Three-Dimensional Cell Culture Models to Investigate the Epithelial Barrier in Eosinophilic Esophagitis
Summary
Here, a protocol for the culture of human esophageal organoids and air-liquid interface culture is provided. Esophageal organoids’ air-liquid interface culture can be used to study the impact of cytokines on the esophageal epithelial barrier.
Abstract
The squamous epithelium of the esophagus is directly exposed to the environment, continuously facing foreign antigens, including food antigens and microbes. Maintaining the integrity of the epithelial barrier is critical for preventing infections and avoiding inflammation caused by harmless food-derived antigens. This article provides simplified protocols for generating human esophageal organoids and air-liquid interface cultures from patient biopsies to study the epithelial compartment of the esophagus in the context of tissue homeostasis and disease. These protocols have been significant scientific milestones in the last decade, describing three-dimensional organ-like structures from patient-derived primary cells, organoids, and air-liquid interface cultures. They offer the possibility to investigate the function of specific cytokines, growth factors, and signaling pathways in the esophageal epithelium within a three-dimensional framework while maintaining the phenotypic and genetic properties of the donor. Organoids provide information on tissue microarchitecture by assessing the transcriptome and proteome after cytokine stimulation. In contrast, air-liquid interface cultures allow the assessment of the epithelial barrier integrity through transepithelial resistance (TEER) or macromolecule flux measurements. Combining these organoids and air-liquid interface cultures is a powerful tool to advance research in impaired esophageal epithelial barrier conditions.
Introduction
Esophageal inflammation compromises the epithelial barrier integrity1,2,3,4,5, as observed in eosinophilic esophagitis (EoE), a Th2-dominated chronic inflammatory disease of the esophagus6. EoE was first described in the 1990s7,8 and is predominantly induced by food antigens9,10,11,12,13. The most frequently occurring symptoms of EoE in the adult population are dysphagia and food impaction14. In children, EoE typically manifests with failure to thrive, food refusal, vomiting, and abdominal pain15. Genome-wide association studies (GWAS) have identified EoE risk genes involved in epithelial barrier integrity, moving the epithelium into the focus of EoE research16,17,18. EoE transcriptomics further revealed that an impaired differentiation process and a reactive basal zone hyperplasia cause the compromised barrier function of the esophageal epithelium3,5,19,20,21,22. The early understanding of EoE being a Th2-mediated disease6 led to the discovery of IL-13 as a driving mediator by disturbing epithelial integrity3,4,21,23. Experimental systems allowing the dissection of cytokine-mediated effects on epithelial integrity from intrinsic barrier impairment through genetic predisposition provide the possibility to study the complex interplay between immune cells and the epithelium in EoE. Human esophageal organoids and air-liquid interface (ALI) cultures have been proposed as valuable tools to analyze the consequence of cytokine stimulation on epithelial integrity5.
The first protocol for generating adult tissue-specific stem cell (ASC)-derived esophageal organoids was established five years after the first published reports of intestinal organoids in 2009 using intestinal Lgr5+ ASCs recapitulating the epithelial compartment of the small intestine24. DeWard et al. pioneered generating organoids from murine esophageal epithelial cells25. In 2018, Kasagi et al. generated human esophageal organoids from the immortalized human esophageal squamous epithelium cell line EPC2-hTERT and primary patient-derived cells26. In the same year, Zhang et al. successfully generated induced pluripotent stem cell (iPSC)-derived esophageal organoids. They delineated the significance of TGFβ and bone morphogenetic protein (BMP) inhibition for esophageal progenitor cell (EPC) development and the crucial role of Notch signaling in the differentiation of the stratified squamous epithelium26,27. Trisno and colleagues complemented these findings by identifying Sox2 as a Wnt inhibitor that directs the developmental fate towards esophageal differentiation28. The subsequent refinements of protocols, medium composition, and culture conditions increased the organoid formation rate and made subculturing and recovering organoids after cryopreservation possible26,29,30,31,32. Although these organoids are powerful tools for studying tissue architecture and expression of potential target genes after stimulation with cytokines, esophageal organoids will not offer the possibility to measure transepithelial resistance (TEER) or macromolecule flux as direct measures for barrier integrity. As previously described by Sherrill and colleagues22, ALI cultures modeling epithelial differentiation4 allow direct assessments of epithelial integrity. Combining patient-derived organoids and ALI cultures is a powerful tool for investigating tissue architecture and epithelial barrier integrity in EoE.
Here are procedures with instructions for isolating viable cells from esophageal biopsies and establishing esophageal organoid and ALI cultures that can further be used to study the effects of cytokines on barrier integrity.
Protocol
The procedures were approved by the ethics committee of Northwest and Central Switzerland (EKNZ; Project-ID 2019-00273). All patients provided written informed consent for the experimental use of biopsies before the endoscopic examination. The reagents and equipment used in the study are listed in the Table of Materials.
1. Cell isolation for patient-derived esophageal organoids
NOTE: A list of the medium constituents for culturing human esophageal organoids is provided in Table 1.
- Obtain the biopsies.
NOTE: In the present study, two biopsies from one esophagus segment are obtained during esophagogastroduodenoscopy with biopsy forceps using a gastroscope with a 2.8 mm working channel. - Transfer the biopsies into commercially available keratinocyte serum-free medium (KSFM; Ca2+ 0.09 mM, 1 ng/mL EGF, 50 µg/mL BPE).
NOTE: Biopsies can be stored on ice for several hours until use. - Replace the KSFM medium with 1 mL of Dispase I (10 U/mL) and incubate the biopsies for 10 min at room temperature.
- Centrifuge the biopsies at room temperature at 300 x g for 2 min.
- Aspirate the Dispase using a 1000 µL pipette without touching the biopsies and the cell debris pellet.
- Rinse the biopsies with 1 mL Dulbecco's Phosphate-Buffered Saline (DPBS).
- Centrifuge the biopsies at room temperature at 300 x g for 2 min.
- Aspirate the supernatant using a 1000 µL pipette.
- Incubate the biopsies with 500 µL of Trypsin-EDTA (0.05%) at 37 °C for 10 min while shaking at 800 rpm.
- Perform mechanical disruption by repeated up-and-down pipetting until a single-cell suspension is obtained.
- Filter the cells through a 70 µm cell strainer using the rubber plunger head of a tuberculin syringe.
- Wash the strainer with 2-4 mL of soybean trypsin inhibitor (250 µg/mL).
- Filter the cells through a 35 µm cell strainer with a snap-on cap on a 5 mL round-bottomed polystyrene tube.
- Transfer the single-cell solution to a 15 mL conical tube.
- Centrifuge at 300 x g for 5 min at 4 °C.
- Aspirate the supernatant using a 1000 µL pipette.
- Resuspend the cells in 100 µL of KSFM medium.
- Mix 10 µL of trypan blue with 10 µL of cell suspension.
- Count the cells using an automated cell counter.
2. Patient-derived organoid culture
- Add 1-2 mL of KSFM to the cell suspension after counting.
- Centrifuge at 300 x g for 5 min at 4 °C.
- Aspirate the supernatant with a 1000 µL pipette without disturbing the cell pellet.
- Resuspend the cell pellet in Basement membrane extract (BME) hydrogel matrix (40 µL of BME per 20,000 cells).
NOTE: After adding the BME, keep the cells on ice to prevent premature solidification of the BME. - Cut off a 200 µL pipette tip to aspirate the viscous BME-cell suspension mixture.
- Form 40 µL droplets in a pre-warmed (37 °C) suspension cell culture plate.
- Incubate the plate without medium for 20-30 min at 37 °C to ensure solidification of the BME droplets.
- Add pre-warmed KSFM-C medium supplemented with 10 µM of Y27632 (ROCK-inhibitor) for the first two days of culture.
- Replace the medium with new KSFM-C medium (without Y27632 and +/- cytokine of interest) every other day.
- Aspirate the medium and scratch off droplets with the pipet tip while continuously adding 1 mL of Dispase II (1.5 U/mL) to the well.
- Transfer the BME-dispase mixture into a 15 mL centrifuge tube and incubate it for 20 min at 37 °C in a shaking water bath to digest the BME.
- Centrifuge at 250 x g for 3 min at 4 °C and aspirate the Dispase II.
- Proceed according to the protocol of the interest readout (e.g., RNA isolation, protein isolation, or fixation with 4% PFA for histology).
3. Cell isolation for patient-derived air-liquid interface (ALI) cultures
- Obtain the biopsies. During esophagogastroduodenoscopy using a gastroscope with a 2.8 mm working channel, two biopsies are taken from one esophagus segment with biopsy forceps.
- Place the biopsies into commercially available keratinocyte serum-free medium (KSFM; Ca2+ 0.09 mM, 1 ng/mL EGF, 50 µg/mL BPE) and store on ice for several hours until use.
- Exchange KSFM with 1 mL of Dispase (10 U/mL). Afterward, incubate the biopsies for 10 min at room temperature.
- Centrifuge the biopsies at room temperature at 300 x g for 2 min.
- Remove the Dispase using a 1000 µL pipette without touching the biopsies and cell debris pellet.
- Wash the biopsies with 1 mL of DPBS.
- At room temperature, spin down the biopsies at 300 x g for 2 min.
- Aspirate the supernatant with a 1000 µL pipette.
- Incubate the biopsies in 500 µL of Trypsin-EDTA (0.05%) at 37 °C for 10 min and continuously mix at 800 rpm during incubation.
- Disrupt the biopsies mechanically with repeated up-and-down pipetting till a single-cell suspension is obtained.
- Pass the dissociated cells through a 70 µm cell strainer with a rubber plunger head from a tuberculin syringe and collect the cells in a 50 mL conical tube.
- Wash the strainer with 2-4 mL of soybean trypsin inhibitor (250 µg/mL) to remove the remaining cells from the strainer.
- Filter the cells with a 35 µm cell strainer snap cap into a 5 mL round bottom polystyrene tube.
- Transfer cells into a 15 mL conical tube.
- Centrifuge at 300 x g at 4 °C for 5 min.
- Remove the supernatant with a 1,000 µL pipette.
- Resuspend the cell pellet in 4 mL of KSFM medium (Ca2+ 0.09 mM), including 10 µM Y27632.
- Transfer the cells into a T25 cell culture flask.
- To expand the primary keratinocytes, culture the cells till 60%-80% confluency for approximately 1 week.
NOTE: Passage (P)0 forms islet-like cell conglomerates and is not a monolayer. Primary keratinocytes form monolayers from P1 on. - Passage at 60%-80% confluency and reseed P1 in 2-3 T25 or 1-2 T75 cell culture flasks.
- Passage P1 again when keratinocytes form a monolayer with 60%-80% confluency.
4. Patient-derived air-liquid interface (ALI) culture
- Seed P2 primary keratinocytes onto transwell inserts for the ALI culture.
NOTE: Seed 400,000 cells in 12 well-inserts (200,000 keratinocytes per 0.6 cm2) in 500 µL of KSFM (Ca2+ 0.09 mM, 1 ng/mL EGF, 50 µg/mL BPE).- Seed 150,000 cells in 24 well-inserts (155,000 keratinocytes per 0.5 cm2) in 100 µL of KSFM (Ca2+ 0.09 mM, 1 ng/mL EGF, 50 µg/mL BPE).
- Alternatively, freeze in the KSFM medium (Ca2+ 0.09 mM + 10% DMSO) at -80 °C for 24 h and then transfer the frozen cells to liquid nitrogen for later use.
NOTE: When using frozen vials, passage once more after thawing before using the cells for the ALI culture.
- Add medium to the lower well beneath the insert of the transwell culture plate.
NOTE: For 12 well plates: 1.5 mL of KSFM (Ca2+ 0.09 mM, 1 ng/mL EGF, 50 µg/mL BPE). For 24 well-plates: 600 µL of KSFM (Ca2+ 0.09 mM, 1 ng/mL EGF, 50 µg/mL BPE). - Replace the medium to high calcium KSFM (Ca2+ 1.8 mM, 1 ng/mL EGF, 50 µg/mL BPE) after 2 days.
- Change the high calcium KSFM medium every second day until day 7.
- Perform airlift on day 7 by aspirating the medium from the upper chamber and replacing the medium in the lower chamber with high calcium KSFM (Ca2+ 1.8 mM, 1 ng/mL EGF, 50 µg/mL BPE) containing 10 ng/mL KGF (=FGF7), 75 µg/mL ascorbic acid (AA).
- Optional: Add cytokine of interest at the desired concentration to the medium.
- Change the medium every second day until day 14.
5. Transepithelial electrical resistance (TEER) measurement
- Sterilize the electrode of the TEER meter by immersing it in a well of a 24-well plate with 5% sodium hypochlorite for 10-15 min.
- Wash off the 5% sodium hypochlorite by immersing the electrode into 4 subsequential wells with sterile ddH2O and letting the electrode air dry.
- Set the blank by placing one electrode in the well and the second electrode into the transwell insert, filling it with PBS, and measuring the TEER.
NOTE: Recommended volumes for TEER measurements: For 12 well-plate (1900 µL in the well and 900 µL in the insert). For 24 well-plate (750-1000 µL in the well and 250 µL in the insert). - Replace the medium of the ALI cultures with room-temperature sterile PBS.
NOTE: Remove the medium from the lower well and, in a second step, from the transwell insert. Similarly, add PBS first to the insert and then to the lower well to prevent detachment of the ALI culture from the transwell membrane. - Place the TEER meter electrodes in the experimental wells with ALI cultures and perform the TEER measurement1.
NOTE: Perform TEER measurements every second day before the medium is changed.
6. Macromolecular flux
- Dilute the FITC-Dextran (3-5 kDa) stock solution to a working 1 mg/mL concentration.
NOTE: Always protect FITC-Dextran from light exposure. - Prepare a dilution row with decreasing FITC concentration (1000 µg/mL to 0.25 µg/mL) as a standard for the readout.
- Add 500 µL of FITC-dextran solution (1 mg/mL) to the upper chamber of the transwell and 1.5 mL medium (+/- cytokine of interest) into the lower compartment and place the plate in the incubator.
- Collect 120 µL of medium from the lower compartment at the respective time points (e.g., 0 min, 15 min, 30 min, 60 min, 90 min, 120 min, 150 min, and 180 min).
- Pipette duplicates (50 µL/well) of each timepoint into a black 96-well transparent flat bottom plate.
- Excite the FITC-dextran at 490 nm and read the emission at a wavelength of 520 nm using a plate reader.
- Calculate the amount of macromolecular flux according to the standard.
Representative Results
Esophageal organoids will grow from primary cells extracted from patient biopsies according to the instructions of the provided protocol, as documented with an inverted brightfield microscope (Figure 1). Epithelial ASCs start forming cell clusters in a self-organizing manner within the first two days of culture after seeding the isolated cells in the basement membrane extract, serving as a scaffold. The size and number of cell clusters, noticeable with an inverted brightfield microscope, increase continuously within the first week (Figure 2). However, at this point, the cell clusters lack the onion-like structure characteristic of the multilayered squamous epithelium of an ASC-derived esophageal organoid. On day 9 of culture, the onion-like multilayered structure and a growing keratinized core in the center of the organoid become apparent (Figure 2A). The organoid culture can be sustained for up to 21 days26. However, it has been demonstrated that the organoids attain full maturation and maximum size between day 11 and day 14. From day 11, the culture process is predominantly characterized by advancing keratinization and reducing metabolically active, viable cells5,26. Thus, the optimal time to retrieve organoids for experimentation depends on the desired differentiation stage and falls between 5 and 11 days.
Up to 3 passages are possible before the organoids stop growing because the increasing keratin core reduces the number of viable cells. The organoids can be harvested for RNA or protein isolation on the desired days to analyze gene/protein expression or for histology and biobanking.
The initial cell isolation steps for the ALI and organoid culture are identical. Before the ALI culture can be established, the freshly isolated primary cells must be expanded in contrast to the organoid protocol. After the second passage, the primary keratinocytes are seeded on transwell inserts and cultured in a medium with low calcium concentration to induce monolayer formation and prevent premature differentiation of the isolated primary keratinocytes. After 48 h, when a confluent monolayer is formed, the calcium concentration of the medium is raised to 1.8 mM, inducing differentiation and stratification of the epithelial layer. After 7 days, the airlift exposes the apical side of the multilayered epithelium to air to cause keratinization. The ALI cultures can usually be maintained for up to 14 days (Figure 2B). TEER measurements and macromolecular flux experiments will monitor the epithelium's functionality. Here, TEER measurements were performed in ALI cultures from primary esophageal keratinocytes, in primary keratinocyte ALI cultures with an NIH/3T3 fibroblast feeding layer on the bottom of each well, and in primary keratinocyte ALI cultures with an NIH/3T3 fibroblast feeding layer on each insert (Figure 2C).
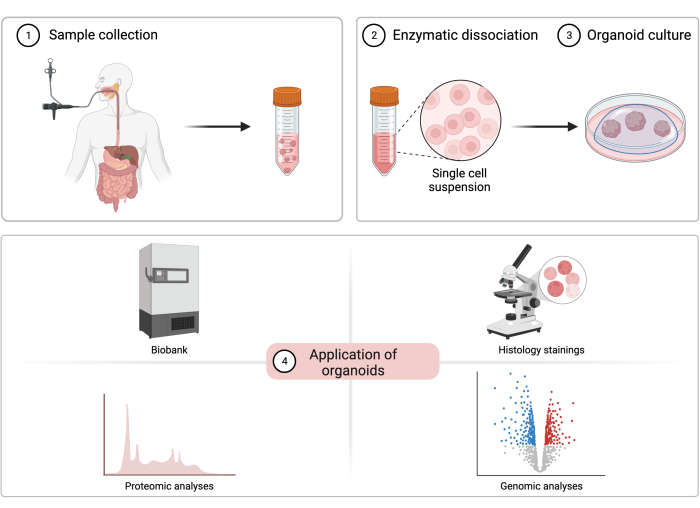
Figure 1: The process of patient-derived esophageal organoid cultivation. A diagram illustrating the sample collection procedure, cell isolation, organoid cultivation, and potential experimental applications of patient-derived esophageal organoids. Please click here to view a larger version of this figure.
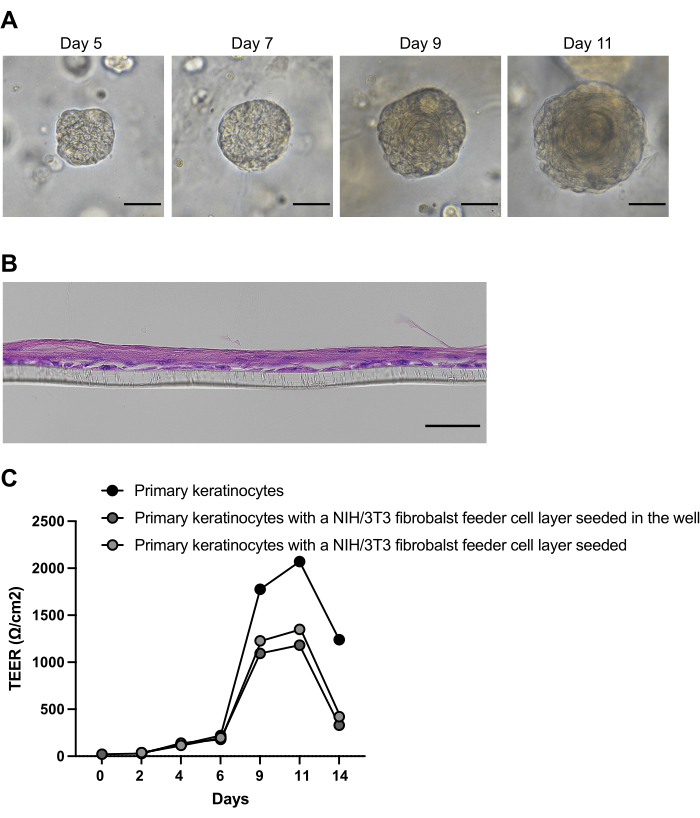
Figure 2: Formation and differentiation of patient-derived esophageal organoids. (A) Representative brightfield images of esophageal organoids cultivated from patient biopsy-derived primary cells. (B) Representative brightfield images of esophageal ALI cultures cultivated from biopsy-derived primary cells at day 14. Scale bars: 50μM. (C) Transepithelial electrical resistance (TEER) measurements at indicated days in air-liquid interface (ALI) cultures from primary esophageal keratinocytes, ALI cultured from primary keratinocytes with an NIH/3T3 fibroblast feeding layer on the bottom of each well, and ALI cultured from primary keratinocytes with an NIH/3T3 fibroblast feeding layer on each insert beneath the keratinocyte layer. Please click here to view a larger version of this figure.
| Medium | KSFM | KSFM-C | |
| Base medium | Keratinocyte-SFM | Keratinocyte-SFM | |
| Supplements | Penicillin/Streptomycin | 1% | 1% |
| Bovine pituitary extract (BPE) | 50 μg/mL | 50 μg/mL | |
| Epidermal growth factor (EGF) | 1 ng/mL | 1 ng/mL | |
| Calcium chloride (CaCl2) | 0.09 mM | 0.6 mM | |
| Y-27632 (ROCK-inhibitor) | – | 10 μM | |
Table 1: Primary cell-derived esophageal organoid media. List of the medium constituents for culturing human esophageal organoids. SFM: Serum-free medium.
Discussion
The provided procedures allow the cultivation of patient-derived organoids and ALI cultures with high prospects of success. The organoid protocol has been adapted from the first published protocol reporting the generation of human esophageal organoids26 and from a recently published protocol32. Sherill and colleagues have described the ALI model22. Organoids and ALI culture models assist each other in studying the impact of cytokines and other mediators on the esophageal epithelial barrier in contexts of esophageal diseases, such as EoE5,26.
One advantage of the organoid protocol is its simplicity in the culture process and the composition of the cell culture medium. Unlike other ASC- and PSC-based protocols, which mostly require the addition of multiple expensive supplements and growth factors24,25,27,28,33, the KSFM medium is relatively inexpensive. It requires only the addition of human recombinant epithelial growth factor (hEGF) and bovine pituitary extract (BPE). KSFM is a base medium optimized for keratinocyte culture and eliminates the need for a fibroblast feeder layer34. It is necessary to titrate the Ca2+ concentration of the KSFM from 0.09 mM to 0.6 mM using CaCl2 to obtain the KSFM-C medium used for organoid culture. ALI cultures require a higher Ca2+ concentration of 1.8 mM than the organoid model. The high Ca2+ concentration is essential to initiate keratinocyte differentiation, subsequently facilitating the formation of the typical onion-like configuration of multilayered squamous epithelium present within esophageal organoids26,34,35 and the epithelial stratification in ALI cultures22.
Preventing premature keratinocyte differentiation, organoid, and ALI culture generation requires avoiding high Ca2+ concentrations during cell isolation. Early differentiation diminishes colony formation and impedes the self-organizing formation of organoids and the formation of a confluent monolayer of undifferentiated keratinocytes as the foundation of ALI cultures. Using serum-containing media to stop the enzymatic activity of trypsin during cell isolation impairs successful organoid generation by triggering premature differentiation, similar to using a Ca2+-enriched medium during cell isolation36. Instead, a soybean trypsin inhibitor or any other serum-free trypsin inhibitor is recommended to stop the enzymatic reaction after biopsy digestion with trypsin. The culture medium is supplemented with the Rho-kinase (ROCK) inhibitor Y27632 during the first two days of culture to enhance the colony formation rate. Inhibition of ROCK has anti-apoptotic effects, prevents keratinocyte senescence, and thus improves organoid formation rate37,38.
Seeding approximately 20,000 cells in a 40 µL of BME droplet is recommended to ensure a thriving organoid culture and 150,000 to 400,000 cells for the ALI culture. An adequate cell seeding count will guarantee sufficient material for subsequent transcriptomics, proteomics, and histology. When utilizing cell lines such as the human esophageal epithelial cell line EPC2-hTERT, the seeding cell count can be decreased due to the heightened organoid formation rate exhibited by this cell line26. This phenomenon is likely attributed to the homogeneity and superior cell cycle synchronicity demonstrated by cell lines compared to biopsy-derived primary cells. The immortalized EPC2-hTERT cell line also preserves its proliferative capacity, enabling infinite passaging of organoids.
Conversely, human organoids derived from esophageal biopsies cannot be maintained for long-term culture26,28. The decline of dividing, viable cells within fully developed esophageal organoids and a decreasing rate of organoid formation with each passage26 indicates that the loss of proliferative capacity in primary cell-derived organoids is due to the terminal differentiation and keratinization of progenitor cells. Previous attempts to maintain the proliferative capacity by supplementing the culture medium with Noggin, Wnt3a, or A83-01 were unsuccessful26,28,32. Hence, long-term biobanking of patient-derived esophageal organoids remains a challenge that must be addressed in future studies.
The advent of organoid culture has brought new experimental possibilities that have greatly enabled the discovery of key inflammatory mediators and environmental factors responsible for epithelial alterations in EoE5,39,40,41,42. Nonetheless, it is essential to note that ASC- and PSC-derived esophageal organoids have limitations as they do not perfectly recapitulate all the cellular compartments of the esophagus. ASC-derived organoids recapitulate the epithelial compartment24,26,27,33, while PSC-derived organoids recapitulate the epithelial, endothelial, and mesenchymal compartments28,43,44. However, all current organoid models lack the immunological microenvironment. In a recent study, human esophageal organoids and ALI cultures have been stimulated with cytokines from the IL-20 cytokine family to decipher a new aspect of the epithelial barrier dysfunction in EoE5. The combination of both three-dimensional culture models evaluated indirect and direct measures of epithelial barrier integrity. More publications will likely use esophageal organoids in combination with ALI cultures to elucidate the effects of defined cytokines on the epithelium. While this approach reduces the complexity of the cross-talk between the immune system and the epithelium to reveal the function of specific mediators on the epithelium, it neglects many other potentially involved factors. Combining the organoid and organ-on-a-chip technology may provide a solution to this limitation. The microfluidics-based organ-on-a-chip technology can replicate tubular organs' luminal and basolateral microenvironment, making in- and efflux of immune cells and other circulatory and luminal components possible. This enables the investigation of interactions between the epithelium and immune cells in homeostasis and disease45,46. The successful cultivation of physiologically polarized kidney tubule organoids exhibiting an intact epithelial barrier and a functional transepithelial ion transport on an organ-on-a-chip platform47 supports the expanded use of patient-derived organoid-on-a-chip technology. Moreover, another progress of utilizing the organoid-on-a-chip technology is demonstrated through the extended cultivation of organoids originating from primary cells by implementing a luminal flow through the microfluidic channels on the apical side of tubular organoids to eliminate shedding epithelial cells46. Such advancements highlight the potential for this technology to replicate in vivo environments in vitro.
The ability to culture 3D organoids that resemble everything from the epithelial compartment to a near-complete mini-organ with most cell types urges for possible clinical applications5,26,46. While considerable progress has been made in recent years, additional obstacles must be overcome before organoids can be used in clinical practice. A significant hurdle for the clinical implementation of organoids is the presence of animal-derived proteins in the culture medium and the scaffolding ECM products. While animal protein-free media for keratinocyte culture are already available and BPE-free KSFM variants have recently emerged, producing animal protein-free ECM alternatives with similar properties and culture outcomes to traditional ECM products appears more challenging. Recently, a potentially promising fully chemical synthesized alternative has been reported48. Nonetheless, currently, there is no commercially accessible animal-origin-free ECM product.
Esophageal organoids and ALI cultures are vital instruments for studying EoE. The culture of organoids and ALI cultures with the provided protocols here served as the foundation for new revelations regarding the involvement of the epithelial compartment in eosinophilic esophagitis5,26,39,41,42,49,50. The simple implementation of these protocols and the continuous advancement of three-dimensional cell culture technologies facilitates experimental approximation to the in vivo environment, resulting in enhanced translatability of discoveries and possibly a gradual reduction of animal experimentation.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The SNSF grant 310030_219210 to J.H.N. supported the publication of this manuscript without restrictions. Figure 1 has been created with the help of BioRender.com.
Materials
| 1250 µL Griptip – Filter | Integra | 4445 | |
| 300 µL Griptip – Filter | Integra | 4435 | |
| 70 µM cell strainer | Sarstedt | 83.3945.070 | |
| Ascorbic Acid | Sigma-Aldrich (Merck) | A4544 | |
| Bovine pituitary extract | Gibco (Thermo Fischer Scientific) | 3700015 | |
| Calcium chloride | Sigma-Aldrich (Merck) | 21115 | |
| Cell Culture Multiwell Plates CELLSTAR for suspension cultures | Greiner Bio-One | 7.657 185 | |
| Cultrex Basement Membrane Extract (BME), Type 2, Pathclear | R&D Systems (Bio-Techne) | 3532-010-02 | |
| Dimethyl sulfoxide (DMSO), >99,5% BioScience Grade | Carl Roth | A994 | |
| Dispase I | Corning | 354235 | |
| Dispase II | Sigma-Aldrich (Merck) | D4693 | |
| Dulbeccos Phosphate Buffered Saline (DPBS) | Sigma-Aldrich (Merck) | D8537 | |
| EVE Automated Cell Counter | NanoEntek | EVE-MC | |
| EVE Cell counting slide | NanoEntek | EVS-050 | |
| Falcon 5 mL Round Bottom Polystyrene Test Tube, with Cell Strainer Snap Cap | Falcon | 352235 | |
| Fluorescin isothiocyanate (FITC)-dextran | Sigma-Aldrich (Merck) | FD4 | average mol wt 3000-5000 |
| Heraeus – Megafuge 40R | Thermo Fisher Scientific | 75004518 | |
| Human recombinant epidermal growth factor | Gibco (Thermo Fischer Scientific) | 3700015 | |
| Keratinocyte-SFM | Gibco (Thermo Fischer Scientific) | 17005042 | |
| Penicillin-Streptomycin | Gibco (Thermo Fischer Scientific) | 15140122 | |
| Recombinant Human KGF/FGF-7 Protein | R&D Systems (Bio-Techne) | 251-KG-010/CF | |
| Screw cap tube, 15 mL | Sarstedt | 62.554.502 | |
| Single Channel EVOLVE 100-1000 µL | Integra | 3018 | |
| Single Channel EVOLVE 20-200 µL | Integra | 3016 | |
| Syringe 1 mL | 1134950 | ||
| ThermoMixer C | Eppendorf | 5382000015 | |
| Trypsin inhibitor from Glycine max (soybean) | Sigma-Aldrich (Merck) | T9128 | |
| Trypsin-EDTA | SAFC Biosciences (Merck) | 59418C | |
| Y27632 dihydrochloride | Tocris (Bio-Techne) | 1254 |
References
- Wu, L., et al. Filaggrin and tight junction proteins are crucial for IL-13-mediated esophageal barrier dysfunction. Am J Physiol Gastrointest Liver Physiol. 315 (3), G341-G350 (2018).
- Davis, B. P., et al. Eosinophilic esophagitis-linked calpain 14 is an IL-13-induced protease that mediates esophageal epithelial barrier impairment. JCI Insight. 1 (4), e86355 (2016).
- Blanchard, C., et al. Coordinate interaction between IL-13 and epithelial differentiation cluster genes in eosinophilic esophagitis. J Immunol. 184 (7), 4033-4041 (2010).
- Kc, K., Rothenberg, M. E., Sherrill, J. D. In vitro model for studying esophageal epithelial differentiation and allergic inflammatory responses identifies keratin involvement in eosinophilic esophagitis. PLoS One. 10 (6), e0127755 (2015).
- Kaymak, T., et al. IL-20 subfamily cytokines impair the oesophageal epithelial barrier by diminishing filaggrin in eosinophilic oesophagitis. Gut. 72 (5), 821-833 (2023).
- Straumann, A., Bauer, M., Fischer, B., Blaser, K., Simon, H. U. Idiopathic eosinophilic esophagitis is associated with a T(H)2-type allergic inflammatory response. J Allergy Clin Immunol. 108 (6), 954-961 (2001).
- Straumann, A., Spichtin, H. P., Bernoulli, R., Loosli, J., Vogtlin, J. Idiopathic eosinophilic esophagitis: a frequently overlooked disease with typical clinical aspects and discrete endoscopic findings. Schweiz Med Wochenschr. 124 (33), 1419-1429 (1994).
- Attwood, S. E., Smyrk, T. C., Demeester, T. R., Jones, J. B. Esophageal eosinophilia with dysphagia. A distinct clinicopathologic syndrome. Dig Dis Sci. 38 (1), 109-116 (1993).
- Kelly, K. J., et al. Eosinophilic esophagitis attributed to gastroesophageal reflux: improvement with an amino acid-based formula. Gastroenterology. 109 (5), 1503-1512 (1995).
- Fogg, M. I., Ruchelli, E., Spergel, J. M. Pollen and eosinophilic esophagitis. J Allergy Clin Immunol. 112 (4), 796-797 (2003).
- Wolf, W. A., Jerath, M. R., Dellon, E. S. De-novo onset of eosinophilic esophagitis after large volume allergen exposures. J Gastrointestin Liver Dis. 22 (2), 205-208 (2013).
- Moawad, F. J., et al. Correlation between eosinophilic oesophagitis and aeroallergens. Aliment Pharmacol Ther. 31 (4), 509-515 (2010).
- Woo, W., Aceves, S. S. The role of the allergist in the management of eosinophilic esophagitis. Curr Opin Gastroenterol. 37 (4), 390-396 (2021).
- Dellon, E. S., et al. Updated International Consensus diagnostic criteria for eosinophilic esophagitis: Proceedings of the AGREE conference. Gastroenterology. 155 (4), 1022-1033 (2018).
- Liacouras, C. A., Spergel, J., Gober, L. M. Eosinophilic esophagitis: Clinical presentation in children. Gastroenterol Clin North Am. 43 (2), 219-229 (2014).
- Sleiman, P. M., et al. GWAS identifies four novel eosinophilic esophagitis loci. Nat Commun. 5, 5593 (2014).
- Kottyan, L. C., et al. Genome-wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat Genet. 46 (8), 895-900 (2014).
- Kottyan, L. C., et al. Replication and meta-analyses nominate numerous eosinophilic esophagitis risk genes. J Allergy Clin Immunol. 147 (1), 255-266 (2021).
- Sherrill, J. D., et al. Analysis and expansion of the eosinophilic esophagitis transcriptome by RNA sequencing. Genes Immun. 15 (6), 361-369 (2014).
- Collins, M. H., et al. Newly developed and validated eosinophilic esophagitis histology scoring system and evidence that it outperforms peak eosinophil count for disease diagnosis and monitoring. Dis Esophagus. 30 (3), 1-8 (2017).
- Rochman, M., et al. Profound loss of esophageal tissue differentiation in patients with eosinophilic esophagitis. J Allergy Clin Immunol. 140 (3), 738-749 (2017).
- Sherrill, J. D., et al. Desmoglein-1 regulates esophageal epithelial barrier function and immune responses in eosinophilic esophagitis. Mucosal Immunol. 7 (3), 718-729 (2014).
- Blanchard, C., et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 120 (6), 1292-1300 (2007).
- Sato, T., et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 459 (7244), 262-265 (2009).
- DeWard, A. D., Cramer, J., Lagasse, E. Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population. Cell Rep. 9 (2), 701-711 (2014).
- Kasagi, Y., et al. The esophageal organoid system reveals functional interplay between Notch and cytokines in reactive epithelial changes. Cell Mol Gastroenterol Hepatol. 5 (3), 333-352 (2018).
- Zhang, Y., et al. 3D modeling of esophageal development using human PSC-derived basal progenitors reveals a critical role for notch signaling. Cell Stem Cell. 23 (4), 516-529 (2018).
- Trisno, S. L., et al. Esophageal organoids from human pluripotent stem cells delineate sox2 functions during esophageal specification. Cell Stem Cell. 23 (4), 501-515 (2018).
- Kijima, T., et al. Three-dimensional organoids reveal therapy resistance of esophageal and oropharyngeal squamous cell carcinoma cells. Cell Mol Gastroenterol Hepatol. 7 (1), 73-91 (2019).
- Karakasheva, T. A., et al. Generation and characterization of patient-derived head and neck, oral, and esophageal cancer organoids. Curr Protoc Stem Cell Biol. 53 (1), e109 (2020).
- Zheng, B., et al. A new murine esophageal organoid culture method and organoid-based model of esophageal squamous cell neoplasia. iScience. 24 (12), 103440 (2021).
- Nakagawa, H., et al. Modeling epithelial homeostasis and reactive epithelial changes in human and murine three-dimensional esophageal organoids. Curr Protoc Stem Cell Biol. 52 (1), e106 (2020).
- Sato, T., et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 141 (5), 1762-1772 (2011).
- Boyce, S. T., Ham, R. G. Calcium-regulated differentiation of normal human epidermal keratinocytes in chemically defined clonal culture and serum-free serial culture. J Invest Dermatol. 81, 33-40 (1983).
- Bertolero, F., Kaighn, M. E., Gonda, M. A., Saffiotti, U. Mouse epidermal keratinocytes. Clonal proliferation and response to hormones and growth factors in serum-free medium. Exp Cell Res. 155 (1), 64-80 (1984).
- Bertolero, F., Kaighn, M. E., Camalier, R. F., Saffiotti, U. Effects of serum and serum-derived factors on growth and differentiation of mouse keratinocytes. In Vitro Cell Dev Biol. 22 (7), 423-428 (1986).
- Witkowski, T. A., et al. Y-27632 acts beyond ROCK inhibition to maintain epidermal stem-like cells in culture. J Cell Sci. 136 (17), (2023).
- Chapman, S., Liu, X., Meyers, C., Schlegel, R., McBride, A. A. Human keratinocytes are efficiently immortalized by a Rho kinase inhibitor. J Clin Invest. 120 (7), 2619-2626 (2010).
- Sasaki, M., et al. Lysyl oxidase regulates epithelial differentiation and barrier integrity in eosinophilic esophagitis. bioRxiv. , (2023).
- Doyle, A. D., et al. Detergent exposure induces epithelial barrier dysfunction and eosinophilic inflammation in the esophagus. Allergy. 78 (1), 192-201 (2023).
- Hara, T., et al. CD73(+) epithelial progenitor cells that contribute to homeostasis and renewal are depleted in eosinophilic esophagitis. Cell Mol Gastroenterol Hepatol. 13 (5), 1449-1467 (2022).
- Kasagi, Y., et al. Fibrostenotic eosinophilic esophagitis might reflect epithelial lysyl oxidase induction by fibroblast-derived TNF-alpha. J Allergy Clin Immunol. 144 (1), 171-182 (2019).
- Spence, J. R., et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 470 (7332), 105-109 (2011).
- Takebe, T., et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 499 (7459), 481-484 (2013).
- Bhatia, S. N., Ingber, D. E. Microfluidic organs-on-chips. Nat Biotechnol. 32 (8), 760-772 (2014).
- Nikolaev, M., et al. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature. 585 (7826), 574-578 (2020).
- Schutgens, F., et al. Tubuloids derived from human adult kidney and urine for personalized disease modeling. Nat Biotechnol. 37 (3), 303-313 (2019).
- Sorrentino, G., et al. Mechano-modulatory synthetic niches for liver organoid derivation. Nat Commun. 11 (1), 3416 (2020).
- Azouz, N. P., et al. The antiprotease SPINK7 serves as an inhibitory checkpoint for esophageal epithelial inflammatory responses. Sci Transl Med. 10 (444), 9736 (2018).
- Azouz, N. P., et al. Functional role of kallikrein 5 and proteinase-activated receptor 2 in eosinophilic esophagitis. Sci Transl Med. 12 (545), 7773 (2020).

