Maintaining and Assessing Various Tissue and Cell Types of the Eye Using a Novel Pumpless Fluidics System
Summary
Real-time analysis of live tissue yields important functional and mechanistic data. This paper describes the protocols and critical variables to ensure accurate and reproducible generation of data by a novel and pump-free multi-channel fluidics system that maintains and assesses a wide range of tissue and cell models.
Abstract
Many in vitro models used to investigate tissue function and cell biology require a flow of media to provide adequate oxygenation and optimal cell conditions required for the maintenance of function and viability. Toward this end, we have developed a multi-channel flow culture system to maintain tissue and cells in culture and continuously assess function and viability by either in-line sensors and/or collection of outflow fractions. The system combines 8-channel, continuous optical sensing of oxygen consumption rate with a built-in fraction collector to simultaneously measure production rates of metabolites and hormone secretion. Although it is able to maintain and assess a wide range of tissue and cell models, including islets, muscle, and hypothalamus, here we describe its operating principles and the experimental preparations/protocols that we have used to investigate bioenergetic regulation of isolated mouse retina, mouse retinal pigment epithelium (RPE)-choroid-sclera, and cultured human RPE cells. Innovations in the design of the system, such as pumpless fluid flow, have produced a greatly simplified operation of a multi-channel flow system. Videos and images are shown that illustrate how to assemble, prepare the instrument for an experiment, and load the different tissue/cell models into the perifusion chambers. In addition, guidelines for selecting conditions for protocol- and tissue-specific experiments are delineated and discussed, including setting the correct flow rate to tissue ratio to obtain consistent and stable culture conditions and accurate determinations of consumption and production rates. The combination of optimal tissue maintenance and real-time assessment of multiple parameters yields highly informative data sets that will have great utility for research in the physiology of the eye and drug discovery for the treatment of impaired vision.
Introduction
Perifusion systems have a long history in the life sciences. In particular, for the study of secretory function by islets, they have been used to characterize the kinetics of insulin secretion in response to secretagogues1. In addition to collecting outflow fractions for subsequent assay of hormones and metabolites, real-time sensors have been incorporated, predominantly for the detection of oxygen consumption2,3,4. Widespread efforts to better understand mechanisms mediating diseases of the eye have been limited by a lack of physiologically relevant methods to assess metabolic regulation and dysregulation of the various isolated components of the eye, including the retina, retinal pigment epithelial (RPE)-choroid-sclera and cultured RPE cells. Static systems designed for cultured cells have been adapted for tissue5, but tissue requires flow for adequate oxygenation. Flow systems have been successful at accurately and reproducibly measuring real-time responses in oxygen consumption rate (OCR) by the retina and RPE-choroid-sclera, and the tissues stay metabolically stable for more than 8 h allowing highly informative protocols involving multiple test compounds4,6,7,8,9. Nonetheless, the operation of fluidics systems has historically required a custom-made apparatus and trained technical staff in non-standardized methodologies. Such systems have not been adopted as the standard methodology in most laboratories. The BaroFuse is a newly developed fluidics system that does not rely on pumps, but rather on gas pressure to drive flow through multiple channels and tissue chambers (Figure 1). Each channel is continuously monitored for OCR, and the outflow is collected with a plate-based fraction collector for subsequent assay of contents. Importantly, the tissue perifusion chambers for the instrument are designed to accommodate tissues of various geometries and sizes.
The heart of the instrument is the fluidics system, where flow is driven from a sealed, pressurized reservoir through small inner diameter (ID) tubing (contributing the most significant flow resistance in the fluid circuit) up into the glass tissue chambers that house the tissue. Pressure to the media reservoir module (MRM) is supplied by low-pressure and high-pressure regulators connected to a gas cylinder containing a mixture of gases (typically 21% O2, 5% CO2, balance N2), and the reservoir is sealed from the top by the perifusion chamber module (PCM) that holds the tissue chamber assemblies (TCAs). Flow rate is controlled by the length and ID of the resistance tubes and the pressure setting of a low-pressure regulator. Outflow tubes connected to the top of tissue chambers deliver fluid to either a waste receptacle (that is continuously weighed for automatic determination of flow rate) or into wells of a 96-well plate controlled by the fraction collector. The O2 detection system measures the lifetime of an O2-sensitive dye painted on the inside of each of the glass tissue chambers downstream of the tissue. This information is then used to continuously calculate OCR. The entire fluidics system resides in a temperature-controlled enclosure and the gas tank, fraction collector and computer are the major components of the instrument (Figure 2A). Finally, software that runs the instrument serves to control its operation (including the preparation and timing of injected test compounds, flow measurement system, and fraction collector timing), as well as processing and graphing the OCR data and other supplemental measurements.
In this paper we describe the protocols for using the fluidics system to perifuse and assess OCR and lactate production rate (LPR) for various isolated components of the eye. LPR is a parameter reflecting glycolytic rate that is highly complementary to OCR, where the pair accounts for the two major branches of energy generation from carbohydrates in the cell10. As preparation of the tissue and loading it into the tissue chambers is best learned by watching the procedure, the video will help illustrate several of the critical steps that are performed during set up and operation that are not easily conveyed by text alone.
The description of the protocol is divided up into 8 sections that correspond to different phases of the experiment (Figure 2B): 1. pre-experimental preparation; 2. preparation/equilibration of the perifusate; 3. instrument set up; 4. tissue equilibration; 5. experimental protocol; 6. instrument break down; 7. data processing; and 8. assays of outflow fractions.
Protocol
All procedures for harvesting tissue from rats and mice were approved by the University of Washington Institutional Animal Care and Use Committee.
1. Pre-experimental preparation
NOTE: The following tasks are completed at least a day in advance of the experiment.
- Designing the experimental protocol
- Assignment of tissue placement in channels: Choose tissue or cell model to be placed in 3 of the 4 channels on each side of the MRM. One tissue chamber on each side is run without tissue to be used for baseline correction.
- Arrange the samples using one of the two typical designs – different test compound protocols on each side (e.g., channels on one side of the MRM receive test compounds while the channels on the other side act as a control); same test compound injection protocol on both sides of the MRM, but different tissues or tissue model versus control on either side of the MRM.
- Selection of flow rate and tissue amount for optimal OCR measurement: Adjust the flow rate until the change in lifetime ratio times 100 is approximately 3.
NOTE: Typical amounts of tissue and corresponding flow rates are shown in Table 1 for components of the eye, where the instrument functions best at flow rates between 6-80 µL/min/channel. - Calculation of required media/buffer volume: Calculate the volume of media to be added to each MRM insert at the beginning of the experiment as
VolumeMRM = 30 mL + Duration of Protocol (in min) x Flow Rate (in mL/min) x 4 channels (Eq.1)
For example, at 0.01 mL/min, a 60 mL starting VolumeMRM will allow for a 12.5 h protocol (where 30 mL will be depleted, while 30 mL will be remaining), whereas at 0.04 mL/min, 90 mL starting VolumeMRM will allow for a 6 h protocol (with 30 mL remaining). - Test compounds injection protocol: Select test compounds to assess, the concentration to be tested (typically chosen to yield near maximal response or as concentration dependencies) and the duration of exposure. Consider solubility and make up stocks in desired solvent such as water, DMSO or ethanol.
- Select the timing of injections and subsequent injections so that the response reaches a steady state prior to adding a subsequent agent. When repeating protocols, match the timing of injections so that multiple time courses can be averaged.
NOTE: The compounds used here are from a previous mitochondrial (Mito) stress test 11 and both oligomycin and carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) require DMSO both in the stock solutions, as well as the final perifusate. - Outflow sampling times: Select the desired fraction collection intervals (from 1-60 min/sample), where faster sampling rates are chosen for rapid changes, and longer time intervals are chosen as steady state is approached. Use adequate well volumes (0.3 to 1.5 mL) to avoid overflow during the sampling interval (choose volumes that are greater than flow rate x the time interval).
NOTE: Sampling times will vary with choice of protocol, but for a Mito stress test, we have commonly used 5 min intervals during the baseline and 15 min intervals during the injections (-15, -10, -5, 0, 15, 30, 45, 60, 75, 90, 105, where each time is the start of the sampling interval). - Enter the selected values for test compounds and fraction collection described above into the user interface (UI), which generates graphical representations of this information. Export and disseminate files for group evaluation and discussions (Supplementary Figure 1).
- Set out the accessories and consumable parts
- Set out the supplies provided and pre-packaged aseptically by the manufacturer which include: TCAs (pack of 8), outflow tubing assemblies (pack of 8 tubes), test compound injection tubing (2), forceps, tubing clamps (3), MRM, MRM inserts (2), stir bars (2), and purge tubing assembly (placed in a biological safety cabinet).
- Do not reuse disposable parts that come in contact with liquid, as this will lead to an increase in experimental failure. Reuse the forceps and stir bars by cleaning and autoclaving them between experiments.
2. Preparation and equilibration of perifusate (Time: 30 min not including incubation time)
- Prepare the media or Krebs-Ringer bicarbonate buffer (KRB) the day before based on calculations from equation 1, typically 200 mL, and then incubate overnight in a 39 °C/5% CO2 incubator in T225 tissue culture flasks with no more than 90 mL in each flask.
- If using commercially prepared KRB or media (warmed to room temperature), prepare the perifusate on the morning of the experiment and place in the 5% CO2 incubator for at least 1 h. Prepare all solutions aseptically.
NOTE: All liquids and parts of the fluidics system that come in contact with liquid are sterile at the outset of the experiment. However, assembly of the system and loading of the tissue is performed open to the air.
3. Equilibration of temperature and dissolved gas to set up the instrument (Time: 75 min)
- Attachment of tubing assemblies to the MRM
- Place the MRM and a fluidics package onto the bench next to the instrument. Ensure the tubing clamps (3), stir bars (2), and forceps are already on the tool tray.
- Place an unused MRM insert with a stir bar in each side of the MRM (see Figure 3).
- Attach the TCAs to injection ports on both ends of the MRM so that the end of the tubing is positioned directly over the stir bar. Ensure that the longer of the two test compound injection assemblies is on the back of the MRM.
- Next, attach the gas inflow feed line and purge tubing assembly to the back and front vacant ports, respectively (see Figure 4A).
- Placing the MRM/tubing assemblies in the enclosure
- Place the MRM (with the tubing assemblies attached) into the MRM heater (Figure 4B).
- Position the four tubing assemblies into the grooves of the walls in the base of the enclosure (two on each side) so they will protrude through to the exterior of the enclosure once the middle enclosure is put in place.
- Secure the MRM between the clamps by tightening the two wheels on the detector stand.
- Feed the longer test compound injection assembly protruding from the back of the enclosure through the two tube guides on the side of the enclosure so the opening of the tubes faces forward (Figure 4C).
- Clamp each of the closed test compound injection assemblies.
- Assembling the enclosure and activating the temperature controllers
- Toggle the power strip that supplies power to all of the electrical devices within the enclosure to the ON position. The fan on the detector stand will turn ON and the MRM temperature controller should light up displaying a set value of 38 °C (Figure 5).
- Turn ON the stirrers to 70 rpm using the UI to ensure that the stir bars are rotating smoothly. Once proper stirring is observed, turn off the stirrers.
- Place the middle section of the enclosure on top of the base.
- Connect the cable on the middle section of the enclosure to the cable from the electrical box to engage power to the ambient temperature controller lever switch and supply power to the ambient temperature heater.
- Place the lid on the enclosure and the display of the upper temperature controller (the ambient temperature controller) will light up and read 36 °C. Start a timer for 30 min, the time it takes for the MRM heater to reach set point temperature.
- Inserting the TCAs into the PCM
- Use the TCA insertion tool to insert each of the 8 TCAs into the PCM holes by firmly pressing on the adaptor with the face of the insertion tool until the top of the tubing sleeve wrapped around the tissue chamber touches the surface circumscribing the holes in the PCM.
- Completely insert one TCA before inserting the next one. Set the partially assembled PCM aside next to the PCM brace and 6 screws.
NOTE: Incomplete insertion of a TCA will prevent the head space from reaching the pressure set point and perifusate will not flow.
- Filling the two inserts in the MRM with pre-equilibrated perifusate
- To do this, 30 min after the enclosure was assembled and the MRM has reached temperature, transfer the pre-equilibrated perifusate into the pre-heated MRM insert by gently dispensing the liquid down the sides using a 50 mL pipette.
NOTE: These steps as well as the ones in section 3.6, should be carried out immediately to avoid transfer of gas between the perifusate in the MRM and the atmosphere.
- To do this, 30 min after the enclosure was assembled and the MRM has reached temperature, transfer the pre-equilibrated perifusate into the pre-heated MRM insert by gently dispensing the liquid down the sides using a 50 mL pipette.
- Assembling the MRM/PCM to create a gas-tight seal and positioning the O2 detector
- Place the PCM onto the MRM by inserting the resistance tubes of the TCAs emanating from the bottom of the PCM into the MRM inserts, 4 on each side of the MRM Divider. Orient the PCM so that the tissue chambers can rest against the O2 detector once it is positioned.
- Secure the PCM and the PCM Support Brace with the 6 screws using the electric screwdriver.
- Secure the TCAs within the PCM support fins with the elastic band provided by stretching it around the fins of the PCM at the level of the rubber gaskets (Figure 6).
- Position the O2 Detector on the detector stand so that the face of it rests against the fins of the PCM. Check that the LED/photodetector pairs line up with the O2-sensitive dye in the tissue chambers. If needed, adjust the O2 detector lateral guides after loosening the set screws on the side of the O2 detector holder.
- Place the lid on top of the Enclosure.
- Equilibrating the gas in the head space in the MRM with the perifusate
- With the high-pressure valve fully secured and closed, open the gas tank valve by turning the cylinder valve on top of the tank counterclockwise.
- Adjust the high-pressure regulator to a pressure of 10 psi using the knob on the regulator.
- Pressurize the MRM by setting the low-pressure regulator to 1.0 psi (Figure 7A).
- Unclamp the purge tube (Figure 7B) to allow the gas from the tank to replace the air in the MRM headspace (the test compound injectors remain clamped) for 15 min. Confirm gas flow by submerging the end of the purge tube into a beaker of water to observe bubbling.
- Once flow has been confirmed, start the O2 detector as described below in section 3.8.
- After 15 min, turn the stirrer on at 70 rpm and leave running for the rest of the experiment. After an additional 15 min, clamp the purge tubing assembly (Figure 7C).
- Lower the pressure on the low-pressure regulator to the operating pressure that achieves the desired liquid flow rate (as specified by the experimental pack – usually between about 0.5-0.7 psi). If the flow rate is above 20 µL/min temporarily set the pressure to 0.3 psi to allow time to load the tissue without the chambers overflowing. This is not necessary if the tissue is loaded within 15 min of the clamp being placed.
NOTE: Do not let buffer run down the outside of the tissue chamber, since the fluid can interfere with O2 sensing.
- Starting the O2 detector
- Activate the O2 detector software on the laptop by clicking on the icon labeled Oxygen Detector.
- Once the program opens (Supplementary Figure 2), confirm the correct COM port is selected. If needed, the COM port may be identified by unplugging and plugging the O2 detector from the computer so that the port number is displayed. If the COM port is unplugged while the application is running, then the application must be closed and reopened before use.
- Click Start then Record (and save the data in the backup folder). Next, click Graph.
- Change the average value on the bottom left of the lifetime graph to 5 (which instructs the program to calculate a moving average with 5 consecutive points). After a minute has passed and the first data point is displayed on the graphing screen, click Auto Scale.
4. Tissue loading and equilibration period (Time: 90 min)
- Positioning the frits in the tissue chambers
- Remove the lid and middle sections of the enclosure.
- After the perifusate in the tissue chambers have risen above the top of the pre-positioned frit, push the frit down with the frit cue by lightly tapping on the top of the frit to remove any air bubbles that formed below or within the frit.
- Position frits about 0.25 inch above the bottom of the tissue chamber.
- Loading tissue into the tissue chambers
- Once the level of media is 0.5 inch from the top, load the tissue into the chamber.
- Loading Retina or RPE-choroid-sclera: Harvest Retina or RPE-choroid-sclera as described in 6. To load the tissue, use fine point forceps to gently place the tissue into each chamber, being careful not to fold the tissue, while using a tissue wipe to prevent liquid from the tissue chamber dripping onto the O2 sensor. Observe tissue sinking toward and onto the frit.
NOTE: Between the time of harvesting the tissue and loading tissue into the chambers, ensure that trauma to the tissue is avoided by not leaving the tissue in a bicarbonate-based buffer/media out of the incubator for more than 10 min and ensuring the tissue is bathed in enough buffer/media (at least 1 mL/10 mg of tissue) to keep the tissue from becoming hypoxic and preventing exposure to air. - Loading RPE cells on transwell membranes: Prepare RPE cells as described previously 12 and in Supplementary File 1. Passage cells using 0.25% trypsin-EDTA and seed on polyethylene terephthalate, track-etched filters (cell culture inserts, pore size 0.4 mm) at a minimum of 2.0 x 105 cells/cm2. On the day of the experiment, cut the membranes into three strips of equal width and load with forceps into the tissue chambers (see Figure 8A).
- Attaching the outflow tubing assemblies to tissue chambers
- Remove the outflow tubing assemblies from the packaging and place the outflow tubing separator on the lip of the middle section of the enclosure so the outflow tubing adapters are on the inside of the enclosure (Figure 8B,C).
- Being careful not to push too hard on the TCAs (or they will come loose from the MRM), attach the outflow tubing adapters onto the top of the tissue chambers TCAs (Figure 8D). Replace the middle of the enclosure and reconnect the ambient temperature control cable.
- Prior to replacing the lid of the enclosure, confirm that the components of the fluidics system inside the enclosure including the O2 detector, PCM, tissue chambers, outflow tubes, MRM and heater, are all positioned properly as shown in Figure 8E.
- Replace the lid of the enclosure. Feed the eight outflow tubes through the fraction collector guide arm.
- Activating the fraction collector
- Make sure the fraction collector is centered relative to the right wall of the enclosure and the outflow tube holder: the left support of the fraction collector base should rest against the edge of the enclosure wall.
- On the laptop, click the UI shortcut and the experimental info page will open (Supplementary Figure 3 Top).
- Fill in the information in the appropriate boxes on the experimental info page (this can be done prior to the experiment start) and then click the Flow & Fraction Collector page on the top (Supplementary Figure 3 Bottom).
- Setting parameters for automated flow rate measurement
- Select the desired integration time in the sample acquisition time drop-down menu on the top middle that balances the desired accuracy (which is proportional to integration time) and the temporal resolution.
- If no outflow fractions will be collected in the experiment, then click Start and go to section 4.7. If outflow fractions will be collected, then carry out steps in section 4.6.
- Collecting outflow fractions
- In the UI software, check the Collect fractions? box on either the experiment info page or the flow & fraction collector page. Then click the Compute FC Settings button.
- When the new window opens, fill in the time of the first injection in the protocol (defined as time = 0) and the flow rate per channel, as well as the time intervals for each sample. Then, click Compute.
- Once the intervals of the collection are verified, click on Generate and Start.
- Measuring flow rates for individual channels (Optional)
- If flow rates of individual channels are to be measured (which under normal conditions vary by only a few percent), weigh eight (or less) microcentrifuge tubes, and record their weight.
- Set the tube holder containing the pre-weighed microcentrifuge tubes on the plate carriage. Click Measure Flow Rate Manually in the other utilities section.
- Select the measurement duration and then click Generate Template. Close the window and click Start. The fraction collector will collect fluid from the outflow tubes for the measurement duration and then the arm will return to its home position.
- Weigh the microfuge tubes after collection and use the difference in weight divided by the measurement duration to calculate the flow rate (where 1 mg = 1 µL).
- Baseline stabilization
- Once the tissue and/or cells have been loaded into the tissue chambers, allow the system to equilibrate for 90 min to establish a flat baseline of O2 consumption, at which time the first test compound can be injected (considered to be time = 0).
- At 30 min prior to the first injection, enter the average last 3 FR per channel value into the injection page for preparing test compound injections.
5. Experimental protocol (Time: 2-6 h)
NOTE: Once baseline stabilization is underway, the next tasks are injecting the test compounds and changing plates on the fraction collector if more than one will be used.
- Preparing test compound injectate
- Enter the names of the compounds, desired concentrations (final and stock solutions), and injection times of the test compounds into the table in the UI on the Injection page. Confirm the information displayed in the program including volumes in the MRM left at the time of the injections and how much stock solution to inject to achieve the desired concentrations (Supplementary Figure 4).
- To calculate how much perifusate and stock are needed for the injection, fill in the white boxes of the injection table and click Compute. Using the calculated values, dilute the test compound stock solution with perifusate prior to injection so that the volume injected is 5% of the volume in the MRM after injection.
- Prepare each test compound by mixing the stock solution and perifusate. Load the syringes for at least 10 min prior to the injection time and keep in a CO2 incubator (maintained at temperatures between 37-40 °C) until ready to inject.
- Injecting test compounds
- Connect the syringe containing the test compound to the injection lines (Figure 9A). Unclamp the soft-walled pharmed tubing leading to the injection line and slowly inject test compounds (Figure 9B) at a rate of approximately 3 mL/min. Re-clamp the tubing on the injection line and then remove the syringe (Figure 9C); repeat for the other side of the MRM.
- After injecting each test compound, prepare each subsequent test compound to be injected according to the UI injection page.
- Determining inflow O2 signal for each channel
- At the end of every experiment, inject the respiratory inhibitor KCN (3 mM) to determine the inflow lifetime signal for each channel which is used to correct for the variation in baseline sensor lifetime and non-mitochondrial consumption of oxygen.
6. Ending the experiment and breaking down the system (Time: 30 min)
- Saving oxygen data
- Click the Save button on the top left of the graphing window on the oxygen detector software; name the file and store it in the folder where the file will be kept. Click the Stop Recording button on the main window to save the backup file.
- Saving the UI experimental info file
- Click the Save profile button on the top left of the UI's general page; name the file and store it into the folder where the file will be kept. On the fraction & flow page of the UI, click the Tools drop down menu and then Save. Keep the generated name or choose another and store the file where desired. If necessary, there are backup files which can be accessed and saved.
- Breaking down the instrument
- Since KCN is volatile, dispose of fluidics assemblies in a fume hood; pour media from MRM and FC waste tray into a waste container, and rinse the MRM inserts and stir bars thoroughly with water. Pour the contents of the waste container (containing KCN) into a labeled chemical waste container for subsequent disposal by chemical safety. Prior to the next experiment, thoroughly clean and autoclave the stir bars and forceps.
7. Data processing (Time: 15-45 min)
- Open the data processor application on a Mac or PC computer. Select the .csv data file generated by an experiment. If this experiment's protocol is similar to a previously analyzed experiment, then select that settings file and click Next Step. Otherwise just click Next Step to begin entering the experiment's settings.
- Fill in the various settings of the experiment. On the determination of reference time point, select the time point directly before the KCN went into effect and the Lifetime values decreased. Select this point with the use of a slider or by typing the time value into the box.
- Click Calculate to generate OCR graphs according to equation 2:
OCR = ([O2]in– [O2]out) x FR = (217 nmol/mL – [O2]out) x FR/tissue basis (Eq. 2)
where O2 concentration in KRB when in equilibrium with 21% O2 at 37 °C is 217 nmol/mL, FR is the flow rate (in mL/min), tissue basis is the amount of tissue loaded into the chamber (i.e., number of retinas, RPE-choroid-sclera or RPE cells). - Save the graphs as .pdf files by pressing the Export Graph button. Graph OCR as either absolute values, or as a fraction of a steady state value by checking the boxes that correspond to the test compound to be set to 1 in the edit settings page.
8. Assays of outflow fractions
- If samples cannot be assayed immediately after the experiment, place the plates at 4 °C if assayed the next day, or frozen if stored longer. If plates are frozen, then thaw samples at 4 °C (so that samples remain cold).
- Once assays are performed on the selected outflow fractions, the columns of data (one per channel) are entered into a .csv file with time (in min) in the leftmost column, and concentration in the right (in either nmol/mL or ng/mL); a link in the data processing program when pressed uploads a template.
- Upload this file into the data processor to calculate and plot the data.
Representative Results
To illustrate the resolution of the data generated from isolated components of the eye, OCR and LPR was measured with three types of tissue (retina, RPE-choroid-sclera, and RPE cells) following a commonly used protocol (the mitochondrial stress test10; Figure 10, Figure 11, and Figure 12). The amount of tissue used for each tissue is shown in Table 1. Data was processed and graphed using the software package that was developed for the fluidics system. The preparation of retina and RPE-choroid-sclera is relatively straight-forward and takes less than 20 min for each tissue type. OCR was constant during the time that test compounds were injected, indicating stable health and function of the tissue and supporting the validity of the method (Figure 10). Once validated for each tissue type, we have not found it necessary to run controls where no test compounds are injected for each experiment. Consistent with data obtained using more conventional perifusion methods6,8,13, OCR decrease in response to oligomycin and increased OCR in response to FCCP. Changes in LPR were in the opposite direction of those observed for OCR: oligomycin increased LPR, which then decreased (but only slightly) in response to FCCP (Figure 11). To compare the statistical significance of the effect of each sequential test compound, t-tests were performed (which are calculated automatically by the software that comes with the instrument). Since the goal of the paper was to describe how to perform the method, the number of replicates carried was not always high enough to produce statistical significance. In general, though, when the number of replicates were 3 or more, effects of FCCP and oligomycin on both OCR and LPR were significant.
RPE cells have not been previously analyzed with flow systems but responded similarly to RPE-choroid-sclera (consistent with the view that a large fraction of OCR is due to RPE cells; Figure 11). These illustrative examples highlight ability of the system to maintain tissue viability as reflected by the stability of OCR in the control channels, and the high signal to noise ratio for changes in OCR of the magnitude induced by oligomycin and FCCP, which was more than 100 to 1. In addition, assays of outflow fractions can be used to correlate the rate of uptake or production of a wide array of compounds exchanging with the extracellular fluid are complementary to OCR (in this case, LPR). These features of the instrument allowed accurate quantification of characteristic differences in tissue responses between tissue types performed in parallel. OCR by RPE-choroid-sclera and RPE cells are consistently more sensitive to oligomycin than retina (Figure 11 and Figure 12), although for the RPE-choroid-sclera the duration of exposure to FCCP was not long enough to reach steady state. A point to consider arose when using DMSO as a solvent. At higher concentrations, (0.2%) DMSO had a transient effect on OCR by retina (presumably reflecting an effect of a change in osmotic pressure brought about by DMSO's effect on membrane permeability).
Based on the assumption that KCN completely inhibits respiration by its direct action on cytochrome c oxidase, OCR at the end of the KCN exposure is set to 0 and all OCR values are calculated based on the change relative to the KCN value. OCR can occur independent of the respiratory chain and cytochrome c oxidase. However, the magnitude of this contribution to overall OCR is generally not more than a few percent (data not shown) and the extended length of time that tissue is exposed to KCN ensures that substrates of oxidases that are not part of the electron transport chain have been depleted.
Statistical analysis
Single experiments were shown as indicated in the figures, but with multiple channels that were averaged. Data was then graphed as the average ± the standard error (SE; calculated as SD/√n).
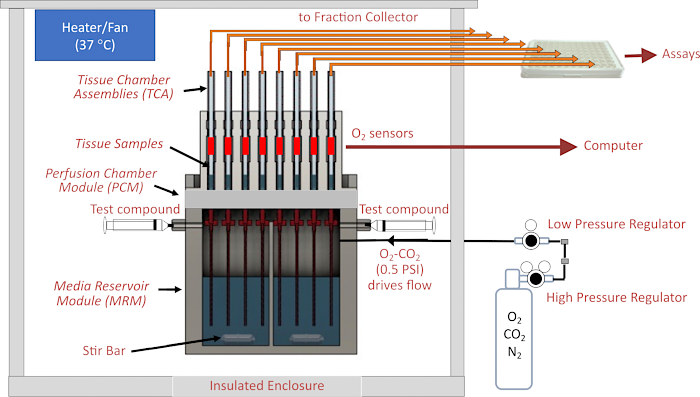
Figure 1. Schematic of the fluidics/assessment system. Major components include the enclosure, temperature control elements, fluidics and tissue chamber systems, regulation of gas pressure in the head space above perifusate, fraction collector/flow rate monitoring, and O2 detectors. Abbreviations: MRM = Media Reservoir Module, PCM = Perifusion Chamber Module, TCA= Tissue Chamber Assemblies. Please click here to view a larger version of this figure.
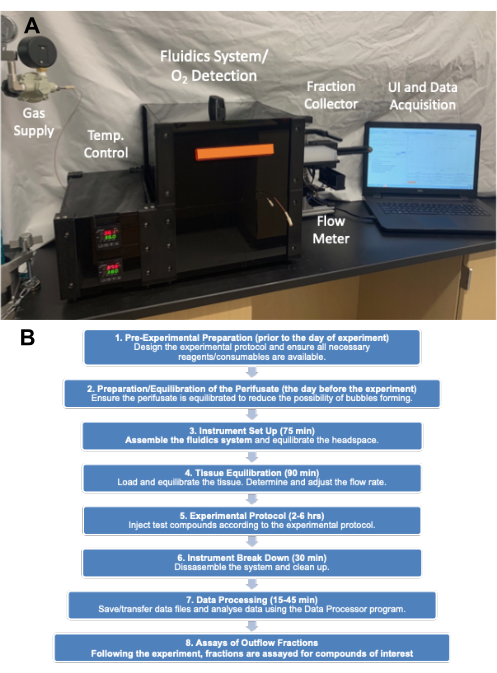
Figure 2. (A) Picture of the major components of the instrument. The major components consist of gas tank (pressure regulators), enclosure, fraction collector and computer. (B) Experimental flow chart showing the major categories of steps and the time it takes to complete them. Please click here to view a larger version of this figure.
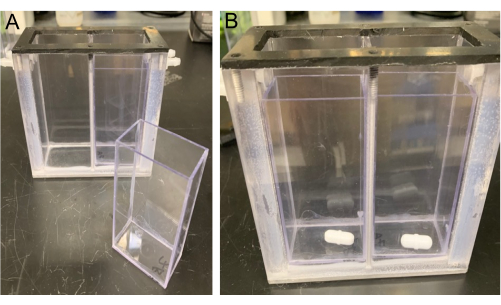
Figure 3. View of the MRM. The MRM is shown with an MRM insert (left) and stir bars (right) placed into the bottom of the MRM inserts (placed in each side of the MRM Divider). Please click here to view a larger version of this figure.
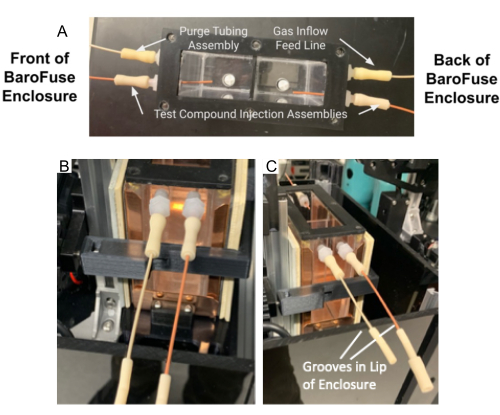
Figure 4. Tubing assembly and purge tubing assembly in the MRM. (A) Test compound injection tubing assembly and purge tubing assembly attached to ports on the MRM. (B–C) The test compound injection assembly and purge tubing assembly (B) are placed in the groove in the front of Enclosure (C). Please click here to view a larger version of this figure.

Figure 5. Powering up the MRM temperature controller. Please click here to view a larger version of this figure.
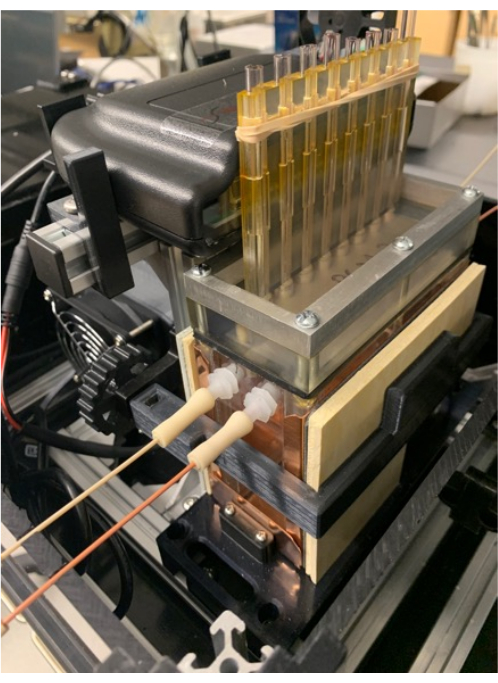
Figure 6. Tissue chambers and gas tank. Positioning the O2 detector on the detector stand (which also supports the MRM and PCM), and placement of the band around the fins of the PCM that help secure the tissue chambers in place. Please click here to view a larger version of this figure.
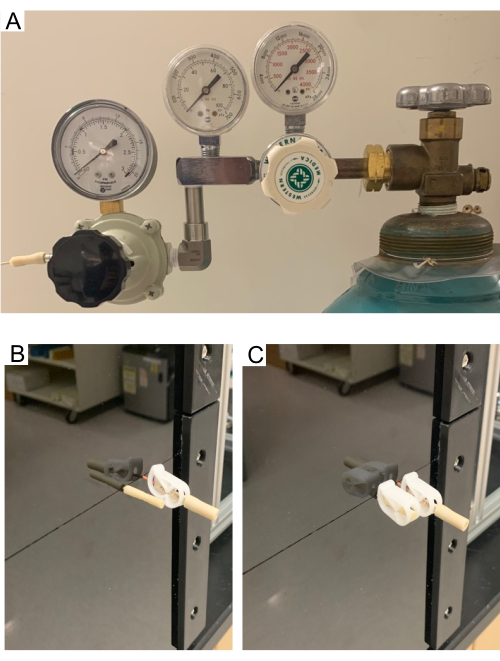
Figure 7. (A) High- and low-pressure regulators on the gas tank. (B–C) Purge tube. Purge tube allows the headspace in the MRM to clear of air and fill with gas from the supply tank. Pictures showing open purge tube (B) and close purge tube (C). Test compound injection assembly stays closed through the purge process. Please click here to view a larger version of this figure.
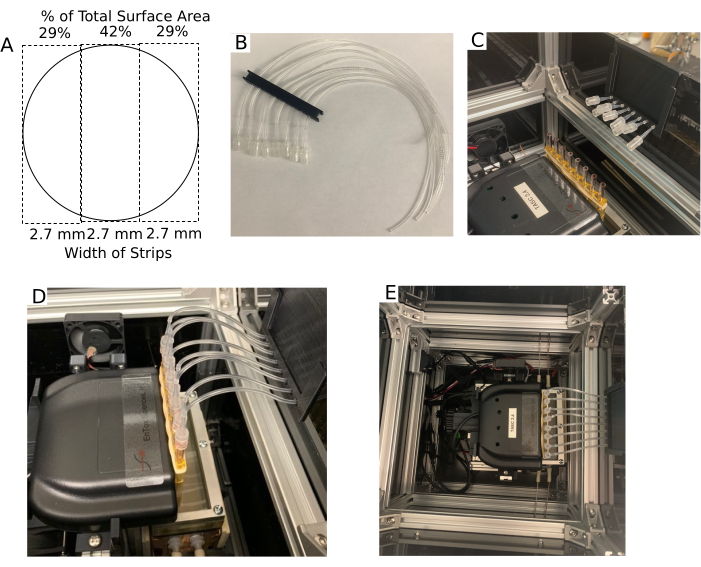
Figure 8. Tissue chamber and the outflow setup. (A) Dimensions of the Transwell membrane after it is cut into three strips of equal width. (B) Outflow multi-tube support. (C) Outflow multi-tube support positioned on the lip of the enclosure with the tubing adapters near the tissue chambers. (D) Picture of outflow tubing assemblies attached to the tissue chambers. (E) Aerial view of the enclosure without the Lid. Please click here to view a larger version of this figure.
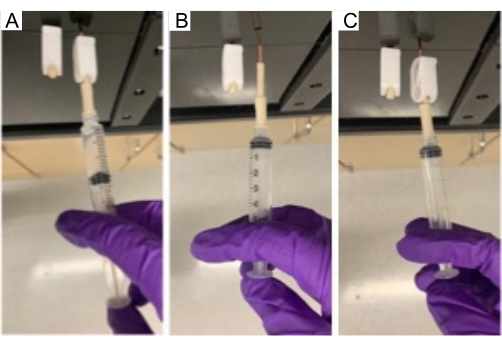
Figure 9. Injection of compound in MRM. Injecting a test compound through the injection port into the MRM using a 5 mL syringe. Please click here to view a larger version of this figure.
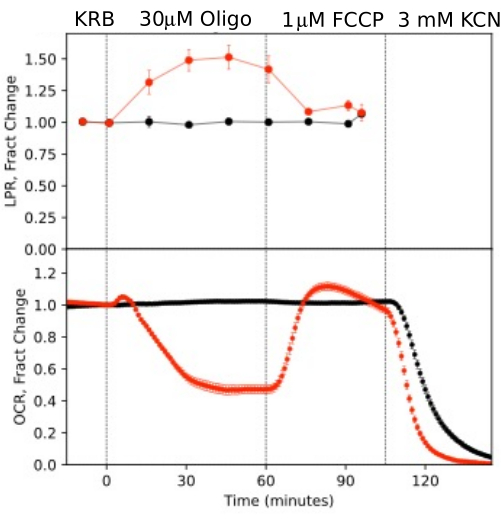
Figure 10. OCR and LPR curves in response to test compounds. OCR and LPR by retina isolated from mice (1 retina/channel) in response to the presence or absence (control) of test compounds as indicated. Each curve is the average of 6 replicates from a single experiment (error bars are SE; p-values are calculated by performing paired t-tests comparing steady state values for each test agent with that of the previous test agent). Please click here to view a larger version of this figure.
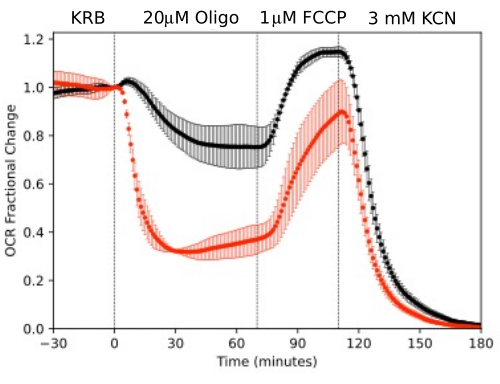
Figure 11. OCR curves. OCR by RPE-choroid-sclera and retina isolated from mice (1 retina or 2 RPE-choroid-sclera/channel) measured in parallel in response to test compounds as indicated. Data is the average of replicates from a single experiment (n = 2 and 4 for RPE-choroid-sclera and retina respectively; p-values are calculated by performing paired t-tests comparing steady state values for each test agent with that of the previous test agent). Please click here to view a larger version of this figure.
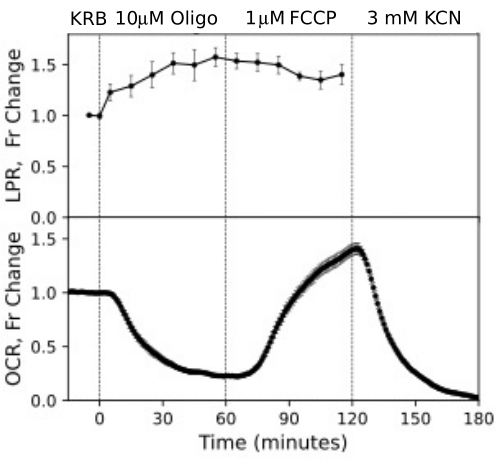
Figure 12. OCR and LPR curves from RPE cells. OCR and LPR from RPE cells attached to transwell membranes that were cut into strips and loaded into the perifusion chambers. Data is the average of replicates from a single experiment (n = 3, with 1.5 membranes/channel (360,000 cells/channel); p-values are calculated by performing paired t-tests comparing steady state values for each test agent with that of the previous test agent). Please click here to view a larger version of this figure.
| TISSUE/CELL | Amount/Channel | FLOW RATE: mL/min |
| Retina (mouse) | 1 | 0.025 |
| RPE-choroid-sclera (mouse) | 2 | 0.02 |
| RPE Cells on Transwell Membranes | 360,000 Cells (4 x 1/3 filter strips) | 0.016 |
Table 1. Recommended operating specifications for different tissue.
Supplementary Figure 1. Graphical representation of experimental design. Timing and composition of exposure to test compounds, and timing of fraction collection. Concentration increment (Conc Inc) is the change in concentration to be implemented. Please click here to download this File.
Supplementary Figure 2. User interface at startup. UI of the startup window of the O2 detection software that monitors the O2 in the tissue chambers inserted into the PCM. Please click here to download this File.
Supplementary Figure 3. User interface for experiment settings. UI for entering experimental information (left) and selecting times for collection of outflow fractions (right). Please click here to download this File.
Supplementary Figure 4. User interface of the injection page. Injection page which calculates injection volumes based on desired concentrations of test compound and volume left in the MRM. Please click here to download this File.
Supplementary File 1: Methods for tissue sample preparation. Please click here to download this File.
Discussion
Due to the importance of bioenergetics in all aspects of cell function and maintenance of various components of the eye, there is a critical need for methods to study its regulation. In particular, neural retina and RPE depend on metabolism for both generation of energy as well as intra- and inter-cellular signaling14,15,16,17. Because of their high oxidative capacity, isolated tissues of the eye are not well-maintained under static conditions18,19 and therefore study of isolated components of the eye require flow systems that can both maintain and assess metabolic processes. The fluidics system was developed to generate OCR and LPR data from a wide range of tissue types and in this paper we presented detailed protocols that were found to produce optimal results.
The major determinant for generating robust data using the flow system includes pre-equilibration of CO2-based media/buffer at 39 °C (to ensure perifusate is not supersaturated with dissolved gas that would degas during the experiment). In particular, media or KRB buffer stored at 4 °C will be supersaturated relative to 37 °C and will degas during the experiment if pre-equilibration times are insufficient. In addition, tissue loaded into the tissue chambers must not be traumatized by improper isolation of tissue due to tearing or incomplete separation of tissue, or by exposing tissue in low amount of bicarbonate-based buffer to atmospheric air for too long. The temperature control, flow stability and reliability of O2 detection have little variability and these factors do not contribute significantly to failure rate.
The instrument has eight flow channels/tissue chambers that run simultaneously which are supplied with perifusate from two reservoirs, four tissue chambers for each reservoir. To get the most accurate time-courses of OCR, kinetic curves are baseline corrected by chambers that are not loaded with tissue. Thus, a typical experimental protocol would involve two groups of three tissue chambers. Protocols in general fall into two categories: one is the different test compound protocols on each side (for instance drug/vehicle on one side of the MRM, and just vehicle on the other); the second is same test compound injection protocol on both sides of the MRM, but different tissue or tissue model on each side of the MRM. In this paper, the effects of oligomycin and FCCP on retina were compared to OCR by tissue that were not exposed to any test compounds, and two tissues were concomitantly assessed under the same protocol and conditions to identify tissue-specific behavior. The latter was illustrated in this study by showing increased dynamic range of metabolic rate by RPE-choroid-sclera relative to retina in parallel in the same experiment. Other reports have described a wider range of study designs including measuring the effects varying O2 levels on OCR and LPR, and concentration dependencies of fuels, drugs, and toxins20,21. In addition, although we have limited the analysis of outflow fractions to the measurement of lactate and calculation of LPR, the information content of an experiment increases greatly if multiple compounds and classes of compounds in the outflow fractions are assayed such as hormones, neurotransmitters, cell signals, and metabolites that can exit the cells20,22,23.
The loading of isolated retina or RPE-choroid-sclera is straightforward, and once isolated these tissues are simply placed into the top of the tissue chambers with forceps and allowed to sink down to the frit. RPE cells cultured on filter inserts develop appropriate polarization and markers of RPE maturity after 4-8 weeks in culture. It is not feasible to remove the RPE for live cell analysis once attached to the transwell membrane, if RPE maturity and polarization are to be maintained24. The perifusion chamber can accommodate strips of the transwell membrane that are cut with a scalpel while submerged in buffer and rapidly inserted into the tissue chambers. Although cutting filter strips has been placed into a static system24, no other fluidics method to assess these important cell types are available. The responses of RPE cells were rapid and more dynamic than either the retina or the RPE-choroid-sclera, likely in part due to immediate access of both the apical and basal aspects of the RPE cells configured as a monolayer on the membrane insert.
Another factor in assuring data has the highest signal to noise is selecting the optimal ratio of tissue loaded into the perifusion chambers relative to the flow rate. Too little tissue relative to the flow rate results in a difference of dissolved O2 concentration between inflow and outflow that is very small and difficult to measure reliably. In contrast, if the flow is too slow, then the concentration of O2 becomes so low that the tissue is affected by hypoxia. Nonetheless, the gas pressure-driven liquid flow can be maintained at flow rates down to 5 mL/min requiring only small amounts of tissue for accurate OCR and LPR measurements. In the experiments shown here, about 20 mL/min/channel was used which was suitable for either one retina, two RPE-choroid-scleras, or 360,000 RPE cells. To minimize the system effects that delay and disperse the exposure of the tissue to the injected test compound, multiple sizes of the tissue chambers are supplied, so that the amount of tissue (and flow rate) is matched with the appropriate size of the chamber.
Data from the analyses shown in this paper were represented in two ways: absolute magnitude with respect to rate, or fractional changes relative to a steady-state or baseline. The focus was on illustration of measurement of responses to test compounds. However, the fluidics system is well-suited to assess and compare effects of tissue treatment prior to the perifusion analysis such as genetic modifications. Testing whether a treatment is different from control is most robust if the effects of the treatment on normalized responses of test compounds is analyzed. If the analysis requires absolute magnitudes, the statistical power of the analyses of specimens that are pretreated is maximized if their assessment and controls are carried out in the same perifusion experiment.
Except for the stirrer, all parts that come in contact with liquid are supplied by the manufacturer as consumables and have been sterilized. These parts should not be reused, as experiments will occasionally be lost due to incomplete cleaning and contaminated surfaces. The system at the outset of setup is sterile. However, media is added to the MRM, and tissue is loaded in the chambers under non-sterile conditions. We have measured OCR in the system that is assembled with parts that are sterile, but where the experiment itself is carried out under non-sterile conditions. It takes about 14 h for bacteria to accumulate to the point of having measurable OCR (unpublished results). If protocols are used that are less than 10 h or so, then accumulation of bacteria and any effects due to these will be negligible.
Many investigators use instruments that are designed to measure OCR under static incubation of a monolayer of cells with a relatively high throughput25,26. In contrast, the fluidics instrument we have tested and described in this paper maintains tissue by ensuring adequate O2 delivery which is critical for the greater diffusion distances that are present in tissue specimens. In addition, it is able to collect fractions allowing assessment of multiple parameters in parallel with OCR which greatly enhances the ability to study relationships between them. Finally, dissolved gas concentrations (such as O2 and CO2) can be controlled, increasing the duration of experiments with bicarbonate-based media and buffer, enabling the user to study the effects of O2. It should be pointed out, a limitation for both methodologies is the inability to study the washout of test compounds, a functionality that other perifusion systems have4,27,28. Another consideration when determining the optimal analysis modality is the fact that fluidics systems use more media and test compounds than static systems. The extra expense is minimized with the current fluidics systems though due to the low flow rates that the system can be used.
Overall, a detailed description of the protocols to perform experiments with a new flow/assessment instrument is described. Data generated with retina and RPE-choroid-sclera recapitulated previous results obtained with systems that are much more difficult to use (and not readily available). It was also demonstrated that the system can maintain and assess RPE cells attached to transwell membranes, a very important cellular model that has not previously been analyzed with flow systems due to the fragility of the cells. The main parts of the protocol consist of a 75 min setup time, followed by a 90 min equilibration period and the experimental protocol making it suitable for routine use by laboratories that do not specialize in the operation of fluidics systems. Although we focused on measuring the acute response of tissue to test compounds, the system is very suitable to comparing tissue from various sources such as animal models or cells models that have been genetically altered or undergone test treatments/conditions. In addition, the scope of assays that can be conducted on the outflow fractions are wide-ranging and include metabolites, cell signaling molecules and secreted hormones/neurotransmitters as well as multi-component analysis generated by mass spectrometry on the fractions as well as the tissue.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This research was funded by grants from the National Institutes of Health (R01 GM148741 I.R.S.), U01 EY034591, R01 EY034364, BrightFocus Foundation, Research to Prevent Blindness (J.R.C.) and R01 EY006641, R01 EY017863 and R21 EY032597 (J.B.H.).
Materials
| BIOLOGICAL SAMPLES | |||
| C57BL/6J mice | Envigo Harlan (Indianapolis, IN) | N/A | |
| REAGENTS | |||
| FCCP | Sigma-Aldrich | C2920L9795 | |
| Glucose | Sigma-Aldrich | G8270G | |
| KCN | Sigma-Aldrich | 60178 | |
| Lactate | MilliporeSigma | L6661 | |
| Oliigomycin A | Sigma-Aldrich | 75351L9795 | |
| CELL CULTURE AND TISSUE HARVESTING | |||
| Beuthanasia-D | Schering-Plough Animal Health Corp., Union, NJ | N/A | |
| Bovine serum albumin | Sigma-Aldrich | A3059 | |
| Euthasol, 390 mg/ml sodium pentobarbital | Virbac | RXEUTHASOL | |
| Fetal bovine serum | Sigma-Aldrich | 12303C | |
| Hank’s Buffered Salt Solution | GIBCO | 14065056 | |
| Krebs Ringer Bicarbonate (KRB) | Thermo Fisher Scientific | J67795L9795 | |
| Matrigel | ThermoFisher | #CB-40230 | |
| Penicillin-streptomycin | ThermoFisher Scientific | 15140122 | |
| ROCKi | Selleck Chemicals | Y-27632 | |
| Trypsin-EDTA | ThermoFisher | #25-200-072 | |
| SUPPLIES | |||
| Gas Cylinders: 21% O2/5% CO2/balance N2 | Praxair Distribution, Inc | N/A | |
| Transwell filters | MilliporeSigma | 3470 | |
| COMMERCIAL ASSAYS | |||
| Amplex Red Glucose/Glucose Oxidase Assay Kit | ThermoFisher | A22189 | |
| Glucose Oxidase from Aerococcus viridans | Invitrogen (Carlsbad, CA) | A22189L9795 | |
| Lactate Oxidase | Sigma-Aldrich | L9795 | |
| EQUIPMENT | |||
| BaroFuse Multi-Channel Perifusion system | EnTox Sciences, Inc (Mercer Island, WA | Model 001-08 | |
| Synergy 4 Fluorometer | BioTek (Winooski, VT) | S4MLFPTA |
References
- Lacy, P. E., Walker, M. M., Fink, C. J. Perifusion of isolated rat islets in vitro: Participation of the microtubular system in the biphasic release of insulin. Diabetes. 21 (10), 987-998 (1972).
- Doliba, N. M., et al. Metabolic and ionic coupling factors in amino acid-stimulated insulin release in pancreatic beta-HC9 cells. American Journal of Physiology. Endocrinology and Metabolism. 292 (6), E1507-E1519 (2007).
- Sweet, I. R., et al. Regulation of ATP/ADP in pancreatic islets. Diabetes. 53 (2), 401-409 (2004).
- Chertov, A. O., et al. Roles of glucose in photoreceptor survival. The Journal of Biological Chemistry. 286 (40), 34700-34711 (2011).
- Kooragayala, K., et al. Quantification of Oxygen Consumption in Retina Ex Vivo Demonstrates Limited Reserve Capacity of Photoreceptor Mitochondria. Investigative Ophthalmology & Visual Science. 56 (13), 8428-8436 (2015).
- Bisbach, C. M., et al. Succinate Can Shuttle Reducing Power from the Hypoxic Retina to the O2-Rich Pigment Epithelium. Cell Reports. 31 (5), 107606 (2020).
- Du, J., et al. Inhibition of mitochondrial pyruvate transport by zaprinast causes massive accumulation of aspartate at the expense of glutamate in the retina. The Journal of Biological Chemistry. 288 (50), 36129-36140 (2013).
- Hass, D. T., et al. Succinate metabolism in the retinal pigment epithelium uncouples respiration from ATP synthesis. Cell Reports. 39 (10), 110917 (2022).
- Kamat, V., et al. Fluidics system for resolving concentration-dependent effects of dissolved gases on tissue metabolism. Elife. 10, e66716 (2021).
- Stryer, L. . Biochemistry. , (1995).
- Gu, X., Ma, Y., Liu, Y., Wan, Q. Measurement of mitochondrial respiration in adherent cells by Seahorse XF96 Cell Mito Stress Test. STAR Protocols. 2 (1), 100245 (2021).
- Engel, A. L., et al. Extracellular matrix dysfunction in Sorsby patient-derived retinal pigment epithelium. Experimental Eye Research. 215, 108899 (2022).
- Zhang, R., et al. Inhibition of Mitochondrial Respiration Impairs Nutrient Consumption and Metabolite Transport in Human Retinal Pigment Epithelium. Journal of Proteome Research. 20 (1), 909-922 (2021).
- Hurley, J. B. Retina Metabolism and Metabolism in the Pigmented Epithelium: A Busy Intersection. Annual Review of Vision Science. 7, 665-692 (2021).
- Xiao, J., et al. Autophagy activation and photoreceptor survival in retinal detachment. Experimental Eye Research. 205, 108492 (2021).
- Okawa, H., Sampath, A. P., Laughlin, S. B., Fain, G. L. ATP consumption by mammalian rod photoreceptors in darkness and in light. Current Biology. 18 (24), 1917-1921 (2008).
- Lakkaraju, A., et al. The cell biology of the retinal pigment epithelium. Progress in Retinal and Eye Research. , 100846 (2020).
- Yu, J., et al. Emerging strategies of engineering retinal organoids and organoid-on-a-chip in modeling intraocular drug delivery: Current progress and future perspectives. Advanced Drug Delivery Reviews. 197, 114842 (2023).
- Arjamaa, O., Nikinmaa, M. Oxygen-dependent diseases in the retina: role of hypoxia-inducible factors. Experimental Eye Research. 83 (3), 473-483 (2006).
- Kamat, V., et al. A Versatile Multi-Channel Fluidics System for the Maintenance and Real-Time Metabolic and Functional Assessment of Tissue or Cells. Cell Reports Methods. In Press. , (2023).
- Neal, A., et al. Quantification of Low-Level Drug Effects Using Real-Time, in vitro Measurement of Oxygen Consumption Rate. Toxicological Sciences. 148 (2), 594-602 (2015).
- Jung, S. R., et al. Reduced cytochrome C is an essential regulator of sustained insulin secretion by pancreatic islets. The Journal of Biological Chemistry. 286 (20), 17422-17434 (2011).
- Rountree, A. M., et al. Control of insulin secretion by cytochrome C and calcium signaling in islets with impaired metabolism. The Journal of Biological Chemistry. 289 (27), 19110-19119 (2014).
- Calton, M. A., Beaulieu, M. O., Benchorin, G., Vollrath, D. Method for measuring extracellular flux from intact polarized epithelial monolayers. Molecular Vision. 24, 425-433 (2018).
- Jarrett, S. G., Rohrer, B., Perron, N. R., Beeson, C., Boulton, M. E. Assessment of mitochondrial damage in retinal cells and tissues using quantitative polymerase chain reaction for mitochondrial DNA damage and extracellular flux assay for mitochondrial respiration activity. Methods in Molecular Biology. 935, 227-243 (2013).
- Perron, N. R., Beeson, C., Rohrer, B. Early alterations in mitochondrial reserve capacity; a means to predict subsequent photoreceptor cell death. Journal of Bioenergetics and Biomembranes. 45 (1-2), 101-109 (2013).
- Cabrera, O., et al. high-throughput assays for evaluation of human pancreatic islet function. Cell Transplantation. 16 (10), 1039-1048 (2008).
- Doliba, N. M., Qin, W., Vinogradov, S. A., Wilson, D. F., Matschinsky, F. M. Palmitic acid acutely inhibits acetylcholine- but not GLP-1-stimulated insulin secretion in mouse pancreatic islets. American Journal of Physiology. Endocrinology and Metabolism. 299 (3), E475-E485 (2010).

