability of key malaria control interventions, which, according to the 2022 WHO World Malaria Report, has increased malaria incidence1. In the last two decades, novel genetic control methods have been developed in laboratory settings to target Anopheles mosquitoes3,4,5,6,7,8,9,10. Among these strategies, those based on gene drive systems (GDSs) and synthetic sex ratio distorters (SDs) seem promising. GDSs rely on the possibility of transmitting, at a very high frequency, a genetic modification that affects female fertility or impairs the parasite life cycle in the mosquito5,11,12. SDs, instead, act by skewing the sex ratio of a mosquito progeny toward males, which leads, over time, to the collapse of a target population due to a lack of females4,6,13. The main components of these genetic systems act primarily on the reproductive organs of the mosquitoes, where the gametes, eggs, and sperm are produced following meiotic division14.
In this protocol, advances in cytogenetic techniques are employed to explore spermatogenesis in An. gambiae focusing on the chromosomes' behavior in situ. The structure of the mosquito testis and the biological processes that take place within it have been previously investigated using a number of cytological methods, such as immunofluorescence, fluorescent reporter transgenes, and DNA and RNA fluorescence in situ hybridization (FISH)15,16,17,18,19,20; the organs show a spindle-like shape, in which the lower pole is attached to a deferent duct connected to the male accessory glands. In the upper pole, the germline stem cells niche proliferates and differentiates into spermatogonia cells embedded inside spermatocysts formed by somatic cells. Following multiple rounds of mitotic division, the spermatogonia differentiate into spermatocytes, which enter meiosis. At prophase, autosome and sex chromosomes pair with their homologs, and crossing over takes place. After the meiotic divisions, round haploid spermatids are generated and enter spermiogenesis, and this process leads to the formation of mature haploid spermatozoa in which the cytoplasm has been removed, the nuclear chromatin is condensed, and flagella emerge at the basal part of the nuclei21,22 (Figure 1 and Figure 2).
In general, spermiogenesis starts around the mid-pupal stage, and mature spermatozoa can be detected in the late pupal stage in the sperm reservoir23. The maturation process of the spermatocysts continues during adult life23,24,25. In Anopheles testes, each step of spermatogenesis can be easily identified by looking at the nuclear morphology of the cells in each spermatocyst (Figure 2). Whole-mount fluorescence in situ hybridization (WFISH), described in this protocol, allows researchers to specifically label a chromosomal region and track it during spermatogenesis while preserving the native structure of the organ and cell nuclei position; this represents an advantage when compared to the standard DNA FISH protocol in which the organ is usually squashed, leading to tissue damage19. In the current protocol, fluorescent probes are used to stain repetitive sequences on the sex chromosomes and, thus, track their behavior during spermatogenesis, from diploid dividing cells to mature haploid spermatozoa. WFISH can be particularly useful for studying sex chromosome meiotic pairing and investigating the cytological phenotypes associated with, for example, synthetic sex ratio distorters, hybrid male sterility, and the knock-out of genes involved in spermatogenesis4,19,26,27.
Given their role as malaria vectors, Anopheles mosquitoes are the target of an increasing number of genetic vector control strategies, which often act in the reproductive organs of these organisms. Several mosquito mutants and cytological phenotypes have been generated that require novel cytological techniques to be investigated26,27,28,29. The method described in this study sheds light on the understanding of spermatogenesis, as well as the cytological mechanisms behind genetic strategies that have the potential to control malaria-transmitting mosquitoes.
Protocol
1. DNA probe labeling
NOTE: Below are the technical steps to generate fluorescent DNA probes that specifically label the sex chromosomes of An. gambiae mosquitoes.
- Probe labeling using PCR
- Extract genomic DNA from pupae or adult males to label the X or Y chromosomes using a commercially available genomic DNA extraction kit (see Table of Materials).
- Prepare the PCR reaction mixture: 200 ng of genomic DNA, 0.05 mM unlabeled nucleotide (dATP, dCTP, dGTP), 0.015 mM of dTTP, and 1 uL of fluorescently labeled dUTP (Cy3, Cy5, or another fluorochrome), 50 pmol of forward and reverse primer (Table 1), 5 µL of 10x PCR-buffer, and 10 U of Taq DNA polymerase (see Table of Materials).
- To label 18S rDNA and satellite AgY53B (Table 1), perform a PCR reaction using the following PCR parameters: one cycle of 95 °C for 10 min; 35 cycles of 95 °C for 30 s, 52 °C for 30 s, and 72 °C for 45 s; one cycle of 72 °C for 5 min; and a final hold at 4 °C.
NOTE: To obtain a good probe concentration with the PCR labeling method (~1 µg in 5 µL), which is critical for a successful WFISH, the PCR reaction should be highly efficient. For this reason, before labeling the probe, testing the efficacy of the primers selected for the amplification is strongly suggested. In addition, including a positive control in the PCR reaction (without the fluorescent dUTP) will help to verify the efficacy of the labeling reaction. - Store the probe at −20 °C in a dark place.
- Obtaining 3' end fluorescent oligonucleotide probes
- Obtain commercially available fluorescent oligonucleotide probes as modified oligos by adding Cy3 or Cy5 fluorochromes (or any other fluorochrome) to the 3' end of the nucleotide sequence (see Table of Materials). See primers/oligonucleotide in Table 1 for the reference sequences used to label the Y-linked satellite AgY477-AgY53B junction region and the X-linked satellite from Contig_240.
NOTE: There are no technical impediments related to the concentration of 3' end-labeled oligonucleotides, as the user can usually choose this before purchase. We suggest diluting ~800 ng of oligo probe solution in the hybridization buffer for efficient labeling using the oligo probe. X- and Y-specific oligo-probes have been previously used for WFISH by Liang and Sharakhov19, and the reference sequence can be found in Table 1.
- Obtain commercially available fluorescent oligonucleotide probes as modified oligos by adding Cy3 or Cy5 fluorochromes (or any other fluorochrome) to the 3' end of the nucleotide sequence (see Table of Materials). See primers/oligonucleotide in Table 1 for the reference sequences used to label the Y-linked satellite AgY477-AgY53B junction region and the X-linked satellite from Contig_240.
2. Preparation of the hybridization solution
NOTE: The fluorescent probes generated in step 1 must be incorporated into a chemical solution that hybridizes with the target sequences.
- Probe precipitation prior to fluorescence in situ hybridization
- To a 1.5 mL tube, add 5 µL of labeled DNA probe (if obtained by the PCR-labeling method) or 2.2 µL of 3' modified oligo probe (~800 ng of probe) from step 1 and 5 µL of salmon sperm DNA (see Table of Materials). Combine the probes specific to different genomic regions in the same tube, and use them as a unique solution in the following steps.
- Precipitate the DNA probe by adding 0.1 volume of 3 M sodium acetate and 2 volumes of 100% ethanol. Keep at −20 °C for at least 2.5 h (increasing the time of incubation at −20 °C will increase the final yield). At this stage, the probes can also be stored overnight before centrifugation.
- Centrifuge at 17,000 x g at 4 °C for 20 min, remove the ethanol, and air-dry the pellet at RT in the dark for ~20 min.
- Hybridization solution
- Before proceeding with the testis dissection (step 3), prepare the hybridization buffer by mixing the following reagents in a 1.5 tube: 500 µL of formamide, 0.2 g of dextran sulfate, 100 µL of 20x sodium saline citrate (SSC), and 200 µL of sterile H20 (see Table of Materials). Vortex the hybridization solution for 1 min, and allow the dextran sulfate to dissolve at 37 °C for 30 min.
- Dissolve the pellet from step 2.1.3 in 20-30 µL of hybridization buffer (vortex for about 1 min, perform a quick spin, and store the tubes at 37 °C in the dark) to obtain the hybridization solution.
3. Testis dissection and fixation
- At room temperature (RT), dissect21 at least ~20 testes from pupae or 1 day old adults in sterile 1x phosphate-buffered saline (PBS) solution, and transfer them to a clean microscope slide containing a fresh drop of 1x PBS solution.
- Transfer the testes using a P1,000 wide-bore filtered tip or the tip of a dissection needle from the 1x PBS drop into an embryo dish containing 3.7% formaldehyde in 1x PBS with 0.1% Tween-20 (PBST), and incubate for 10 min at RT.
- Wash the testes in 1x PBST for 5 min at RT. Incubate the testes in 0.1 mg/mL RNAse A (see Table of Materials) diluted in sterile 1x PBS for 30 min at 37 °C.
- Remove the RNAse solution, add the penetrating solution (1% Triton/0.1 M HCl in 1x PBST), and incubate at RT for 10 min.
NOTE: Proteinase K can be used as a penetrating agent at a final concentration of 10 µg/mL in 1x PBST to increase the permeabilization of the testes. - Wash the testes in 1x PBST two times for 5 min each at RT.
4. Hybridization
NOTE: This section describes the final steps for the in situ hybridization.
- After the washing step (step 3.5), transfer the testes in a 1.5 mL tube with 20-30 µL of hybridization solution containing the previously prepared probe (step 2.2.2). Use the tip of the pipette to mix the solution gently. Gently flick the tube five times before proceeding to the next steps.
- Incubate for 5 min at 75 °C for DNA denaturation.
- Incubate overnight at 37 °C (if possible, with rocking at less than 100 rpm) for DNA-DNA hybridization.
- Transfer the testes back into an embryo dish using a P1,000 wide-bore filtered tip, and wash them in 2x SSC preheated to 50 °C for 5 min.
NOTE: After the hybridization step, a final washing step using 2x SSC is required; this plays an important role in removing any background signal caused by the presence of unhybridized probes inside the organ tissue. If a strong background signal is present, repeating the final washing step is recommended. - Remove the 2x SSC, and mount the testes using a commercially available mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) (see Table of Materials) on a frosted glass slide, seal with coverslip sealant, and incubate for at least 2 h at RT in the dark.
- Perform confocal imaging. The whole testes can be visualized using 40x or 63x oil immersion objectives. If a z-stack is performed, we suggest using a z-step of 1.25 µm.
Representative Results
In this work, WFISH was used to investigate the chromosome behavior during spermatogenesis in An. gambiae. The first crucial step for applying this protocol is obtaining testes that show a low level of morphological alteration after dissection. Basic knowledge of the mosquito anatomy is required to perform a successful testis dissection, and below, some guidances for this procedure are given. In the Anopheles mosquito, mature testes can be found lying in the sixth abdominal segment of the pupal and adult stages21. As shown in Figure 1, the vas deferens connects the testes to the male accessory glands (MAGs), which are located in the last segment of the abdomen. The MAGs are connected to a unique ejaculatory duct that delivers the sperms and seminal fluids to the copulatory organ and the external part of the male genital apparatus21. The entire internal male genital apparatus can be dissected using different approaches depending on the life stage of the mosquito. During the pupal stage, testes can be easily identified using a stereomicroscope throughout the light cuticle by looking at the ventral side of the abdomen in the proximity of the sixth segment (Figure 1). To dissect the testes, the lower part of the abdomen, including the sixth segment, can be isolated from the rest of the body using a pair of needles and transferred into a clean drop of 1x PBS. Following the removal of the last segment, the entire apparatus can be squished out of the abdomen by applying gentle pressure with the dissecting needles. To dissect the testes from adult males, the first step involves isolating the entire abdomen in a fresh drop of 1x PBS and then pulling out the last segment carrying the clasps, which are the male copulatory structures (Figure 1). At this point, the lower part of the MAGs should emerge and be easily identifiable due to their yellow color. The entire male apparatus can then be slowly pulled out with the help of a needle or forceps in a drop of 1x PBS until the pair of testes attached to the vas deferens is visible. Before proceeding with the fixation, it is important to isolate the testes from the other parts of the male apparatus by cutting in the proximity of the lower part of the vas deferens (Figure 1).
The age of the pupae or adult males is an important factor to consider depending on the spermatogenesis stage under investigation. In An. gambiae, spermatogenesis starts at the early/mid-pupal stage, and it continues throughout the entire life of the individual24. Between 3 h to 10 h after pupation, the premeiotic and meiotic stages are more represented in the testis (meiotic prophase, meiotic divisions), the spermatid DNA is relatively uncondensed, and mature sperms have not yet been formed. Late pupae and 1 day old adults offer a good balance between the premeiotic, meiotic, and post-meiotic stages (Figure 2 and Figure 3). In adults that are more than 4 days old, the premeiotic stages and spermatocysts are less represented, and the testes are mainly occupied by mature sperms contained in the sperm reservoir.
To investigate the behavior of the sex chromosomes during different stages of spermatogenesis, WFISH was performed on testes dissected at the late pupal stage to ensure a good representation of the entire process. To follow the behavior of these chromosomes, fluorescent probes specific to repetitive sequences located exclusively on the X or Y chromosome were used. The fluorescent probes can be generated using PCR or obtained commercially as 3' end-labeled oligonucleotides. Using an oligo with a length of >40 bps is recommended to allow good signal detection from the fluorescent oligo probe. In our experience, 3' end-labeled oligos perform better than PCR-labeled probes in terms of signal detection. In addition, the copy number of the target sequence is a factor that may affect the efficacy of WFISH. If labeling is unsuccessful, using the PCR labeling method on a longer fragment or designing several oligos specific to the target region is suggested.
The current methods, based on using a penetrating solution (1% Triton/0.1 M HCl in 1x PBST), allow a good level of testis permeabilization and penetration of the probes, resulting in a successful hybridization reaction. Oligo probes specific to sex chromosome repetitive sequences can be designed based on the extensive characterization of repetitive elements performed by Hall et al.20. In addition, consensus sequences specific to X- or Y-linked repetitive elements can be obtained using a bioinformatic platform such as the RedKmer pipeline30. It is important to notice that sex chromosome probes can target repetitive elements such as satellites and retrotransposons, and they can have a different level of hybridization with the X or the Y chromosomes depending on the species under examination20,31,32. As shown in Figure 3, a good level of hybridization of the probes and low background allowed the visualization of the targeted chromosomes throughout spermatogenesis. The pairing of labeled sex chromosomes could be seen in the premeiotic and meiotic stages. This was followed by detecting either the X or the Y chromosomes in haploid cell nuclei chromosomes resulting from meiotic divisions. Subsequently, X- or Y-bearing spermatids could be followed throughout spermiogenesis, marked by different levels of DNA condensation, to the final step of arrow-shaped mature spermatozoa. In the present experimental setup, confocal Z-stacks were used to acquire information regarding the 3D spatial organization of the cells during this process (Video 1).
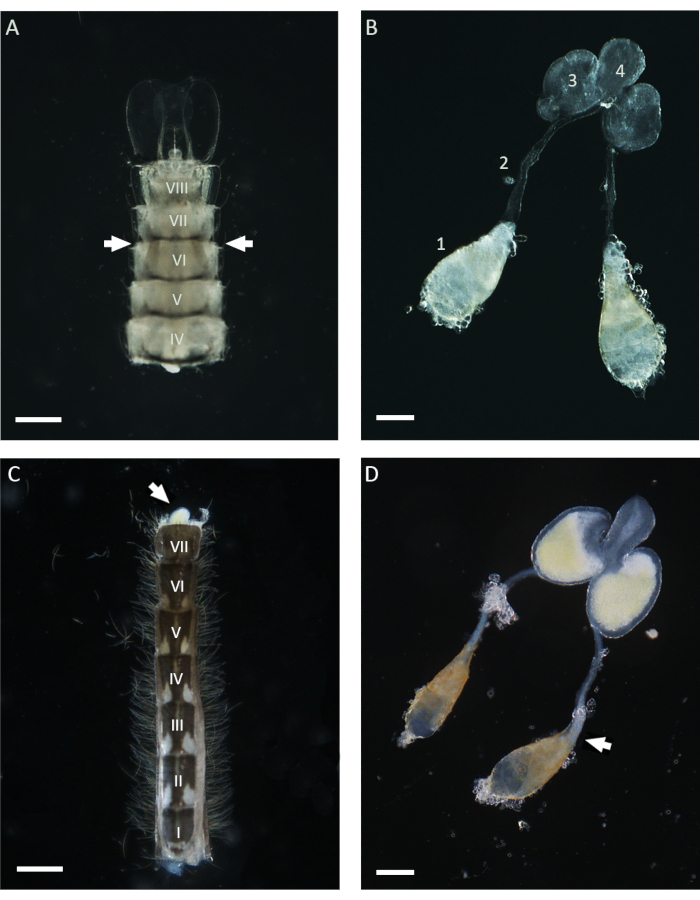
Figure 1: Testes dissected from pupae and 1 day old adult Anopheles gambiae males. (A) An abdomen dissected at the late pupal stage showing the position of the testes in the proximity of the sixth abdominal segment. The testes can be identified throughout the cuticle and appear as brownish structures on both sides of the abdomen (with arrows). (B) Testes dissected at the pupal stage, showing the mature testes (1), vas deferens (2), MAGs (3), and ejaculatory duct (4). (C) An abdomen dissected from a 1 day old adult male after removing the basal clasp segment. The MAGs can be squished from the abdomen by applying gentle pressure (white arrow). (D) Male internal reproductive apparatus dissected from a 1 day old adult male. The white arrow indicates the position occupied by mature sperms, which appear as a white aggregate at the basal pole of the testis. Scale bars: (A,C) 200 µm; (B,D) 100 µm. The Roman numerals from I to VIII indicate the abdominal segments. Please click here to view a larger version of this figure.
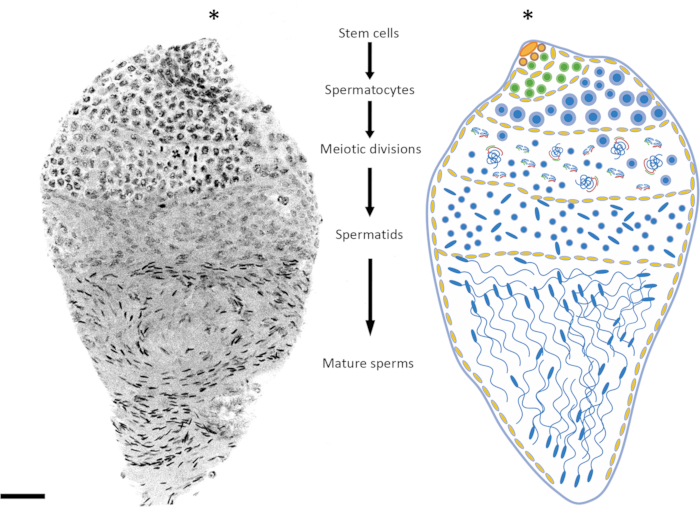
Figure 2: Representation of spermatogenesis in Anopheles gambiae. The image on the left shows an An. gambiae late pupa testis following whole-mount DAPI staining. On the right is a schematic version for better visualization. By observing the nuclear shape and condensation level, it is relatively easy to follow all the spermatogenesis stages from diploid cells to haploid spermatozoa. The stem cell niche is situated in the upper pole of the organ, where differentiation into spermatogonia starts. The spermatogonia cells increase in number after mitotic division (green cells), and the spermatocysts increase in size (yellow cells). The spermatogonia cells differentiate into spermatocytes after multiple rounds of mitotic divisions (blue cells). The spermatocytes, which are characterized by relatively larger nuclei than the cells of the other stages of the process, are the cells that will undergo meiotic division. Cells undergoing meiosis can be detected by looking at the presence of chromosomes at different meiotic stages; chiasmata and metaphase chromosomes can be detected even at low magnification. The premeiotic stages are overrepresented in testes dissected at the early pupal stage. After the first and second meiotic divisions, spermatids are produced and can usually be found lying in the middle of the testis. The nuclei of spermatids show a certain degree of variation in their shape, from a round to an arrow-like shape. The spermatids enter the spermiogenesis process, during which the nuclei start condensing, and their structure changes into arrow-like dots. When mosquitoes mature sexually after emerging, spermatocysts containing mature sperms can occupy most of the testis volume at the expense of spermatocysts at a different stage of development. Scale bar: 20 µm. The asterisk (*) indicates the apical part of the testis. Please click here to view a larger version of this figure.
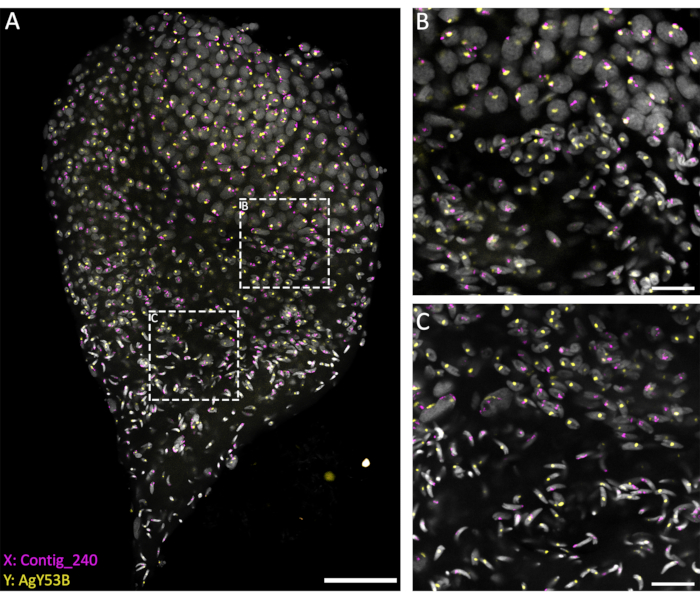
Figure 3: WFISH on an An. gambiae testis dissected from the late pupal stage. WFISH was performed using probes specific for the X (Contig_240) and Y (oligo probe specific for AgY53B) chromosomes. (A) WFISH allows to follow the behavior of the sex chromosome during spermatogenesis from diploid cells to haploid spermatozoa. In this image, it is possible to appreciate the dramatic changes that the nuclei undergo during spermatogenesis. Labeling the sex chromosomes allows for the discrimination between diploid and haploid cells. In diploid cells, the signal from the sex chromosomes is linked to the same nuclei. In haploid cells (spermatids and spermatozoa), the signal of the sex chromosomes is unlinked due to the meiotic reductional division. (B,C) A higher-magnification (63x) image of the testis shown in (A). They were acquired at different positions along the Z-axis. The white dotted frames indicate the area of acquisition. (B) The transition stage between spermatocytes and spermatids, showing the formation of haploid cells and the separation of the signals from the sex chromosomes into separate nuclei. (C) The transition stage between haploid spermatids and mature spermatozoa. This stage shows the changes in the nuclear condensation level; mature spermatozoa show a more condensed and elongated shape than spermatids. Scale bars: (A), 30 µm; (B,C), 10 µm; Gray: DAPI. Please click here to view a larger version of this figure.
| Target sequence | Primer Sequence and Oligo-probe consensus | Reference |
| Contig_240 (X) | 5’-CAATAAATTTCCTTTTTAATGATGC AAAATCTACGTCTCTAGC-3’-[Fluorochrome] |
19 |
| AgY53B (Y) | 5’AGAAGAATAGAATCAGAATAGTCGG TTTCTTCATCCTGAAAGCC-3’-[Fluorochrome] |
This study |
| AgY477- AgY53B junction region (Y) |
5’-TTCTAAGTTTCTAGGCTTTAAGGAT GAAGAAACCGACTATTC-3’-[Fluorochrome] |
19 |
| 18S rDNA (X) | F: AACTGTGGAAAAGCCAGAGC R: TCCACTTGATCCTTGCAAAA |
19 |
| AgY53B (Y) | F: CCTTTAAACACATGCTCAAATT R: GTTTCTTCATCCTTAAAGCCTAG |
19 |
Table 1: List of oligo probes specific to the X or Y chromosome in An. gambiae.
Video 1: A 3D stack on WFISH performed on an An. gambiae testis dissected from the late pupal stage. To obtain a 3D representation of the spermatogenesis process, a confocal 3D stack can be performed on testes showing a low number of structural alterations. In this study, the stacks were performed with an interval of 1.25 µm between two optical sections under a 63x or 40x oil lens in order not to lose information about the 3D spatial organization of the cells. Gray: DAPI, yellow: Contig_240 (X), magenta: AgY53B (Y). Please click here to download this Video.
Discussion
Commonly, FISH protocols require the squashing of the organ of interest to allow for chromosome staining. This causes a loss of information regarding the spatial arrangement of the cells within that organ33. This protocol describes how biological processes, such as spermatogenesis, can be studied in situ while maintaining the intact native structure of the testis and its internal cytological organization. Probes targeting different DNA repetitive elements, which are particularly enriched in sex chromosomes20, can be simultaneously used to reveal the dynamics of sperm maturation. Depending on the timing of the testis dissection, WFISH offers the opportunity to study different stages of spermatogenesis through mosquito development. WFISH is useful for studying specific phenomena such as hybrid incompatibility, which, in Anopheles mosquitoes, is due to the presence of meiotic defects such as premeiotic failure and sex chromosome non-disjunction19,34,35. Besides the biological aspect, spermatogenesis is the target of a number of genetic strategies developed to control pest insects such as Anopheles mosquitoes. In this context, the X-linked rDNA locus of An. gambiae has been used as a target to develop a synthetic sex ratio distorter, which, by damaging X-bearing sperms, biases the progeny toward males4,8,13.
This technology mirrors the action of natural sex ratio meiotic drives that have been identified in several taxa, including mosquitoes, but that still remain poorly understood28,36,37,38,39,40,41. WFISH offers the opportunity to investigate this phenomenon and paves the way for refining or improving sex distortion-based genetic strategies by, for example, providing information on how the cytology of sperm production is affected by the choice of the target sites used for sex chromosome shredding. Although, in our experience, WFISH shows high chances of success, failure can still occur. This might be due to an inefficient level of tissue permeabilization, which can be overcome by increasing the incubation time of the penetrating solution. Alternatively, Proteinase K can be used during the permeabilization step. In some cases, we noticed a non-uniform level of probe penetration, with a higher signal in spermatocytes nuclei and a lower or absent signal in the meiotic and spermiogenesis stages. This might be due to a difference in the permeabilization level depending on the cell stage. In addition, WFISH proved to be valuable when using fluorescent probes designed to target DNA sequences present in high copy numbers. When targeting single-copy genes, the signal detection may not be sufficient. In this case, methods for signal amplification, such as tyramide signal amplification (TSA), must be integrated42.
This protocol could be coupled with immunostaining or with transgenic reporter strains harboring germline-specific fluorescent markers16,18, as this would add information about protein localization and gene expression in situ. In this work, WFISH is described as a technique to investigate spermatogenesis in Anopheles mosquitoes; however, given the shared anatomy of male reproductive organs, this protocol could be applied to other mosquito species that play a role in disease transmission. Similarly, female gametogenesis could be investigated using this technique. In addition, cytological studies in organs or tissues of interest, such as the mosquito midgut, which is a target for parasite invasion, or atypical genetic backgrounds, such as those in hybrid mosquitoes, could be explored43. Moreover, this technique can potentially be transferred to other organisms within the Diptera order.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by a grant from the Bill & Melinda Gates Foundation and Open Philanthropy. We thank the Facility for Imaging by Light Microscopy (FILM) at Imperial College London for the microscopy analysis. Figure 2 was created with Biorender.com.
Materials
| Amersham CyDye Fluorescent Nucleotides, Cy3-dUTP | Cytiva | PA53022 | |
| Amersham CyDye Fluorescent Nucleotides, Cy5-dUTP | Cytiva | PA55022 | |
| ART Wide Bore Filtered Pipette Tips | ThermoFisher Scientific | 2079GPK | |
| CytoBond Removable Coverslip Sealant | SciGene | 2020-00-1 | |
| Dextran sulfate sodium salt from Leuconostoc spp. | Sigma-Aldrich | D8906-5G | |
| DNeasy Blood & Tissue Kits | Qiagen | 69504 | |
| Embryo Dishes | VWR | 70543-30 | |
| Ethanol, molecular grade | Sigma-Aldrich | 51976 | |
| Formamide | ThermoFisher Scientific | 17899 | |
| GoTaq G2 DNA Polymerase | Promega | M7841 | |
| Hydrochloric acid, 37% | Sigma-Aldrich | 320331 | |
| Microscope slides, SuperFrost | VWR | 631-0114 | |
| PBS (10x), pH 7.4 | ThermoFisher Scientific | 70011044 | |
| Pierce 16% Formaldehyde (w/v), Methanol-free | ThermoFisher Scientific | 28906 | |
| ProLong Gold Antifade Mountant with DAPI | ThermoFisher Scientific | P36941 | |
| RNase A/T1 Mix | ThermoFisher Scientific | EN0551 | |
| Set of dATP, dCTP, dGTP, dTTP | Promega | U1330 | |
| Sodium Acetate Solution | ThermoFisher Scientific | R1181 | |
| SP8 inverted confocal microscope | Leica | ||
| Triton X-100 | Sigma-Aldrich | 9036-19-5 | |
| TWEEN 20 | Sigma-Aldrich | P1379 | |
| UltraPure Salmon Sperm DNA Solution | ThermoFisher Scientific | 15632011 | |
| UltraPure SSC 20x | ThermoFisher Scientific | 15557044 | |
| Primer sequences | |||
| 5’-CAATAAATTTCCTTTTTAATGATGC AAAATCTACGTCTCTAGC-3’-[Fluorochrome] |
Eurofins Genomics | Contig_240 (X) | |
| 5’AGAAGAATAGAATCAGAATAGT CGG TTTCTTCATCCTGAAAGCC-3’-[Fluorochrome] |
Eurofins Genomics | AgY53B (Y) | |
| 5’-TTCTAAGTTTCTAGGCTTTAAGGA T GAAGAAACCGACTATTC-3’-[Fluorochrome] |
Eurofins Genomics | AgY477- AgY53B junction region (Y) |
|
| F: AACTGTGGAAAAGCCAGAGC R: TCCACTTGATCCTTGCAAAA |
Eurofins Genomics | 18S rDNA (X) | |
| F: CCTTTAAACACATGCTCAAATT R: GTTTCTTCATCCTTAAAGCCTAG |
Eurofins Genomics | AgY53B (Y) |
References
- World Malaria Report. World Health Organization Available from: https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2022 (2022)
- Bhatt, S., et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature. 526 (7572), 207-211 (2015).
- Hammond, A., et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nature Biotechnology. 34 (1), 78-83 (2016).
- Galizi, R., et al. A synthetic sex ratio distortion system for the control of the human malaria mosquito. Nature Communications. 5 (1), 3977 (2014).
- Kyrou, K., et al. A CRISPR-Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nature Biotechnology. 36 (11), 1062-1066 (2018).
- Simoni, A., et al. A male-biased sex-distorter gene drive for the human malaria vector Anopheles gambiae. Nature Biotechnology. 38 (9), 1054-1060 (2020).
- Gantz, V. M., et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proceedings of the National Academy of Sciences of the United States of America. 112 (49), E6736-E6743 (2015).
- Bernardini, F., Kriezis, A., Galizi, R., Nolan, T., Crisanti, A. Introgression of a synthetic sex ratio distortion system from Anopheles gambiae into Anopheles arabiensis. Scientific Reports. 9 (1), 5158 (2019).
- Hammond, A. M., Galizi, R. Gene drives to fight malaria: Current state and future directions. Pathogens and Global Health. 111 (8), 412-423 (2017).
- Garrood, W. T., et al. Driving down malaria transmission with engineered gene drives. Frontiers in Genetics. 13, 891218 (2022).
- Hoermann, A., et al. Gene drive mosquitoes can aid malaria elimination by retarding Plasmodium sporogonic development. Science Advances. 8 (38), (2022).
- Nash, A., et al. Integral gene drives for population replacement. Biology Open. 8 (1), (2019).
- Galizi, R., et al. A CRISPR-Cas9 sex-ratio distortion system for genetic control. Scientific Reports. 6 (1), 31139 (2016).
- Terradas, G., Hermann, A., James, A. A., McGinnis, W., Bier, E. High-resolution in situ analysis of Cas9 germline transcript distributions in gene-drive Anopheles mosquitoes. G3: Genes, Genomes, Genetics. 12 (1), (2022).
- Durant, A. C., Donini, A. Ammonium transporter expression in sperm of the disease vector Aedes aegypti mosquito influences male fertility. Proceedings of the National Academy of Sciences of the United States of America. 117 (47), 29712-29719 (2020).
- Taxiarchi, C., et al. High-resolution transcriptional profiling of Anopheles gambiae spermatogenesis reveals mechanisms of sex chromosome regulation. Scientific Reports. 9 (1), 14841 (2019).
- Pompon, J., Levashina, E. A. A new role of the mosquito complement-like cascade in male fertility in Anopheles gambiae. PLoS Biology. 13 (9), e1002255 (2015).
- Papathanos, P. A., Windbichler, N., Menichelli, M., Burt, A., Crisanti, A. The vasa regulatory region mediates germline expression and maternal transmission of proteins in the malaria mosquito Anopheles gambiae: A versatile tool for genetic control strategies. BMC Molecular Biology. 10 (1), 13 (2009).
- Liang, J., Sharakhov, I. V. Premeiotic and meiotic failures lead to hybrid male sterility in the Anopheles gambiae complex. Proceedings of the Royal Society B. 286 (1906), 20191080 (2019).
- Hall, A. B., et al. Radical remodeling of the Y chromosome in a recent radiation of malaria mosquitoes. Proceedings of the National Academy of Sciences of the United States of America. 113 (15), E2114-E2123 (2016).
- Clements, A. N. . The Biology of Mosquitoes. Volume 1: Development, Nutrition and Reproduction. , (1992).
- Demarco, R. S., Eikenes, &. #. 1. 9. 7. ;. H., Haglund, K., Jones, D. L. Investigating spermatogenesis in Drosophila melanogaster. Methods. 68 (1), 218-227 (2014).
- Helinski, M. E., Parker, A. G., Knols, B. G. Radiation biology of mosquitoes. Malaria Journal. 8, S6 (2009).
- Huho, B. J., et al. A reliable morphological method to assess the age of male Anopheles gambiae. Malaria Journal. 5 (1), 62 (2006).
- Fabian, L., Brill, J. A. Drosophila spermiogenesis. Spermatogenesis. 2 (3), 197-212 (2012).
- Li, M., et al. Suppressing mosquito populations with precision guided sterile males. Nature Communications. 12 (1), 5374 (2021).
- Thailayil, J., Magnusson, K., Godfray, H. C., Crisanti, A., Catteruccia, F. Spermless males elicit large-scale female responses to mating in the malaria mosquito Anopheles gambiae. Proceedings of the National Academy of Sciences of the United States of America. 108 (33), 13677-13681 (2011).
- Haghighat-Khah, R. E., et al. Cellular mechanisms regulating synthetic sex ratio distortion in the Anopheles gambiae germline. Pathogens and Global Health. 114 (7), 370-378 (2020).
- Yamamoto, D. S., et al. A synthetic male-specific sterilization system using the mammalian pro-apoptotic factor in a malaria vector mosquito. Scientific Reports. 9 (1), 8160 (2019).
- Papathanos, P. A., Windbichler, N. Redkmer: An assembly-free pipeline for the identification of abundant and specific X-chromosome target sequences for X-shredding by CRISPR endonucleases. The CRISPR Journal. 1 (1), 88-98 (2018).
- Sharma, A., Kinney, N. A., Timoshevskiy, V. A., Sharakhova, M. V., Sharakhov, I. V. Structural variation of the X chromosome heterochromatin in the Anopheles gambiae complex. Genes. 11 (3), 327 (2020).
- Krzywinski, J., Sangaré, D., Besansky, N. J. Satellite DNA from the Y chromosome of the malaria vector Anopheles gambiae. Genetics. 169 (1), 185-196 (2005).
- Timoshevskiy, V. A., Sharma, A., Sharakhov, I. V., Sharakhova, M. V. Fluorescent in situ hybridization on mitotic chromosomes of mosquitoes. Journal of Visualized Experiments. (67), e4215 (2012).
- Liang, J., Hodge, J. M., Sharakhov, I. V. Asymmetric phenotypes of sterile hybrid males from reciprocal crosses between species of the Anopheles gambiae complex. Frontiers in Ecology and Evolution. 9, 660207 (2021).
- Slotman, M., Torre, A. D., Powell, J. R. The genetics of inviability and male sterility in hybrids between Anopheles gambiae and An. arabiensis. Genetics. 167 (1), 275-287 (2004).
- Wood, R. J., Newton, M. E. Sex-ratio distortion caused by meiotic drive in mosquitoes. The American Naturalist. 137 (3), 379-391 (1991).
- Cazemajor, M., Joly, D., Montchamp-Moreau, C. Sex-ratio meiotic drive in Drosophila simulans is related to equational nondisjunction of the Y chromosome. Genetics. 154 (1), 229-236 (2000).
- Jaenike, J. Sex chromosome meiotic drive. Annual Review of Ecology and Systematics. 32 (1), 25-49 (2001).
- Courret, C., Chang, C. H., Wei, K. H., Montchamp-Moreau, C., Larracuente, A. M. Meiotic drive mechanisms: Lessons from Drosophila. Proceedings of the Royal Society B. 286 (1913), 20191430 (2019).
- Zanders, S. E., Unckless, R. L. Fertility costs of meiotic drivers. Current Biology. 29 (11), R512-R520 (2019).
- Newton, M. E., Wood, R. J., Southern, D. I. A cytogenetic analysis of meiotic drive in the mosquito, Aedes aegypti (L.). Genetica. 46 (3), 297-318 (1976).
- Carabajal Paladino, L. Z., Nguyen, P., Šíchová, J., Marec, F. Mapping of single-copy genes by TSA-FISH in the codling moth, Cydia pomonella. BMC Genomic Data. 15, S15 (2014).
- Bernardini, F., et al. Cross-species Y chromosome function between malaria vectors of the Anopheles gambiae species complex. Genetics. 207 (2), 729-740 (2017).

