Combining QD-FRET and Microfluidics to Monitor DNA Nanocomplex Self-Assembly in Real-Time
Summary
We present a novel and powerful integration of nanophotonics (QD-FRET) and microfluidics to investigate the formation of polyelectrolyte polyplexes, which is expected to provide better control and synthesis of uniform and customizable polyplexes for future nucleic acid-based therapeutics.
Abstract
Protocol
A. Biotinylation of DNA
Plasmid DNA were covalently biotinylated with guanine-specific biotin labels as described by the manufacturer (Mirus Bio, Madison, WI) but scaled to have ~1-2 biotin labels per DNA. Plasmid DNA (pEGFP-C1, 4.9 kb, Clontech, Mountain View, CA) was labeled in this protocol.
- Dissolve desired amount of pDNA into TE buffer of DNase-free and RNase-free (molecular biology-grade quality) water to make a 1μg/μL DNA solution.
- Conduct the labeling reaction using the following reaction mixtures. Add the LabelIT reagent last.
For 100μg DNA reaction:
| DNase-free and RNase-free water | 75 μL |
| 10X Labeling Buffer A | 20 μL |
| 1μg/μL DNA | 100 μL |
| LabelIT reagent | 5 μL |
| Total Volume | 200 μL |
- Incubate the reaction at 37 °C for 1 hour.
- Purify the labeled sample by ethanol or isopropanol precipitation following standard protocols.
Note: Gel filtration based columns may lead to high UV absorbance or fluorescence background, which may affect the DNA quantification or fluorescence characterization.
Note: The level of biotinylation may be determined by HABA-based tests.
B. Labeling of the Cy5-Cationic Polymer
Chitosan (390 kDa, 83.5% deacetylated, Vanson, Redmond, WA) was used as a model cationic polymer in this study. The free primary amines on the chitosan polymer backbone were labeled with Cy5-NHS (Amersham Biosciences, Piscataway, NJ).
- To facilitate complete conjugation of Cy5 dye, calculate the required amount of Cy5-NHS such that the molar ratio of Cy5 : primary amines is 1 : 200.
- Adjust pH of the chitosan solution (in 25mM acetate buffer) to ~6.5 by addition of NaOH. Note that the NHS reaction is more efficient at basic pH, but the solubility of chitosan here limits the working pH range.
- While stirring, slowly add the calculated amount of Cy5-NHS (1 mg/ml DMSO) to the chitosan solution in a drop-by-drop manner.
- Agitate the mixture in the dark at room temperature overnight.
- To purify, dialyze with 10k MWCO Slide-a-Lyzer (Pierce) for 2 hr against 1% acetate buffer at room temp in the dark.
- Replace buffer and dialyze another 2 hr at room temp in the dark.
- Replace buffer and dialyze overnight at 4 °C in the dark.
- Store purified labeled polymer at -20 °C.
Note: In this study, a standard curve is constructed by measuring the emission intensity of Cy5-NHS ester at 670 nm. Characterize the labeling density by measuring the obtained emission at 670 nm from Cy5-labeled chitosan in the standard curve. Absorbance may also be used to determine the labeling efficiency but was not performed here.
C. Preparation of QD-labeled DNA and Cy5-Polymer
The molar ratio of pDNA to QD was kept in excess (pDNA : QD ≈ 1 : 2) to ensure complete conjugation of QDs to pDNA. The number of QDs labeled onto each pDNA can be estimated through TEM imaging or other equivalent facilities. In our study, the number of QDs per pDNA is estimated to be ~1-3 by TEM and single molecule spectroscopy.1 Use Millipore Milli-Q gradient water (>18.0 MW, 0.2um filtered) during the preparation.
- Calculate the required amount of chitosan for 10μg pDNA according to desired N/P ratio, the theoretical ratio of protonated amines in the chitosan solution to the negative phosphates in the DNA solution.
- Add streptavidin-functionalized 605QDs (Qdot 605 ITK, Invitrogen, Carlsbad, CA) into the biotinylated pDNA solution.
- Incubate the solution at room temperature in the dark for 15 min.
- Add the QD-labeled DNA into 50 mM sodium sulfate solution to make the final volume 200μL.
- Dilute Cy5-chitosan, according to desired N/P ratio, with Milli-Q water to make the final volume 200μL.
Note: Keep the reaction in dark to prevent possible photobleaching.
Important: Be cautious to use the Qdot 605 ITK™ streptavidin conjugate (the ITK series), as Quantum dots in this catalog are designed for the purpose of FRET. The regular Qdot series are conjugated with a PEG layer to prevent non-specific binding, especially for cellular labeling. However, this additional coating enlarges the donor-acceptor distance, resulting in reduced energy transfer efficiency.
D. Fabrication of the SU-8 Masters Using Standard Photolithography
- Si wafer is piranha cleaned and baked at 200 °C for 5 min.
- For the designated master thickness of 25μm, spin coat the negative photoresist (SU-8 2025, Microchem, Newton, MA) on Si wafer at 2000 rpm for 30 sec.
- Soft bake the wafer on a hotplate with a ramp of 65 °C/hr to 95 °C.
- Expose to UV light (365nm) for 250mJ/cm2 through a mask film (CAD/Art Services, Bandon, OR) containing the design of microchannels.
- Post-exposure bake the wafer on a hotplate with a ramp of 65 °C/hr to 95 °C.
- Develop the wafer using SU-8 photoresist developer.
- The patterned wafer is hard baked on a hotplate with a ramp of 65 °C/hr to 200 °C. Maintain the wafer at 200 °C for at least 5 hours, then gradually cool the wafer down to room temperature.
Important: Gradual ramping during the SU-8 master baking process is necessary, otherwise the SU-8 structure may detached from the silicon wafer or cracks on the SU-8 structure may be induced by stress-release.
E. Replica Molding of PDMS from the masters and Bonding to the Cover Glass
- The SU-8 master is placed in a weighing boat.
- Mix silicone elastomer and curing agent (Poly(dimethylsiloxane), PDMS, Sylgard 184, Dow Corning, Midland, MI) in a 10 : 1 ratio.
- Pour the PDMS mixture onto the SU-8 master and leave the weighing boat in a vacuum desiccator to remove bubbles.
- Cure the PDMS at 65 °C for 1-2 hours.
- Peel the PDMS strip from the Si master mold.
- Punch channel inlets and outlets of the fluidic device.
- Clean the PDMS strip and cover glass with ethanol and then air-dry.
- Treat the cleaned PDMS strip and cover glass with oxygen plasma (20W for 1min).
- Immediately bond the PDMS strip with cover glass.
- Leave the bonded microfluidic chip in the oven at 95 °C for overnight.
Important: Plasma treatment and overnight baking are essential to enhanced bonding strength.
F. Monitor the formation of DNA Nanocomplexes In the Microfluidic Device
- Fill the microfluidic channel with water (to ensure there is no bubbles within the microfluidic channel), before loading the reagents to ensure smooth flow during the experiment.
- Load the QD-labeled DNA and Cy5-labeled chitosan solutions into two individual glass syringes, through the tubing described in the video.
- Connect the tubing with the two inlets of microfluidic devices. Be cautious not to introduce any air during the process. Set the flow rate at 20nL/min (PHD-2000 syringe pump, Holliston, MA), under laminar flow conditions.
- Check the microchannels under the microscope.
- When the flow is stable (~15 to 20 minutes), QD-mediated FRET should be observed in the center of the channel.
- Take fluorescence pictures (Cooled CCD, Qimaging, BC, Canada) at different locations along the channel.
- Analyze the fluorescence images with ImageJ and OriginLab.
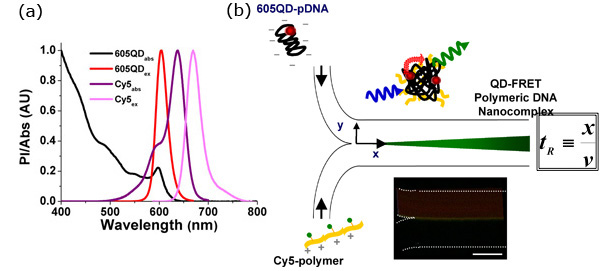
Figure 1. QD-FRET provides a sensitive indication of the onset of DNA Nanocomplexes self-assembly
- Quantum dot-mediated fluorescence resonance energy transfer (QD-FRET) can provide a quantitative and highly sensitive indication of polyplex stability in either extra- or intra-cellular environments, allowing for unambiguous detection of the onset of interactions between DNA and the gene carrier. The FRET pair, 605QD and Cy5, was chosen based on maximizing spectral overlap between the donor and acceptor and minimizing potential cross-talk. For this pair, the Förster distance is 69.4Å.3
- Self-assembly of the QD-FRET DNA nanocomplexes. Anionic plasmid DNA (pDNA) and the cationic gene carrier were labeled with QD (energy donor) and Cy5 (energy acceptor), respectively. QD-FRET nanocomplexes were formed through electrostatic complex coacervation. Upon excitation at 488 nm, QD-FRET-mediated Cy5 emission indicated formation of a compact and intact nanocomplex. The residence time (tR) can be calculated according to the distance (x) which measures from where the two streams meet to the position of reaction under investigation, and the mean flow speed (v). Due to the nature of laminar flow, mixing only takes place at the interface (center of each image), allowing precise calculation of mass transport as a function of tR. Temporal resolution can be adjusted by varying the applied flow rates. (Inset) FRET-mediated signal was observed immediately at the interface when the two streams met, indicating that binding was rapid, occurring within a few milliseconds according to the applied flow rates. Scale Bar: 100μm.
Discussion
- Significance of our work:
- This is the first attempt to monitor polymeric DNA nanocomplex self-assembly kinetics in real-time (millisecond resolution) through QD-FRET responses within a simple microfluidic chip.
- QD-mediated FRET provides a highly sensitive and quantitative indication of the onset of molecular interactions and throughout the self-assembly process, whereas microfluidics offers a well-controlled microenvironment to spatially analyze the process during the DNA nanocomplex synthesis.
- The integration of microfluidics and nanophotonics suggests a new and interesting approach to investigate any type of complexation reactions.
- The resulting QD-FRET polymeric DNA nanocomplexes could be readily applied for establishing structure-function relationships.1,2
Disclosures
The authors have nothing to disclose.
Acknowledgements
Funding support provided by NIH grant HL89764, NSF grants 0546012, 0730503 and 0725528.
Materials
| Material Name | Type | Company | Catalogue Number | Comment |
|---|---|---|---|---|
| SU-8 | MicroChem | SU-8 2025 | ||
| PDMS | Dow Corning | Sylgard 184 | ||
| Plasmid DNA | Clontech | pEGFP-C1 | 4.9 kb, MW = 3.3 x 106 | |
| LabelIT Biotin Labeling Kit | Mirus Bio LLC | MIR 3400 | Standard protocol yields labeling efficiency of approximately one label every 20-60 bp of double-stranded DNA, The density of labeling was adjusted in this work. | |
| Streptavidin 605QD | Invitrogen | Qdot® 605 ITK™ Streptavidin Conjugate | ||
| Cy5-NHS Ester | Amersham Biosciences | PA15101 | ||
| Chitosan | Vanson | 390 kDa | 83.5% deacetylated | |
| Cover Glass | Fisher | 12-545C | No. 1; Size: 40 x 22mm | |
| Gastight Glass Syringe | Hamilton | TLL series | 50μL to 500μL depending on sample volume | |
| Tygon Tubing | SmallParts | 0.02 ID, 100ft | Tygon Tubes Microbore, 0.02 ID, 100ft | |
| Connector | SmallParts | HTX-23R | Customized in length of 0.750″ | |
| Syringe Pump | Harvard Apparatus | PHD-2000 | ||
| CCD | Qimaging | Intensified Retiga Cooled | ||
| Microscope | Olympus | BX-51 | 100W mercury arc lamp | |
| ImageJ | NIH | v1.36b | http://rsb.info.nih.gov/ij | |
| Origin Pro8 | OriginLab | Student Version | ||
| Microscope Filter sets | Omega Optical | 475AF40 | Excitation filter in both channels | |
| Microscope Filter sets | Omega Optical | 595AF60 | Emission filter in 605QD channel | |
| Microscope Filter sets | Omega Optical | 670DF40 | Emission filter in QD-FRET channel | |
| Microscope Filter sets | Omega Optical | 500 DRLP | Long pass dichroic in 605QD channel | |
| Microscope Filter sets | Omega Optical | 595DRLP | Long pass dichroic in QD-FRET channel |
References
- Ho, Y. P., Chen, H. H., Leong, K. W., Wang, T. H. Evaluating the intracellular stability and unpacking of DNA nanocomplexes by quantum dots-FRET. J Control Release. 116, 83-89 (2006).
- Chen, H. H. Quantitative comparison of intracellular unpacking kinetics of polyplexes by a model constructed from quantum dot-FRET. Mol. Ther. 16, 324-332 (2008).
- Zhang, C. Y., Yeh, H. C., Kuroki, M., Wang, T. H. Single-Quantum-Dot-Based DNA Nanosensor. Nat Mat. 4, 826-831 (2005).

