Selection of Aptamers for Amyloid β-Protein, the Causative Agent of Alzheimer's Disease
Summary
Aptamers are short ribo-/deoxyribo-oligonucleotides selected by in-vitro evolution methods based on affinity for a specific target. Aptamers are molecular recognition tools with versatile therapeutic, diagnostic, and research applications. We demonstrate methods for selection of aptamers for amyloid β-protein, the causative agent of Alzheimer’s disease.
Abstract
Alzheimer’s disease (AD) is a progressive, age-dependent, neurodegenerative disorder with an insidious course that renders its presymptomatic diagnosis difficult1. Definite AD diagnosis is achieved only postmortem, thus establishing presymptomatic, early diagnosis of AD is crucial for developing and administering effective therapies2,3.
Amyloid β-protein (Aβ) is central to AD pathogenesis. Soluble, oligomeric Aβ assemblies are believed to affect neurotoxicity underlying synaptic dysfunction and neuron loss in AD4,5. Various forms of soluble Aβ assemblies have been described, however, their interrelationships and relevance to AD etiology and pathogenesis are complex and not well understood6. Specific molecular recognition tools may unravel the relationships amongst Aβ assemblies and facilitate detection and characterization of these assemblies early in the disease course before symptoms emerge. Molecular recognition commonly relies on antibodies. However, an alternative class of molecular recognition tools, aptamers, offers important advantages relative to antibodies7,8. Aptamers are oligonucleotides generated by in-vitro selection: systematic evolution of ligands by exponential enrichment (SELEX)9,10. SELEX is an iterative process that, similar to Darwinian evolution, allows selection, amplification, enrichment, and perpetuation of a property, e.g., avid, specific, ligand binding (aptamers) or catalytic activity (ribozymes and DNAzymes).
Despite emergence of aptamers as tools in modern biotechnology and medicine11, they have been underutilized in the amyloid field. Few RNA or ssDNA aptamers have been selected against various forms of prion proteins (PrP)12-16. An RNA aptamer generated against recombinant bovine PrP was shown to recognize bovine PrP-β17, a soluble, oligomeric, β-sheet-rich conformational variant of full-length PrP that forms amyloid fibrils18. Aptamers generated using monomeric and several forms of fibrillar β2-microglobulin (β2m) were found to bind fibrils of certain other amyloidogenic proteins besides β2m fibrils19. Ylera et al. described RNA aptamers selected against immobilized monomeric Aβ4020. Unexpectedly, these aptamers bound fibrillar Aβ40. Altogether, these data raise several important questions. Why did aptamers selected against monomeric proteins recognize their polymeric forms? Could aptamers against monomeric and/or oligomeric forms of amyloidogenic proteins be obtained? To address these questions, we attempted to select aptamers for covalently-stabilized oligomeric Aβ4021 generated using photo-induced cross-linking of unmodified proteins (PICUP)22,23. Similar to previous findings17,19,20, these aptamers reacted with fibrils of Aβ and several other amyloidogenic proteins likely recognizing a potentially common amyloid structural aptatope21. Here, we present the SELEX methodology used in production of these aptamers21.
Protocol
Part 1: Protein preparation and cross-linking
Initially, the protein used for SELEX is pretreated with 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP) to obtain homogeneous, aggregate-free preparations, as described previously23. This step is necessary because pre-formed aggregates induce rapid aggregation of amyloidogenic proteins, resulting in poor experimental reproducibility24, and are undesirable for selection of aptamers for unaggregated, non-fibrillar forms of the protein.
- Weigh out ~800 μg (~180 nmol) pure Aβ40 using a microbalance. Transfer the dry lyophilized peptide into labeled, silicon-coated, low-adsorbent 1.6-ml microfuge tubes.
- Dissolve the peptide in 100% 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP, 400 μl) to obtain 0.5 mM peptide solution as described previously23.
- Divide this solution into four 100-μl aliquots such that each tube contains ~45 nmol Aβ40 nominally. Proceed to removal of HFIP as described previously23.
- Before solubilizing the HFIP-treated peptides in buffer for cross-linking reactions, prepare the cross-linking and quenching reagents. Weigh out ammonium persulfate (APS, MW 228.2 g/mol) and prepare a solution of 40 mM in 10 mM sodium phosphate, pH 7.4. Mix using a vortex until the solution is clear.
- Prepare 2 mM solution of Tris(2,2-bipyridyl)dichlororuthenium(II) hexahydrate (RuBpy, MW 748.63 g/mol) in 10 mM sodium phosphate, pH 7.4. Mix using a vortex and verify complete dissolution. Protect the tube from light using aluminum foil.
- Prepare the quenching reagent. Weigh out dithiothreitol (DTT, MW 154.5 g/mol) and dissolve in deionized water or in 10-mM sodium phosphate, pH 7.4, to 1 M.
- Dissolve the HFIP-treated peptide exactly as described previously23,25 but aim to obtain ~60 μM peptide solution.
- Perform PICUP to generate a mixture of oligomeric Aβ40 as described previously23. A typical PICUP reaction is performed in a volume of 20 μl where the final concentrations of protein, RuBpy, and APS are 30 μM, 0.05 mM, and 1 mM, respectively23. Here, the mixture used for PICUP contains 108 μl protein, 6 μl RuBpy, 6 μl APS, and 1 μl DTT and the concentration of protein, RuBpy, and APS are all doubled relative to the typical reaction. This reduces the number of PICUP reactions to be performed and increases the protein concentration for desalting.
Part 2: Desalting the protein preparation
Before using the protein for SELEX, desalting is performed to remove the cross-linking reagents used for PICUP. This PICUP reaction mixture contains the cross-linking reagents and ~55 μM protein (nominal concentration).
- Remove the top cap off a 5-ml desalting column, support the column by a stand, place a beaker below the column outlet, and remove the outlet plug. Let the storage buffer flow through and collect into the beaker.
- Equilibrate the column in 10 mM ammonium acetate, pH 8.3. Add 5 resin-bed volumes (25 ml) of this buffer in 3-ml aliquots to the column and allow it to flow through.
- Meanwhile, label 8 low-adsorbent, silicon-coated 1.6-ml tubes and place them in a tube rack.
- After the column is equilibrated, apply a 0.5-0.7-ml aliquot of the PICUP reaction mixture per column per single desalting use (columns can be washed, stored, and equilibrated for later uses). Allow the protein mixture to soak into the column resin and discard the flow-through in the beaker.
- Place the first collection tube below the column outlet. Add 0.5 ml acetate buffer to the column and collect the first 0.5-ml fraction flowing through.
- Repeat steps 2.4 and 2.5, and collect up to eight 0.5-ml fractions in the corresponding tubes.
- Number another set of 8 low-adsorbent, silicon-coated, small 0.6-ml tubes and place them in a rack. Label another tube as “blank.” Take out 150 μl of each fraction and transfer into the labeled 0.6-ml tubes. Use 20 μl for SDS-PAGE analysis (Figure 1) as shown previously23.
- Transfer 150 μl 10 mM ammonium acetate, pH 8.3, into the blank tube. Use this tube to set the blank in a spectrophotometer.
- Measure the absorbance in 130 μl of each fraction at λ=280 nm in a Quartz cuvette.
- After the absorbance is measured, combine the fractions with the highest protein content (typically fractions 3-5), gently mix using a pipette, and keep a 10-μl aliquot for amino acid analysis to determine the actual protein concentration (not shown).
- Divide the sample into multiple aliquots of ~2 nmol protein per tube and lyophilize the samples in a lyophilizer.
- After completion of lyophilization, treat the samples with 100% HFIP as before.
- Store the tubes at -20 °C. Use one tube for each SELEX cycle (below).
Part 3: Amplification of the synthetic random ssDNA library by PCR
The synthetic ssDNA library used here for SELEX included 49 randomized nucleotides (A:T:G:C=25:25:25:25%) flanked by constant regions comprising cloning sites (BamHI, EcoRI) and a T7 promoter, as described previously26.
- To amplify the library, set up a standard PCR reaction as follows: 202 μl deionized water, 40 μl 10x Taq DNA polymerase buffer, 2 μl ssDNA template (0.5 nmol), 120 μl 25 mM MgCl2, 28 μl 10 mM deoxynucleoside triphosphate mix (dNTPs), 1 μl forward primer (300 pmol), 1 μl reverse primer (300 pmol), and 4 μl Taq polymerase.
- Perform the PCR reaction using a thermal cycler with the following settings: 5 min at 94 °C for initial denaturation, 20 cycles each of 94 °C for 30 sec, 52 °C for 30 sec for annealing, 72 °C for 30 sec for extension, and final extension at 72 °C for 7 min.
- After the completion of the PCR reaction, purify the amplified DNA product using the Qiagen QiaQuick PCR purification kit as per the manufacturer’s instructions. Generally, in these experiments the concentration and yield of DNA are 160 ± 10 ng/μl in 50 μl.
- Verify the amount of DNA after PCR by 2% agarose gel electrophoresis.
Part 4: Generation of 32P-labeled RNA by in-vitro transcription
- Set up the transcription reaction in an O-ring-capped, 1.6-ml tube according to the manufacturer’s instructions with some modifications as follows: 20 μl 5x T7 transcription buffer, 7.5 μl each of 100 mM rATP, rGTP, rUTP, 1 μl 100 mM rCTP, 2 μl α32P-CTP (3000 Ci/mmol), 5-10 μg purified DNA template (~30-40 μl of purified DNA product), 10 μl enzyme mix, and make up the final volume to 100 μl by adding nuclease-free water.
- Mix the solution gently by a pipette, centrifuge the mixture, and incubate at 37 °C overnight.
- At the end of the reaction, the DNA template has to be removed. Add RQ1 RNase-free DNase to a concentration of 1 U/μg of template DNA and incubate for 4 h at 37 °C to digest the DNA template.
- After 4 h, extract the RNA by adding 1 volume of citrate-saturated phenol:chloroform:isoamyl alcohol (125:24:1, pH 4.7). Mix by a vortex for ~1 min and centrifuge at 16,000 g for 2 min.
- Transfer the upper, aqueous phase to a fresh tube or discard the bottom phase by aspiration using a micropipette. Add 1 volume of chloroform:isoamyl alcohol (24:1), mix by a vortex for 1 min and centrifuge as described in 4.4.
- Transfer the upper, aqueous phase to a fresh tube or discard the bottom phase by aspiration using a micropipette. Residual chloroform can be removed by performing a quick spin (10 seconds) in a microcentrifuge and removal of the bottom phase with a micropipette. In this step, it is easier to remove the bottom phase rather than the supernate.
- To precipitate the RNA, add 0.1-volume equivalent of 3M sodium acetate, pH 5.2, and 1-volume equivalent of 2-propanol. Mix and place in a -20 °C freezer for 15 min.
- After 15 min, spin at top speed, preferably in a refrigerated microcentrifuge at 4 °C, for 20-30 min to precipitate the RNA product.
- Aspirate the supernate carefully, wash the RNA pellet with 0.5 ml of 70% ethanol, centrifuge at 4 °C and discard the ethanol by aspiration.
- Transfer the tube containing the RNA pellet to a heat block and dry the pellet at 37 °C for 5 min.
- Dissolve the RNA sample in 150 mM STE buffer, pH 8.0 (provided with the illustra ProbeQuant G-50 microcolumns) or nuclease-free water to a volume identical to that of the in-vitro transcription reaction i.e., 100 μl (step 4.1).
- Heat the tube containing the RNA product at 70 °C for 10 min in a heat block and mix by a vortex to facilitate RNA dissolution.
- Centrifuge at top speed for 1 min at room temperature.
- Keep a 1-μl aliquot of RNA in a labeled 0.6-ml tube for scintillation counting and TBE-urea polyacrylamide gel electrophoresis (Part 6).
Part 5: Removal of unincorporated nucleotides, RNA desalting, and scintillation counting
To remove the unincorporated nucleotides, use two G-50 columns according to the manufacturer’s instructions.
- Invert columns and mix by a vortex to resuspend the resin.
- Snap off the bottom closure of the columns at perforation using the plastic tool provided in the kit and make sure to leave the outlet untouched. Loosen the cap a quarter turn and place the columns into clean collection tubes provided in the G-50 kit.
- Spin the columns in the collection tubes at 730 g for 1 min to remove the storage buffer.
- Transfer columns to two new O-ring-capped, 1.6-ml tubes and load 50 μl of 32P-labeled RNA sample per column directly onto the resin without perturbing the resin.
- Spin at 730 g for 2 min to collect the purified labeled RNA. After this step, discard the columns.
- Transfer 1 μl of G-50-purified RNA to a 0.6-ml tube for scintillation counting and electrophoresis (Part 6). Store stock RNA at -20 °C until use for SELEX. It is desirable that RNA is used within 2 days after labeling to avoid its degradation and loss of activity.
- Use the two 1-μl RNA aliquots from steps 4.14 and 5.6 to measure the counts per min (cpm/μl) using a scintillation counter. Here, we use the Triathler bench-top scintillation counter.
- Transfer the tubes containing the RNA sample inside the plastic adaptor used for 32P counting, provided with Triathler. Put the tube and the adaptor inside the counting chamber, close the lid of the chamber, choose 32P counting, and press start to begin counting.
- Calculate % label incorporation i.e., % incorporation of α32P-CTP = (cpm/μl for “G-50” sample ÷ cpm/μl for “TOTAL” sample) x 100. TOTAL sample is the sample before G-50 purification.
- After scintillation counting and calculation of percent incorporation, calculate the yield of RNA and specific activity of RNA. For example, if the counts for a pool of RNA before (237370 cpm/μl) and after G-50 purification (191330 cpm/μl) yield 81% incorporation, then the following are calculated before obtaining the yield of RNA:
From step 4.1, the amount of α32P-CTP at 3000 Ci/mmol and 10 μCi/μl is: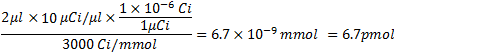 .
.
The amount of unlabeled CTP used is: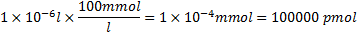 .
.
Total amount of CTP used is 100006.7pmol. The molecular weight of RNA is calculated as follows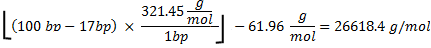
where 321.45 g/mol27 is the average mass of rNTPs, 100 bp is the number of bases in the sequence, and 17 bp is the number of bases in the T7 promoter that are not transcribed. Subtraction of 61.96 g/mole from the oligonucleotide molecular weight takes into account removal of HPO2 (63.98) and the addition of two hydrogen atoms (2.02)27. Because, of the 4 rNTPs, CTP is the limiting reagent, if all the CTP were to be incorporated the theoretical mass of RNA produced would have been: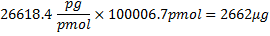 .
.
However, because of 81% incorporation, this mass is now 2156 μg. Counts per μg of RNA would then be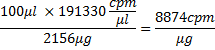
where 100 μl is the transcription reaction volume, and in terms of moles: 26618.4 μg/μmol x 8874 cpm/μg=2.4×108 cpm/μmol. From here, the amount of RNA in nmol and its appropriate volume used for incubation with protein can be calculated e.g., 5 μl RNA will contain ~4 nmol RNA, i.e.,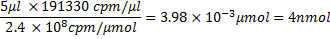
In these experiments, 300 pmol to 1 nmol protein and 4 nmol to 100 nmol RNA were used21.
Part 6: Characterization of the RNA product by electrophoresis and autoradiography
- To prepare the samples for electrophoresis, use the 1-μl aliquots of RNA before and after G-50 purification from steps 4.14 and 5.6, respectively. Add 4 μl STE buffer and 5 μl 2x Novex TBE-Urea Sample Buffer.
- Heat the samples at 70 °C for 5 min as usually recommended for RNA electrophoresis (heating was found to be unnecessary because the gel resolution of the RNA samples with and without heating is the same in this experimental setup).
- Assemble a 6% TBE-urea-polyacrylamide gel in the gel-running apparatus, fill the inside and outside chambers with 1x Novex TBE-Urea Running Buffer. Rinse the wells of the precast gel using the buffer by a 1-ml pipette.
- Centrifuge the RNA tubes and load the samples (10 μl total) using gel-loading tips. Run the gel at 180 V for 50 min.
- After 50 min, disassemble the gel by breaking apart the plastic mold, remove and discard only the shorter backing of the gel mold, but leave the longer backing as a support for the gel.
- Clean the surface of the work area with Decon (or other appropriate radioactivity decontaminant) making sure all the contaminating radioactive spots are removed.
- Lay out two layers of plastic Saran wrap; wrap the gel and the plastic backing in the two-plied plastic wrap.
- Expose the gel wrapped in plastic to a sheet of autoradiography x-ray film inside an exposure cassette in the dark room, under safe light. Leave the cassette at -20 °C for 60-90 min.
- Develop the film in the dark room under safe light after 60-90 min using an automatic film developer (Figure 2).
Part 7: RNA protein incubation and filter binding
First, the RNA and protein are incubated in solution and then the RNA sequences that bind to the protein are separated from the non-binders. As SELEX cycles progress, filter binding will give an indication of protein RNA binding enrichment.
- Incubate the RNA at 90 °C for 10 min for denaturation and then at room temperature for 10 min for slow renaturation.
- Dissolve the protein from step 2.12 in 8 μl 60 mM NaOH and add 36 μl nuclease-free deionized water. Sonicate the mixture for 1 min and add 36 μl 2x RNA binding buffer (20 mM Tris, 300 mM NaCl, 10 mM MgCl2, pH 7.5).
- Mix the appropriate amount of RNA with 20 μl 10x RNA binding buffer and make up the volume to 200 μl by adding nuclease-free water. Label the tube “negative control.”
- Mix 20 μl protein and the desired amount of RNA with 20 μl 10x RNA binding buffer and make up the volume to 200 μl with nuclease-free water. Label the tube “reaction”.
- Mix and incubate the tubes for 30 min at room temperature. Meanwhile prepare the filters and the filter-binding setup for the next step.
- Attach a 125-ml side-arm flask to a vacuum inlet. Place a pre-cleaned, porous glass support for the filter on the side-arm flask.
- Equilibrate 3 filter membranes in 2-3 ml of 1x RNA binding buffer in a 35×10-mm Petri dish for 10-15 min. The first filter will be used for adjusting the vacuum suction, the second will be used for RNA-alone, negative-control filter binding, and the third will be used for filter binding of the RNA-protein mixture.
- After 30 min, centrifuge the binding reaction and the control tubes at top speed for 5 min at room temperature.
- Turn on the vacuum and place the first membrane on the porous glass. Using a micropipette, drip 0.5 ml of RNA binding buffer onto the membrane and adjust the vacuum to allow slow flow of each buffer drop through the membrane.
- Place the second membrane onto the porous glass and using the same flow rate, apply the negative-control onto the membrane.
- Apply 4×0.5-ml aliquots of 1x RNA binding buffer to wash the membrane and discard the flow-through. Note that if pre-clearing is desired, the flow-through is kept for RNA extraction. Pre-clearing removes RNA sequences that bind to the filter.
- Remove the negative-control membrane and place into a correspondingly-labeled 1.6-ml tube for scintillation counting.
- Replace the porous glass with a pre-cleaned second porous glass support.
- Place the third disk onto the porous glass and apply the reaction mixture. Wash the filter as in step 7.11 and discard the flow-through. The number of washes can be increased in later SELEX cycles to increase the stringency of SELEX conditions.
- Remove the filter disk and place into a 1.6-ml tube labeled “reaction mixture” and keep for scintillation counting.
- Perform scintillation counting as in step 5.8 and note down the counts for the respective filters.
- Calculate the level of the filter-bound radioactivity compared to the total amount of radioactivity applied to the membranes (% binding). This will give an indication of binding enrichment as SELEX progresses.
Part 8: RNA extraction from the filters
RNA is extracted from the filters to obtain the sequences that bind to the protein. These sequences are amplified for the next SELEX cycle.
- After scintillation counting, remove the binding-reaction filter from the tube (from step 7.15) and place into a clean, dry, 35×10-mm Petri dish.
- Use a clean scalpel and a pair of tweezers to cut the membrane in small pieces.
- Using the tweezers, replace the cut pieces of the membrane in the same labeled tube from step 8.1.
- Add 400 μl elution buffer (7 M urea, 3 mM EDTA, 100 mM sodium citrate, pH 5.0) and incubate the tube at 95 °C for 10 min.
- Centrifuge the tube at top speed at room temperature, aspirate, and collect the extraction butter into a new labeled tube.
- Measure the remaining radioactivity counts in the tube containing the membrane pieces by scintillation counting to assess the extraction efficiency.
- Repeat the extraction process (steps 8.4-8.6) thrice. The efficiency after 3 extractions is usually ~95-96%.
- In the tubes containing the RNA extracts, add 1 volume (400 μl) of citrate-saturated (pH 4.7) phenol:chloroform:isoamyl alcohol (125:24:1). Mix by a vortex for ~1 min and centrifuge at 16,000 g for 2 min.
- Transfer the upper, aqueous phase to a fresh tube or discard the bottom phase by aspiration using a micropipette.
- Add 1 volume of chloroform:isoamyl alcohol (24:1), mix by a vortex for 1 min and centrifuge at 16,000 g for 2 min.
- Transfer the upper, aqueous phase to a fresh tube or discard the bottom phase by aspiration using a micropipette.
- To precipitate the RNA, add 0.1 volume of 3 M sodium acetate, pH 5.2, 3-4 μl glycogen (10 μg/μl) as RNA coprecipitant, and 1 volume equivalent of 2-propanol. Mix and place in a -20 °C freezer overnight.
- Spin at top speed, preferably in a microcentrifuge at 4 °C, for 20-30 min to precipitate the RNA from step 8.12.
- After centrifugation, the RNA separates in a liquid phase that is barely visible. Aspirate the supernate carefully without dislodging this barely visible precipitant phase at the bottom of the tube.
- Wash the RNA “pellet” with 0.5 ml of 70% ethanol, centrifuge for 5 min at top speed at 4 °C and discard the ethanol by aspiration without dislodging the barely visible precipitant phase.
- Dissolve the RNA pellet in 50 μl STE buffer and proceed to G-50 purification as in step 5.
Part 9: Reverse transcription and PCR for continuing SELEX cycles
To proceed to the next cycle of SELEX, RNA has to be reverse-transcribed to DNA and amplified by PCR.
- In 5 labeled 0.6-ml tubes, mix 3 μl of the purified, G-50-desalted RNA with 2 μl of 8-fold-diluted reverse primer. Label one tube ‘negative control.’
- Incubate the mixture at 70 °C for 5 min, and then on ice for another 5 min to allow hybridization of the primer to the RNA.
- To this mixture on ice, add 6.4 μl nuclease-free water, 4 μl ImProm-II 5x reaction buffer, 1.6 μl 25 mM MgCl2, 1 μl 10 mM dNTP mix, 1 μl RNasein Ribonuclease Inhibitor, 1 μl ImProm-II Reverse Transcriptase making up a total of 15 μl.
- For the negative control, add 7.4 μl nuclease-free water and leave out the reverse transcriptase. Label the tubes accordingly. The negative control is included to verify that contaminating DNA from a previous cycle is not amplified for the next SELEX cycle. This step tests the effectiveness of the digestion of DNA template by RQ1 RNase-free DNase in step 4.3.
- Use a thermal cycler to incubate the reaction mixture at 25 °C for 5 min, 42 °C for 1 h for annealing and extension of first-strand DNA, respectively, followed by 70 °C for 15 min to inactivate the reverse transcriptase.
- After the reverse-transcription reaction, set up the PCR mix. Add 30 μl nuclease-free water, 10 μl 10x Taq buffer, 30 μl 25 mM MgCl2, 7 μl 10 mM dNTP mix, 1 μl of each primer, and 1 μl Taq polymerase in all the tubes.
- Run the PCR program as in Part 3 for 9-14 cycles.
- Purify the DNA product as in step 3.3.
- Mix 8 μl of DNA product with 2 μl DNA loading dye and electrophorese on 2% agarose at 100 V for 15-20 min (Figure 3).
- DNA product is ready to be transcribed to labeled RNA as in Part 4 and used for the next cycle of SELEX.
Part 10: Representative Results
In SELEX experiments, the nature of Aβ40 oligomers used as the target, the quality of RNA used for each cycle, and successful RNA extraction and amplification after each cycle are important. We used PICUP to generate an oligomeric Aβ40 mixture for SELEX after purification and removal of the cross-linking reagents. The desalting experiments described in Part 2 usually lead to 50-55% protein loss. The protein amount and quality can be assessed using absorbance measurements (λ=280) and SDS-PAGE (Figure 1). The average absorbance profile of Aβ40 eluates from 5 individual experiments overlaying a typical SDS-PAGE profile of Aβ40 eluted in one of those experiments are shown in Figure 1. The data show that the protein consistently elutes off the column in fractions 3-5 and the cross-linking reagents elute after fraction 6 (increased absorbance in fraction 7, Figure 1). SDS-PAGE shows the typical Aβ40 oligomer distribution22. This distribution was reproducible after the protein fractions were lyophilized (2.11), treated with HFIP (2.11), resolubilized (7.1), and re-analyzed by SDS-PAGE.
Integrity of RNA for each SELEX cycle is also important for iterative progression of SELEX, especially when nuclease-susceptible ribo-oligonucleotides are used. After RNA amplification and labeling (Part 4), the quality of labeled RNA can be assessed by TBE-urea-polyacrylamide gel electrophoresis. A typical profile of an intact labeled RNA product before and after G-50 purification (Part 5) is shown in Figure 2.
After each SELEX cycle, RNA is extracted from the membrane after filter binding (Part 8) and reverse-transcribed (Part 9) to DNA for PCR amplification (Step 9.5). The DNA template from a previous cycle is then used to generate 32P-labeled RNA (Part 4) for the next cycle. If some contaminating DNA template from a previous cycle persists in the labeled RNA product after in-vitro transcription reactions (Part 4), the efficiency of SELEX cycles will be reduced, demanding more cycles. To control for this, after each SELEX cycle and the corresponding reverse-transcription PCR reaction, agarose electrophoresis is performed (9.12). Absence of DNA product in the negative-control reaction tubes (9.4) indicates successful removal of the DNA template originating from a previous SELEX cycle (Figure 3). If DNA amplification by PCR is observed in the negative-control tube, it is advised that the duration of incubation with RQ1 DNase (4.3) be prolonged. The manufacturer-recommended incubation duration with RQ1 DNase is 15 min, however, we found that longer incubations (4-5 h) were required to remove the template DNA completely (Figure 3).
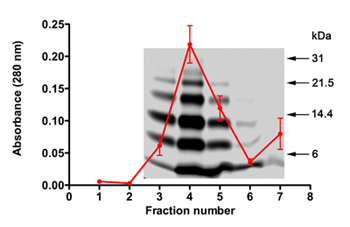
Figure 1. SDS-PAGE and absorbance profile of PICUP-generated, desalted Aβ40 oligomers. The absorbance profile from 5 individual desalting columns was averaged and overlain on a representative gel. Molecular weight markers are shown on the right.

Figure 2. TBE-urea-polyacrylamide gel electrophoresis of RNA product before and after G-50 purification. The migration direction of RNA product is from cathode to anode as indicated.
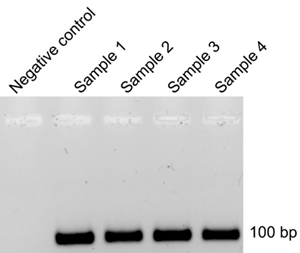
Figure 3. Agarose gel electrophoresis of DNA product after reverse-transcription and PCR amplification.
Discussion
The starting point of the SELEX process is synthesis of a random oligonucleotide library typically containing 1012-1015 sequences. In DNA SELEX, this library is used directly after a ssDNA pool is generated, whereas in RNA SELEX, demonstrated here, the ssDNA library is converted first to an RNA pool enzymatically by in-vitro transcription. Then, SELEX is performed iteratively whereby each cycle comprises exposure and binding of oligonucleotides to the intended target, partitioning of binders from non-binding sequences, and elution of binding sequences. In later cycles, the stoichiometry of the target and RNA and/or the number of washes can be altered, or competitive inhibitors can be added in the binding or wash buffer to increase the stringency of the SELEX conditions. Filter binding is a classic, simple, and fast method used for SELEX10, though numerous methods have been described28 for binding and partitioning. Filter binding is ideal for capture and selection of RNA-target interactions in solution. When the selection targets are small molecules or peptides, the pore size of the membrane used in filter binding becomes a limiting factor and an important consideration.
Following elution, the target-binding oligonucleotides are amplified and used for further cycles. When using DNA SELEX, the enriched pool can be amplified by PCR directly and used for the next cycle after digestion of the complementary sequence and DNA labeling. When using RNA SELEX, this pool has to be converted to DNA by reverse-transcription and then amplified by PCR, in-vitro transcribed, and labeled again for continuation to the next cycle. Usually, 8-20 such cycles are required to obtain an enriched aptamer pool, which is cloned subsequently and individual aptamers are sequenced and characterized. In studies using amyloid proteins, characterization of aptamers is particularly important because the inherent affinity of oligonucleotides for fibrillar amyloid structures potentially hinders development of aptamers specific for non-fibrillar amyloid proteins under physiological conditions21. This inherent, sequence-independent affinity of oligonucleotides may have led to generation of fibril-cross-reactive aptamers in studies aiming to generate aptamers for non-fibrillar amyloidogenic proteins17,19-21. Recently, Takahashi et al. reported generation of RNA aptamers against an oligomeric model of Aβ40 and showed reactivity with monomeric Aβ with micromolar affinity29. However, cross-reactivity of these aptamers with fibrillar Aβ40 or with other fibrillar amyloidogenic proteins was not determined.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by grants AG030709 from NIH/NIA and 07-65798 from the California Department of Public Health. We acknowledge Margaret M. Condron for peptide synthesis and amino acid analysis, Dr. Elizabeth F. Neufeld for helping and supporting the initial steps of the project, Dr. Chi-Hong B. Chen for providing support and reagents, and Dr. Andrew D. Ellington for helpful discussions.
Materials
| Material Name | Type | Company | Catalogue Number | Comment |
|---|---|---|---|---|
| Aβ40 | UCLA Biopolymers Laboratory | Lyophilized powder | ||
| MX5 Automated-S Microbalance | Mettler Toledo | |||
| Silicon-coated, 1.6-ml tubes | Denville Scientific | C19033 or C19035 | ||
| 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP) | TCI America | H0424 | Use in a fume hood. | |
| Ammonium persulfate | Sigma | A-7460 | Vortex until the solution is clear. APS is prepared freshly each time and should be used within 48 h. | |
| Tris(2,2-bipridyl)dichlororuthenium(II) hexahydrate | Sigma | 224758-1G | Vortex until the solution is clear. Cover the RuBpy tube with foil to protect the reagent from ambient light. RuBpy is prepared freshly each time and should be used within 48 h. | |
| Dithiothreitol (DTT) | Sigma | 43815 | ||
| D-Salt™ Excellulose™ desalting columns | Thermo Scientific | 20449 | ||
| Ammonium acetate | Fisher Scientific | A637-500 | ||
| Silicon-coated, 0.6-ml tubes | Denville Scientific | C19063 | ||
| Novex Tricine Gels (10–20%) | Invitrogen | EC6625B0X | 10-well; mini size (8 cm X 8 cm); 25 μl loading volume per well; separation range 5 kDa to 40 kDa | |
| Quartz cuvette | Hellma | 105.250-QS | ||
| Beckman DU 640 spectrophotometer | Beckman | |||
| ssDNA library | Integrated DNA Technologies | Custom-ordered | The library was designed to contain 49 random nucleotides flanked by two constant regions containing primer-binding and cloning sites: 5′-TAA TAC GAC TCA CTA TAG GGA ATT CCG CGT GTG C (N:25:25:25:25%) (N)49 G TCC GTT CGG GAT CCT C-3′ | |
| Taq DNA polymerase | USB Corporation | 71160 | Recombinant Thermus aquaticus DNA Polymerase supplied with 10× PCR Buffer and a separate tube of 25 mM MgCl2 for routine PCR. | |
| PCR Nucleotide Mix, 10 mM solution | USB Corporation | 77212 | (10 mM each dATP, dCTP, dGTP, dTTP) | |
| Forward primer | Integrated DNA Technologies | Custom-ordered | 5′-TAA TAC GAC TCA CTA TAG GGA ATT CCG CGT GTG C-3′ | |
| Reverse primer | Integrated DNA Technologies | Custom-ordered | 5′-GAG GAT CCC GAA CGG AC-3′ | |
| Thermal cycler | Denville Scientific | Techne TC-312 | ||
| QIAquick PCR Purification Kit (50) | QIAGEN | 28104 | ||
| Agarose | Denville Scientific | CA3510-8 | ||
| Conical, sterile 1.6-ml tubes with caps attached with O-rings | Denville Scientific | C19040-S | ||
| RiboMAX™ Large Scale RNA Production System–T7 | Promega | P1300 | The kit contains: 120 μl Enzyme Mix (RNA polymerase, recombinant RNasin® ribonuclease inhibitor and recombinant inorganic pyrophosphatase); 240 μl transcription 5 buffer; 100 μl each of 4 rNTPs, 100 mM; 110 U RQ1 RNase-free DNase, 1 U/μl; 10 μl linear control DNA, 1 mg/ml; 1 ml 3M sodium acetate (pH 5.2); 1.25 ml nuclease-Free water | |
| α-32P-cytidine 5′-triphosphate, 250 μCi (9.25 MBq), | Perkin Elmer | BLU008H250UC | Specific Activity: 3000 Ci (111 TBq)/mmol, 50 mM Tricine (pH 7.6) | |
| Citrate-saturated phenol:chloroform:isoamyl alcohol (125:24:1, pH 4.7) | Sigma (Fluka) | 77619 | ||
| Chloroform:Isoamyl alcohol (24:1) | Sigma | C0549 | ||
| Absolute ethanol for molecular biology | Sigma | E7023 | ||
| Z216-MK refrigerated microcentrifuge | Denville Scientific | C0216-MK | ||
| illustra ProbeQuant™ G-50 Micro Columns | GE Healthcare | Obtained from Fisher Scientific (45-001-487) | Prepacked with Sephadex™ G-50 DNA Grade and pre-equilibrated in STE buffer containing 0.15% Kathon as Biocide | |
| Triathler Bench-top Scintillation counter | Hidex Oy, Turku, Finland | Triathler LSC Model: 425-034 | ||
| Novex® TBE-Urea Sample Buffer (2×) | Invitrogen | LC6876 | ||
| 6% TBE-Urea Gels 1.0 mm, 10 wells | Invitrogen | EC6865BOX | ||
| Novex® TBE Running Buffer (5×) | Invitrogen | LC6675 | ||
| Radioactivity decontaminant | Fisher Scientific | 04-355-67 | ||
| Gel-loading tips | Denville Scientific | P3080 | ||
| XCell SureLock Mini-Cell | Invitrogen | EI0001 | XCell SureLock Mini-Cell | |
| Autoradiography film | Denville Scientific | E3018 | Use in complete darkness | |
| Autoradiography film, Hyperfilm™ ECL | Amersham Biosciences | RPN3114K | Can be used under red safe light. | |
| Membrane discs | Millipore | GSWP02500 | Mixed cellulose ester, hydrophilic, 0.22-μm disc membranes | |
| Fritted glass support base for 125-ml flask | VWR | 26316-696 | ||
| Petri dishes | Fisher Scientific | 08-757-11YZ | ||
| Urea | Fisher Scientific | AC32738-0050 | ||
| EDTA | Fisher Scientific | 118430010 | ||
| Glycogen | Sigma | G1767 | ||
| 2-Propanol for molecular biology | Sigma | I9516 | ||
| Recombinant RNase inhibitor | USB Corporation | 71571 | ||
| ImProm-II™Reverse Transcription System | Promega | A3802 | ||
| Recombinant RNase inhibitor | USB Corporation | 71571 | ||
| RapidRun™ Loading Dye | USB Corporation | 77524 |
References
- Monien, B. H., Apostolova, L. G., Bitan, G. Early diagnostics and therapeutics for Alzheimer’s disease-how early can we get there. Expert. Rev. Neurother. 6, 1293-1306 (2006).
- Nestor, P. J., Scheltens, P., Hodges, J. R. Advances in the early detection of Alzheimer’s disease. Nat. Med. 10, S34-S41 (2004).
- Kawas, C. H. Clinical practice. Early Alzheimer’s disease. N. Engl. J. Med. 349, 1056-1063 (2003).
- Haass, C., Selkoe, D. J. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid β-peptide. Nat. Rev. Mol. Cell. Biol. 8, 101-112 (2007).
- Kirkitadze, M. D., Bitan, G., Teplow, D. B. Paradigm shifts in Alzheimer’s disease and other neurodegenerative disorders: the emerging role of oligomeric assemblies. J. Neurosci. Res. 69, 567-577 (2002).
- Rahimi, F., Shanmugam, A., Bitan, G. Structure-function relationships of pre-fibrillar protein assemblies in Alzheimer’s disease and related disorders. Curr. Alzheimer Res. 5, 319-341 (2008).
- Jayasena, S. D. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 45, 1628-1650 (1999).
- Bunka, D. H., Stockley, P. G. Aptamers come of age – at last. Nat. Rev. Microbiol. 4, 588-596 (2006).
- Ellington, A. D., Szostak, J. W. In vitro selection of RNA molecules that bind specific ligands. Nature. 346, 818-822 (1990).
- Tuerk, C., Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 249, 505-510 (1990).
- Lee, J. F., Stovall, G. M., Ellington, A. D. Aptamer therapeutics advance. Curr. Opin. Chem. Biol. 10, 282-289 (2006).
- Weiss, S. RNA aptamers specifically interact with the prion protein PrP. J. Virol. 71, 8790-8797 (1997).
- Bibby, D. F. Application of a novel in vitro selection technique to isolate and characterise high affinity DNA aptamers binding mammalian prion proteins. J. Virol. Methods. 151, 107-115 (2008).
- Rhie, A. Characterization of 2′-fluoro-RNA aptamers that bind preferentially to disease-associated conformations of prion protein and inhibit conversion. J. Biol. Chem. 278, 39697-39705 (2003).
- King, D. J., Safar, J. G., Legname, G., Prusiner, S. B. Thioaptamer interactions with prion proteins: sequence-specific and non-specific binding sites. J. Mol. Biol. 369, 1001-1014 (2007).
- Proske, D. Prion-protein-specific aptamer reduces PrPSc formation. ChemBioChem. 3, 717-725 (2002).
- Murakami, K., Nishikawa, F., Noda, K., Yokoyama, T., Nishikawa, S. Anti-bovine prion protein RNA aptamer containing tandem GGA repeat interacts both with recombinant bovine prion protein and its β isoform with high affinity. Prion. 2, 73-80 (2008).
- Luhrs, T., Zahn, R., Wuthrich, K. Amyloid formation by recombinant full-length prion proteins in phospholipid bicelle solutions. J. Mol. Biol. 357, 833-841 (2006).
- Bunka, D. H. Production and characterization of RNA aptamers specific for amyloid fibril epitopes. J. Biol. Chem. 282, 34500-34509 (2007).
- Ylera, F., Lurz, R., Erdmann, V. A., Furste, J. P. Selection of RNA aptamers to the Alzheimer’s disease amyloid peptide. Biochem. Biophys. Res. Commun. 290, 1583-1588 (2002).
- Rahimi, F., Murakami, K., Summers, J. L., Chen, C. H., Bitan, G. RNA aptamers generated against oligomeric Aβ40 recognize common amyloid aptatopes with low specificity but high sensitivity. PLoS ONE. 4, e7694-e7694 (2009).
- Bitan, G., Lomakin, A., Teplow, D. B. Amyloid β-protein oligomerization: prenucleation interactions revealed by photo-induced cross-linking of unmodified proteins. J. Biol. Chem. 276, 35176-35184 (2001).
- Rahimi, F., Maiti, P., Bitan, G. Photo-induced cross-linking of unmodified proteins (PICUP) applied to amyloidogenic peptides. J. Vis. Exp. , (2009).
- Bitan, G., Fradinger, E. A., Spring, S. M., Teplow, D. B. Neurotoxic protein oligomers-what you see is not always what you get. Amyloid. 12, 88-95 (2005).
- Bitan, G. Structural study of metastable amyloidogenic protein oligomers by photo-induced cross-linking of unmodified proteins. Methods Enzymol. 413, 217-236 (2006).
- Chen, C. H., Chernis, G. A., Hoang, V. Q., Landgraf, R. Inhibition of heregulin signaling by an aptamer that preferentially binds to the oligomeric form of human epidermal growth factor receptor-3. Proc. Natl. Acad. Sci. USA. 100, 9226-9231 (2003).
- Adams, D. S. . Lab math: a handbook of measurements, calculations, and other quantitative skills for use at the bench. , (2003).
- Gopinath, S. C. Methods developed for SELEX. Anal. Bioanal. Chem. 387, 171-182 (2007).
- Takahashi, T., Tada, K., Mihara, H. RNA aptamers selected against amyloid β-peptide (Aβ) inhibit the aggregation of Aβ. Mol. Biosyst. 5, 986-991 (2009).

