Imaging of mtHyPer7, a Ratiometric Biosensor for Mitochondrial Peroxide, in Living Yeast Cells
Summary
Hydrogen peroxide (H2O2) is both a source of oxidative damage and a signaling molecule. This protocol describes how to measure mitochondrial H2O2 using mitochondria-targeted HyPer7 (mtHyPer7), a genetically encoded ratiometric biosensor, in living yeast. It details how to optimize imaging conditions and perform quantitative cellular and subcellular analysis using freely available software.
Abstract
Mitochondrial dysfunction, or functional alteration, is found in many diseases and conditions, including neurodegenerative and musculoskeletal disorders, cancer, and normal aging. Here, an approach is described to assess mitochondrial function in living yeast cells at cellular and subcellular resolutions using a genetically encoded, minimally invasive, ratiometric biosensor. The biosensor, mitochondria-targeted HyPer7 (mtHyPer7), detects hydrogen peroxide (H2O2) in mitochondria. It consists of a mitochondrial signal sequence fused to a circularly permuted fluorescent protein and the H2O2-responsive domain of a bacterial OxyR protein. The biosensor is generated and integrated into the yeast genome using a CRISPR-Cas9 marker-free system, for more consistent expression compared to plasmid-borne constructs.
mtHyPer7 is quantitatively targeted to mitochondria, has no detectable effect on yeast growth rate or mitochondrial morphology, and provides a quantitative readout for mitochondrial H2O2 under normal growth conditions and upon exposure to oxidative stress. This protocol explains how to optimize imaging conditions using a spinning-disk confocal microscope system and perform quantitative analysis using freely available software. These tools make it possible to collect rich spatiotemporal information on mitochondria both within cells and among cells in a population. Moreover, the workflow described here can be used to validate other biosensors.
Introduction
Mitochondria are essential eukaryotic cellular organelles that are well known for their function in producing ATP through oxidative phosphorylation and electron transport1. In addition, mitochondria are sites for calcium storage, the synthesis of lipids, amino acids, fatty acid and iron-sulfur clusters, and signal transduction2,3. Within cells, mitochondria form a dynamic network with characteristic morphology and distribution, which varies according to cell type and metabolic state. Furthermore, although mitochondria can fuse and divide, not all the mitochondria in a cell are equivalent. Numerous studies have documented the functional heterogeneity of mitochondria within individual cells in attributes such as membrane potential and oxidative state4,5,6. This variation in mitochondrial function is due in part to damage to the organelle from mtDNA mutations (which occur at a higher rate than in nuclear DNA) and to oxidative damage by reactive oxygen species (ROS) generated both within and outside the organelle7,8,9. Damage to the organelle is mitigated by mitochondrial quality control mechanisms that repair the damage or eliminate mitochondria that are damaged beyond repair10.
Hydrogen peroxide (H2O2) is a reactive oxygen species that is a source of oxidative damage to cellular proteins, nucleic acids, and lipids. However, H2O2 also serves as a signaling molecule that regulates cellular activities through the reversible oxidation of thiols in target proteins11,12. H2O2 is produced from electrons that leak from the mitochondrial electron transport chain and by specific enzymes, such as NADPH oxidase and monoamine oxidases13,14,15,16,17,18,19,20. It is also inactivated by antioxidant systems, including those based on thioredoxin and glutathione21,22,23. Thus, analysis of mitochondrial H2O2 levels is critical for understanding the role of this metabolite in normal mitochondrial and cellular function and under conditions of oxidative stress.
The overall goal of this protocol is to detect mitochondrial H2O2 using a genetically encoded ratiometric H2O2 biosensor, HyPer7, that is targeted to the organelle (mtHyPer7). mtHyPer7 is a chimera consisting of the mitochondrial signal sequence from ATP9 (the Su9 presequence), a circularly permuted form of green fluorescent protein (GFP), and the H2O2-binding domain of the OxyR protein from Neisseria meningitidis24 (Figure 1). In circularly permuted GFP, the N- and C-termini of native GFP are fused and new termini are formed near the chromophore, which impart greater mobility to the protein and greater lability of its spectral characteristics compared to native GFP25. The interaction of mtHyPer7's OxyR domain with H2O2 is high-affinity, H2O2-selective, and leads to reversible oxidation of the conserved cysteine residues and disulfide bridge formation. Conformational changes associated with the oxidation of OxyR are transferred to the circularly permuted GFP in mtHyPer7, which results in a spectral shift in the excitation maximum of the mtHyPer7 chromophore from 405 nm in the reduced state to 488 nm in the H2O2-oxidized state26. Thus, the ratio of fluorescence from mtHyPer7 in response to excitation at 488 nm versus 405 nm reflects the oxidation of the probe by H2O2.
Ideally, a biosensor should provide a real-time, absolute, quantitative readout of its target molecule. Unfortunately, however, this is not always possible in real-world measurements. In the case of oxidation sensors, such as mtHyPer7, the real-time readout is affected by the rate of reduction of the disulfide bridge. The reduction systems used by ROS biosensors differ, and these can dramatically alter probe response dynamics-as shown by the comparison of HyPer7, reduced by the thioredoxin system, and roGFP2-Tsa2ΔCR, reduced by glutathione-in yeast cytosol27. Thus, to draw a conclusion about the relative H2O2 concentration from mtHyPer7 ratios, one must assume that the reduction system maintains a constant capacity during the experiment. Notwithstanding these considerations, HyPer7 and related probes have been used in various contexts to obtain information about H2O2 in living cells25,28,29.
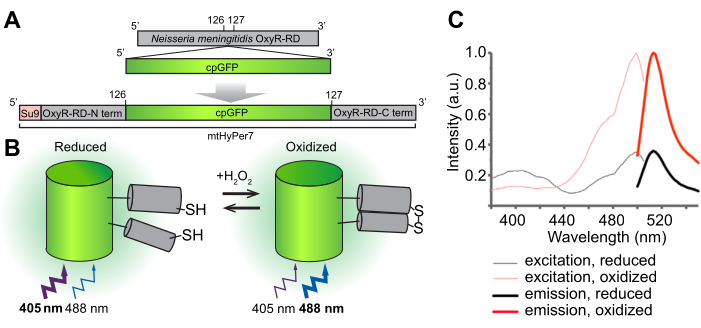
Figure 1: Design, molecular mechanism, and excitation/emission spectra of the H2O2 biosensor mtHyPer7. (A) The mtHyPer7 probe is derived by inserting circularly permuted GFP into the OxyR-RD domain from Neisseria meningitidis. It contains the mitochondrial targeting sequence from subunit 9 of the ATP synthase from Neurospora crassa (Su9). (B) Illustration of the H2O2-sensing mechanism of mtHyPer7. Oxidation of cysteines in the RD domain increases fluorescence emission upon excitation at 488 nm and decreases emission produced by excitation at 405 nm. (C) Excitation and emission spectra of HyPer7 in oxidized and reduced forms. This figure is reprinted with permission from Pak et al.24. Abbreviations: GFP = green fluorescent protein; cpGFP = circularly permuted GFP. Please click here to view a larger version of this figure.
This ratiometric imaging of mtHyPer7 offers important benefits for mitochondrial H2O2 quantitation24,27; it provides an internal control for probe concentration. In addition, the shift in excitation peak produced by H2O2 exposure is not complete, even in saturating concentrations of H2O2. Thus, ratio imaging can increase the sensitivity by incorporating two spectral points in the analysis.
The mtHyPer7 probe used here has a very high affinity for H2O2 and relatively low sensitivity to pH24, and has been successfully targeted to Caenorhabditis elegans mitochondria30. This protein has also been used in yeast27,31. However, previous studies relied on plasmid-borne expression of mtHyPer7, which results in cell-to-cell variability in probe expression27. In addition, the construct described in this protocol was integrated into a conserved, gene-free region on chromosome X32 using a CRISPR-based approach for marker-free integration. Expression of the integrated biosensor gene is also controlled by the strong constitutive TEF1 promoter (Figure 2). As a result, there is more stable, consistent expression of the biosensor in yeast cell populations compared to that observed using plasmid-borne biosensor expression, and cells bearing the biosensor can be propagated without the need for selective media.
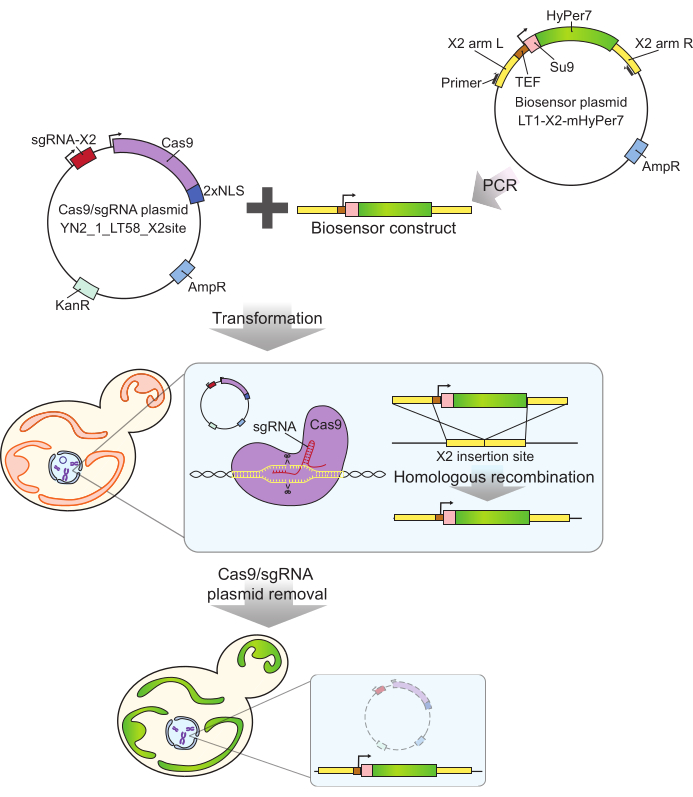
Figure 2: Generation of mtHyPer7-expressing cells by CRISPR. The Cas9 and sgRNA-containing plasmid (YN2_1_LT58_X2site) and PCR-amplified mtHyPer7-containing biosensor construct are introduced into budding yeast cells by lithium acetate transformation. The gene-free insertion site on chromosome X (X2) is recognized and cut by Cas9 protein with the sgRNA, and the biosensor is integrated into the genome by homologous recombination. After identification of the successful transformants by microscopic screening, colony PCR, and sequencing, the Cas9 plasmid is removed (cured) by growth in non-selective media. Abbreviations: sgRNA = single guide RNA; TEF = transcription enhancer factor. Please click here to view a larger version of this figure.
Finally, mtHyPer7 offers advantages over other ROS biosensors. For example, organic dyes used to detect ROS (e.g., dihydroethidium [DHE]2 and MitoSOX3) can produce uneven or nonspecific staining and are often delivered in solvents such as ethanol or dimethyl sulfoxide, which require additional controls for solvent effects. Another class of ROS biosensors are fluorescence resonance energy transfer (FRET)-based biosensors (e.g., Redoxfluor for cellular redox state4, and the peroxide sensors HSP-FRET5, OxyFRET6, and PerFRET6). These probes are genetically encoded and highly sensitive in principle and can be quantitatively targeted to mitochondria using well-characterized mitochondrial signal sequences. However, there are challenges in the use of FRET-based probes, including background due to cross-excitation and bleed-through, and stringent requirements for the proximity and orientation of the fluorophores for FRET to occur33,34. In addition, FRET probes consist of two fluorescent proteins that require larger constructs for expression in cells of interest compared to spectra-shifting probes. The protocol described here was developed to take advantage of the strengths of the HyPer7-based biosensor, and to use this compact, ratiometric, high-affinity, genetically encoded probe for quantitative imaging of peroxide in mitochondria in living yeast.
Protocol
1. Generation of the biosensor plasmid, integration into the yeast genome, and assessment of the targeting of mtHyPer7 to mitochondria and effects on mitochondrial morphology, cellular growth rates, or oxidative stress sensitivity
NOTE: Refer to Supplemental File 1, Supplemental Table S1, Supplemental Table S2, and Supplemental Table S3 for biosensor plasmid construction, strains, plasmids, and primers, respectively, used for biosensor construction and characterization. See the Table of Materials for details related to all materials, reagents, and instruments used in this protocol.
- Amplify the biosensor construct from the biosensor plasmid using the primers Y290 and Y291 (Figure 2) via polymerase chain reaction (PCR).
- Mix 500 ng of the Cas9/guide RNA plasmid (YN2_1_LT58_X2site32) with 50 µL of the PCR product from step 1.1 (the biosensor construct). Transform the mixture into budding yeast using the lithium acetate method35 and select transformants on YPD plates containing 200 mg/mL G418.
- Screen candidate transformants by PCR amplification of genomic DNA using the primers Y292 and Y293. For transformants bearing an insert of the expected size (positive: 3.5 kb; negative: 0.3 kb), sequence the inserted region for further validation.
- Evaluate the effect of the biosensor on growth and mitochondrial function (Figure 3). If the biosensor appears to affect the function of cells or mitochondria, try to minimize the effect by changing the promoter, integration site, or parent strain.
- Determine the effect of the biosensor construct on growth rate in the presence and absence of an oxidative stressor (e.g., paraquat), as described previously36.
- Dilute mid-log phase cells to a 600 nm optical density (OD600) of 0.0035 in 200 µL of corresponding media with and without treatment into each well of a 96-well flat-bottom plate. Measure the optical density of the culture every 20 min for 72 h using a plate reader. Calculate the maximum growth rate (slope) as the maximum change in OD over a 240 min interval during the 72 h course.
- Visualize the cells by fluorescence microscopy and evaluate the brightness and mitochondrial morphology as described previously37.
- Grow the cells to the mid-log phase, stain the mitochondria with 250 nM MitoTracker Red for 30 min, and wash twice before imaging. Capture Z stacks of 6 µm depth at 0.3 µm intervals on a wide-field or confocal fluorescence microscope and inspect visually. Look for mitochondria forming elongated tubular structures.
- Determine the effect of the biosensor construct on growth rate in the presence and absence of an oxidative stressor (e.g., paraquat), as described previously36.
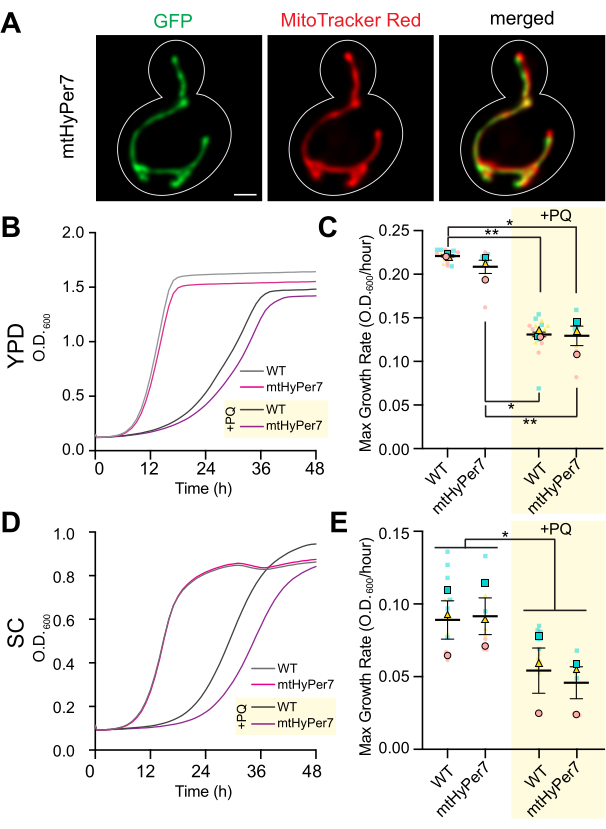
Figure 3: mtHyPer7 is targeted to mitochondria and does not affect mitochondrial morphology, cell growth, or sensitivity to oxidative stress. (A) Mitochondrial morphology in living yeast cells expressing mtHyPer7. Left panel: mtHyPer7 visualized with 488 nm excitation. Middle panel: mitochondria labeled with 250 nM MitoTracker Red. Right panel: merged images. Maximum-intensity projections are shown. The cell outline is shown in white. Scale bar = 1 µm. (B,C) Growth curves and maximum growth rate of wild-type cells and cells expressing mtHyPer7 grown in the presence (+PQ) or absence of 2.5 mM paraquat in YPD media. (D,E) Growth curves and maximum growth rate of wild-type and mtHyPer7-expressing cells grown in the presence (+PQ) or absence of 2.5 mM paraquat in SC media. All growth curves are the means of three independent replicates. Maximum growth rates are represented as mean ± standard error of the mean (SEM). Growth curve analysis was done by diluting mid-log phase cells to an OD600 of 0.0035 in 200 µL of corresponding media into each well of a 96-well flat-bottom plate. The OD of the culture was measured every 20 min for 72 h using a plate reader. Each strain/condition was plated in triplicate and the averaged growth rate was plotted. The maximum growth rate (slope) was calculated using the changes in OD over a 240 min interval over the course of 72 h. Abbreviations: WT = wild type; PQ = paraquat. Please click here to view a larger version of this figure.
2. Cell culture and preparation for imaging (Figure 4)
- Propagate the cells in synthetic medium to the mid-log phase for imaging. A 5 mL mid-log phase culture provides enough cells for four or five slides and a total of >100 budded cells for analysis.
- In the morning the day before the experiment, prepare liquid precultures by inoculating 5 mL of synthetic complete (SC) medium in a 50 mL conical-bottom tube with a single colony of yeast cells.
- Incubate the preculture in an orbital shaker at 200 rpm and 30 °C for 6-8 h. Measure the OD600 of the preculture, which should be at the mid-log phase: ~0.5-1 × 107 cells/mL, OD600 ~0.1-0.3.
- Prepare a mid-log phase culture using the preculture for imaging the next day. Calculate the amount of preculture needed for a mid-log phase culture after overnight growth (8-16 h). When the cells are grown in SC medium, the doubling time is ~2 h. Therefore, the volume (V) of preculture for inoculation of a 5 mL overnight culture is calculated using equation (1):
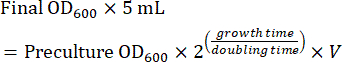 (1)
(1) - Inoculate 5 mL of SC medium in a 50 mL conical-bottom tube with the amount of preculture calculated in step 2.1.3. Grow in an orbital shaker at 200 rpm and 30 °C for 8-16 h.
- On the day of imaging, confirm that the culture generated in step 2.1.4 is at the mid-log phase (OD600 ~0.1-0.3). Concentrate the cells from 1 mL of the mid-log phase culture by centrifugation at 6,000 × g for 30 s. Remove the supernatant, leaving 10-20 µL of the supernatant in the tube. Resuspend the cell pellet by gently mixing with residual media using a micropipette.
- Use an air blower or lint-free tissue to remove dust from a glass microscope slide and add 1.8 µL of the cell suspension to the slide. Cover the cells with a #1.5 (170 µm thick) glass coverslip, lowering the coverslip slowly at an angle to avoid introducing bubbles.
- Image immediately and discard the slide after 10 min of imaging.
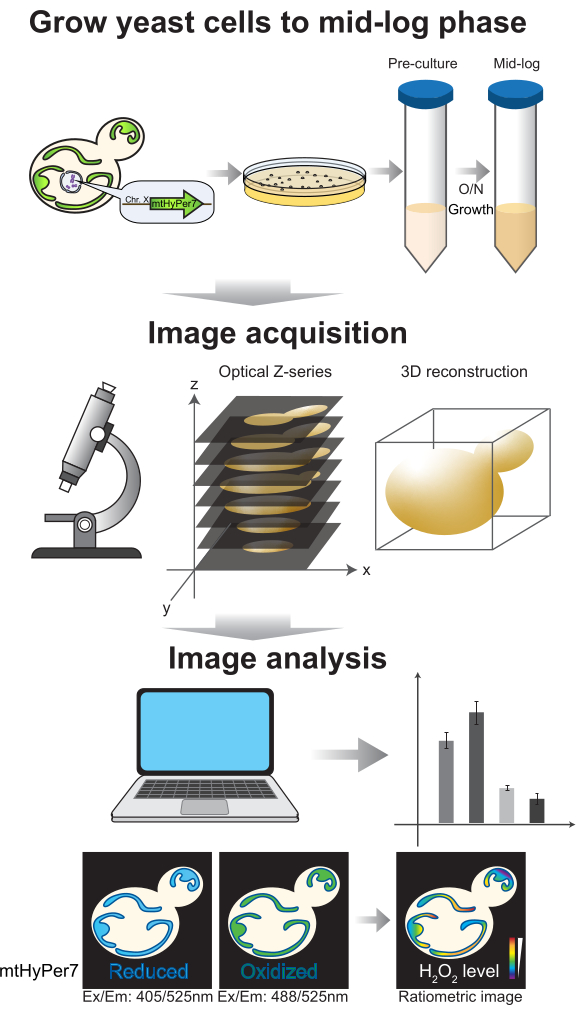
Figure 4: Cell growth and imaging. Budding yeast cells expressing mtHyPer7 are grown to the mid-log phase. Z series images are collected by spinning-disk confocal microscopy and then subjected to 3D reconstruction and analysis. See protocol sections 2-3. Please click here to view a larger version of this figure.
3. Optimization of imaging conditions and image collection (Figure 5)
- Select an imaging mode. For optical sectioning without postprocessing, a spinning-disk confocal instrument is preferred. However, if the signal is low, or this instrument is unavailable, use wide-field imaging, ensuring that the wide-field images are deconvolved to remove out-of-focus blur, as described previously38.
- Select optics. Use a high-numerical aperture oil-immersion lens, such as 100x/1.45 Plan-Apochromat.
- Select excitation and emission wavelengths. For spinning-disk confocal imaging, use a laser excitation at 405 nm and 488 nm and a standard GFP emission filter. For wide-field imaging, use a light emitting diode (LED) or lamp excitation, but ensure that the filter setup allows variation of the excitation while passing emission through the standard GFP filter.
NOTE: This can be accomplished, for example, by removing the excitation filter from a GFP cube and using an LED or filter wheel to select the excitation. - Select exposure time and illumination intensity.
- Establish acquisition conditions that yield a readily detectable signal and acceptable resolution in each fluorescence channel. For example, on a spinning-disk confocal microscope with an sCMOS camera, use 2 x 2 binning, 20-40% laser power, and 200-600 ms exposure.
- Examine the image histogram. In a 12-bit image (4,096 possible gray levels), make sure that the total dynamic range of the pixel values is at least several hundred gray levels, without saturation (Figure 5A). In addition, make sure the range is one order of magnitude higher than the noise level. Calculate the noise level as the standard deviation of pixel values in a cell-free area of an image, measured as described in step 4.1.3.1.
- Test for photobleaching or imaging-induced mitochondrial stress under the selected imaging conditions. If excessive photobleaching or an increase in mitochondrial H2O2 is observed, reduce the laser power and increase the exposure or binning.
- Collect a time-lapse series of images, with no delay between acquisitions, to assess the effects of the imaging conditions on signal stability and oxidative stress in mitochondria. Using the microscope acquisition software or ImageJ, measure the average pixel value in each fluorescence channel to confirm signal stability (<5% change; Figure 5B). If the experiment does not involve time-lapse imaging, make sure that the fluorescence is stable over two or three repeated Z stacks (25-35 exposures).
NOTE: A decrease in fluorescence of both channels may indicate photobleaching; however, a decrease in 405 nm-excited fluorescence accompanied by an increase in the 488 nm-excited channel may indicate increased mitochondrial H2O2 and imaging-induced oxidative stress in the organelle.
- Collect a time-lapse series of images, with no delay between acquisitions, to assess the effects of the imaging conditions on signal stability and oxidative stress in mitochondria. Using the microscope acquisition software or ImageJ, measure the average pixel value in each fluorescence channel to confirm signal stability (<5% change; Figure 5B). If the experiment does not involve time-lapse imaging, make sure that the fluorescence is stable over two or three repeated Z stacks (25-35 exposures).
- Acquire images. If collecting Z stacks, ensure that the Z interval is the same for all images in the dataset, and include the entire cell. For budding yeast, image with a Z interval of 0.5 µm and a total stack depth of 6 µm. Acquire transmitted-light images to document cell boundaries.
- Analyze images manually or semi-automatically via the scripts available in Supplemental File 2 and at https://github.com/theresaswayne/biosensor, using the Fiji distribution of ImageJ39 and the statistical software R (Figure 6). In the software instructions, hierarchical menu commands are shown in boldface with "|", indicating a sequence of menu selections. Options and buttons are shown in boldface.
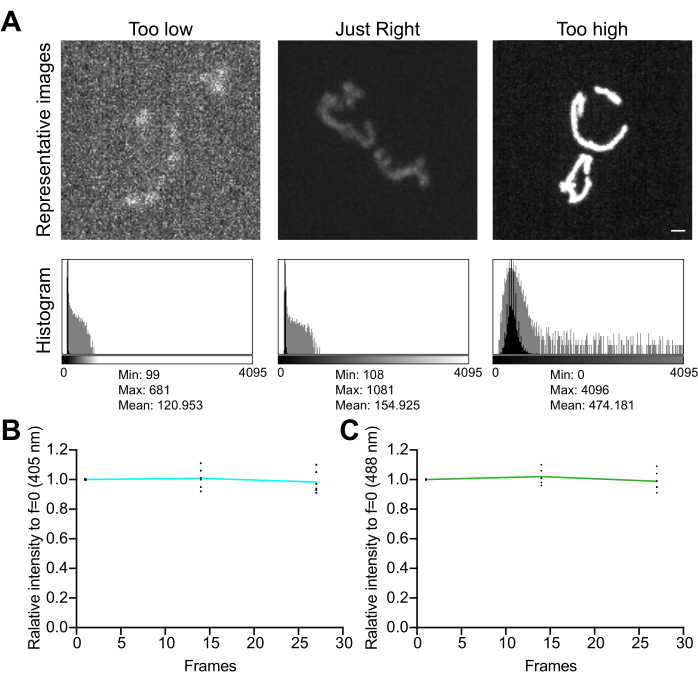
Figure 5: Optimization of imaging. (A) Evaluation of images and histograms for the correct intensity range. Maximum-intensity projections of spinning-disk confocal images are shown. The histograms are shown with both a linear Y axis (black) and a log-scale Y axis (gray) to illustrate the image dynamic range. Left panels: a noisy image with low intensity and dynamic range. Center panels: an image with an acceptable dynamic range (~1,000 gray levels) and intensity. Right panels: an image improperly contrast-enhanced to produce excessive saturation. Scale bar = 1 µm. (B,C) Analysis of photobleaching of mtHyPer7. Successive Z stacks were collected with no delay, Z stacks were summed, and the mean intensity of mitochondria was measured and normalized to the first time point. Results from the first three time points (27 exposures) are shown. (B) Excitation wavelength: 405 nm. (C) Excitation wavelength: 488 nm. The values shown are the averages of three cells from each of three independent trials. Please click here to view a larger version of this figure.
4. Semiautomated analysis with scripts
- Generate and measure ratio images using a biosensor analysis script (e.g., biosensor.ijm or biosensor-image-subtraction.ijm).
- Select the script to use. The protocol for using each script is similar; any differences will be specified in the text.
- biosensor.ijm: In this script, background and noise are corrected using user-selected image areas, fixed values, or no subtraction. Select methods for background and noise correction independently.
- biosensor-image-subtraction.ijm: In this script, background and noise are both handled by the same user-selected method, which may be one of the following: blank image, user-selected area within the image, fixed value, or no correction.
- In Fiji, open a multi-channel Z stack image acquired in step 3.5. Open the desired biosensor analysis script file. Run the script by clicking Run in the Script Editor window. In the dialog window that appears, enter the requested information.
- Select the channel numbers of the numerator and denominator for ratio calculation. For mtHyPer7, the numerator is the channel excited at 488 nm, and the denominator is the channel excited at 405 nm. Select the channel number of the transmitted-light image, if present, or 0 if there is none.
- Select the desired background subtraction method from the following four options. If the background is relatively uniform, choose Select an image area, which directly measures the background level in the image. If the background varies substantially across the field, use the biosensor-image-subtraction.ijm script and select Blank image (to collect a blank image for correction, capture a multichannel Z stack with identical acquisition conditions in a cell-free field of view, or from a slide made with cell-free growth medium). Fixed value allows entry of a previously measured background value. None leaves the background uncorrected, but may reduce the precision of the measurement.
- Select the desired noise subtraction method. The noise value is used as a lower threshold on the background-subtracted, masked channel images to reduce the effect of random variation in the detector readout. Opt for Select an image area or Fixed value to allow entry of a previously measured noise level, which usually works well as the noise levels are constant if the imaging conditions are kept constant. Alternatively, choose None and use a value of 1 as the lower threshold, which may increase the variability of the measurements.
- Select a thresholding algorithm to detect mitochondria accurately and consistently; Otsu or MaxEntropy are recommended. Ideally, use the same algorithm for all images in an experiment, but ensure the accurate recognition of mitochondria. Use a different thresholding method if there are changes in morphology during an experiment.
- Select the number of regions of interest (ROIs) per cell. For example, if measuring mother-bud differences, select 2.
- Select the output folder where measurements and ratio images will be saved.
- Follow the prompts for background and noise correction.
- Area selection (if applicable): If Select an image area was chosen for background or noise measurement, follow the prompts to draw a background area (outside any cells or fluorescent artifacts) using the rectangle ROI tool. After drawing the area, click OK.
- Fixed value entry (if applicable): If Fixed value was chosen for background or noise measurement, follow the prompts to enter background and/or noise values for each fluorescence channel.
- Blank reference image (if applicable): In the biosensor-image-subtraction.ijm script, if Blank image was chosen for background or noise correction, follow the prompts to select a blank image file.
- Mark ROIs for measurement. In mid-log phase cultures, restrict the analysis to budded cells.
- Draw ROIs corresponding to individual cells or subcellular regions based on the bright-field image. The ROI does not have to exactly match the cell outline, as only thresholded mitochondria within the ROI will be measured. Alternatively, open a previously saved ROI set: in the ROI Manager, click More, then select the ROI file.
- Press T after creating each ROI to add the selected ROI to the ROI Manager. In the ROI Manager, check Show All to document the cells that were marked. Each added region will appear as a numbered item in the ROI Manager list. If analyzing more than one ROI per cell (e.g., mother and bud), mark the ROIs in the same order for each cell analyzed.
- After all the desired ROIs have been added to the ROI Manager, click OK in the Mark cells dialog window.
- Choose the measurement table format. In the MultiMeasure dialog window that appears, check Measure all slices. Check the One row per slice option to produce a table with the desired format. Use the process_multiROI_tables.R script to process tables created with the One row per slice option; do not check Append Results. Repeat this step 3x (for measurement of the numerator [488], denominator [405], and ratio [488/405] images).
- The script will save the output files to the folder selected in step 4.1.2.6.
- Select the script to use. The protocol for using each script is similar; any differences will be specified in the text.
- Calculation of weighted mean ratios for each region or cell
- Using the results obtained in the previous step, calculate the weighted mean ratios for each region or cell using equation (2). Calculate either "pixelwise" or "regionwise" ratios (see the discussion for details).
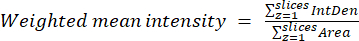 (2)
(2)
NOTE: The area and integrated density (IntDen) values include only the pixels within the thresholded mitochondria. This calculation can be automated using the process_multiROI_tables.R script and R statistical software.
- Using the results obtained in the previous step, calculate the weighted mean ratios for each region or cell using equation (2). Calculate either "pixelwise" or "regionwise" ratios (see the discussion for details).
- Generate a colorized ratio image. As changes in hue (color) are more obvious to the human eye than changes in intensity, convert the ratio value to a color scale for easier visual interpretation. The colorize_ratio_image.ijm script will colorize a masked ratio image.
- In Fiji, open a ratio Z stack image generated in step 4.1.6. Open the colorize_ratio_image.ijm script file. Run the script by clicking Run in the Script Editor window. In the dialog window that appears, enter the requested information.
- Colorization method: In the Unmodulated option, all mitochondrial pixels appear at the same brightness. However, some images may appear noisy as both dim and bright pixels contribute to the ratio image. To reduce this effect, use the Intensity modulated option.
- Minimum and maximum displayed value: These values control the range of ratio values that will be colorized. Choose values near the average minimum and maximum values observed in an experiment (based on the weighted mean ratios calculated in step 4.2.1). Within an experiment, be sure to acquire all images with identical imaging conditions and display all images with identical minimum and maximum values.
- Projection mode: Z stacks are displayed as projections to show the entire mitochondrial population. The projection is created before colorization. Select Max for a maximum-intensity projection (preserving the maximum ratio and intensity values) or Average for an average-intensity projection.
- Output folder: Select the folder in which to save the colorized images.
- If the Unmodulated option was selected, select a color scheme (lookup table; LUT) in the dialog window that appears. Use the Fire or Rainbow RGB LUTs built into Fiji, or any desired LUT in ImageJ's .lut format (Import from LUT file). The Rainbow Smooth40 LUT is recommended (included in Supplemental File 2 and on GitHub).
- The script saves the output files to the folder selected in step 4.3.1.4. These include the colorized ratio image (Color_RGB.tif) and the colorized ratio image with a calibration bar showing the correspondence between ratio values and image color (Color_with_bar.tif).
- In Fiji, open a ratio Z stack image generated in step 4.1.6. Open the colorize_ratio_image.ijm script file. Run the script by clicking Run in the Script Editor window. In the dialog window that appears, enter the requested information.
5. Manual analysis
NOTE: This approach takes more time but allows flexibility in preprocessing and setting the threshold.
- Correct for background.
- Set options in Fiji.
- Click on Analyze | Set Measurements. In the dialog window that appears, check the boxes for Area, Mean, StdDev, IntDen, and Display Label. Set the decimal places to 3.
- Click on Edit | Options | Input/Output. In the dialog window that appears, check the boxes for Save row numbers and Save column headers.
- To increase the sensitivity of the ratio calculation, subtract the background from each fluorescence channel. If the background is relatively uniform, measure the mean intensity of a cell-free area in an image, and subtract this value from the entire image (step 5.1.2.1). Alternatively, if the background varies substantially across the field, use a blank image for correction (step 5.1.2.2.).
- To subtract a user-selected region, click on Image | Color | Split channels, and keep all the channel images open, since they will be needed later. For each fluorescence channel, draw an ROI on the background (outside any cells) and click on Analyze | Measure, or for stacks, click Image | Stacks | Measure Stack. Observe the mean value of the ROI that appears in the Results table. Click on Edit | Select None, then Process | Math | Subtract. In the dialog window that appears, enter the measured mean background value, rounded to the nearest integer.
NOTE: Rounding prevents errors with binary operations later. - To subtract a blank image from a data file, collect a blank image from a completely cell-free field of view or from a slide prepared with cell-free growth medium. Click on Process | Image Calculator. In the dialog window that appears, set the operation to Subtract, and set Image1 and Image2 to the cell image and the blank image, respectively. Check Create New Window and 32-bit Result.
- To subtract a user-selected region, click on Image | Color | Split channels, and keep all the channel images open, since they will be needed later. For each fluorescence channel, draw an ROI on the background (outside any cells) and click on Analyze | Measure, or for stacks, click Image | Stacks | Measure Stack. Observe the mean value of the ROI that appears in the Results table. Click on Edit | Select None, then Process | Math | Subtract. In the dialog window that appears, enter the measured mean background value, rounded to the nearest integer.
- Set options in Fiji.
- To limit the analysis to mitochondria, perform segmentation. Because each channel may change intensity depending on the biosensor's state, use the sum of the two channels to define the area of mitochondria consistently.
- To create a sum image, click on Process | Image Calculator. In the dialog window that appears, set the operation to Add, and set Image1 and Image2 to the two background-subtracted fluorescent channels in any order. Check Create New Window and 32-bit Result and wait for a sum image to appear.
- Set the threshold to define mitochondria on the sum image.
- Click on Image | Adjust | Threshold. In the Threshold window that appears, check Dark Background and Stack Histogram. Set Method to the desired algorithm (e.g., Otsu or MaxEntropy). Employ automated thresholding for reproducibility, but if no automatic method is suitable, manually adjust the threshold.
NOTE: Ideally, the same algorithm should be used for all images in an experiment, but changes in morphology during an experiment might necessitate a different thresholding method in some conditions. - When the threshold is satisfactory, click Apply. In the dialog window that appears, choose Convert to Mask. In the dialog window that appears, check Black background and leave the other boxes unchecked. Save the resulting mask image for evaluation of the segmentation accuracy.
- Click on Image | Adjust | Threshold. In the Threshold window that appears, check Dark Background and Stack Histogram. Set Method to the desired algorithm (e.g., Otsu or MaxEntropy). Employ automated thresholding for reproducibility, but if no automatic method is suitable, manually adjust the threshold.
- Convert the mask values from 0 and 255 to 0 and 1, respectively, by clicking on Process | Math | Divide. Set the value to 255, and when prompted, select the option to process all images.
- Apply the mask to the background-subtracted fluorescence channels by clicking on Process | Image Calculator. In the dialog window that appears, set the operation to Multiply. Set Image1 and Image2 to the numerator channel and the mask, respectively. Check Create New Window and 32-bit Result; repeat the mask multiplication with the denominator channel.
- Prepare each masked channel for ratio calculation by setting the background pixels to NaN ("not a number").
- Select the masked numerator channel and click on Image | Adjust | Threshold. In the Threshold window, click Set, and in the dialog window that appears, set the minimum to the calculated noise level or to 1, and leave the maximum as it is.
- In the Threshold window, click Apply. In the next dialog box, choose Set to NaN. Repeat for the masked denominator channel.
- Generate the ratio image.
- Click on Process | Image Calculator. In the dialog window that appears, set operation to Divide. Set Image1 and Image2 to the background-subtracted masked numerator and denominator channels, respectively. For mtHyPer7, the numerator is the oxidized form excited at 488 nm, and the denominator is the reduced form excited at 405 nm. Therefore, higher ratios indicate higher H2O2.
- Check Create New Window and 32-bit Result. Save the ratio image for analysis.
- Mark and quantify areas of interest. Each cell or subcellular region is selected and stored as an ROI in the ROI Manager. The ROIs do not have to perfectly match the cell outlines, as only the masked mitochondria regions will be measured.
- Using the transmitted-light image to prevent bias, create ROIs corresponding to individual cells (or subcellular regions). In mid-log phase cultures, restrict the analysis to yeast cells bearing buds, which are actively engaged in cell division. Press T after creating each ROI to add it to the ROI Manager. Check Show All to document the cells that were marked.
- Click on the ratio image created above, and in the ROI Manager, click More … | MultiMeasure. In the dialog window that appears, check Measure all slices. Do not check Append Results. Check the One row per slice option to produce a table with the desired format. The process_multiROI_tables.R script will process tables created with the One row per slice option in the R statistical software.
- Save the results. Click on the Results window, then File | Save, and save the results table in .csv or .xls format. Set the filename to match the image name.
- Save the ROIs. In the ROI Manager, first make sure that no ROIs are selected: click Deselect, then More | Save. Open the saved ROIs along with the original image in Fiji to cross-reference the measurements with the image.
- Calculate the weighted mean ratios per cell or region by using the results obtained in the previous step and equation (3). Calculate either "pixelwise" or "regionwise" ratios (see the discussion for details).
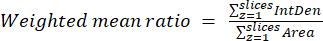 (3)
(3)
NOTE: The area and integrated density values include only the pixels within the thresholded mitochondria. This calculation can be automated using the process_multiROI_tables.R script in the R statistical software. - Generate a colorized ratio image.
NOTE: Colorized images can be unmodulated, where all mitochondrial pixels appear at the same brightness, or intensity-modulated, where pixel intensity in the original image is used to set intensities in the colorized image.- To produce an unmodulated colorized image:
- Open the ratio image generated above. Click on Image | Duplicate to generate a copy of the image, then close the original image.
- If the image is a Z stack, either select a single slice, or generate a Z projection to view all mitochondria. Use either a maximum-intensity (retaining the maximum ratio and intensity values at each XY pixel coordinate) or an average-intensity projection.
- Click on Image | Lookup Tables and set the LUT to the desired color scheme (e.g., Rainbow RGB or Fire [available by default in ImageJ]). Alternatively, click on File | Import LUT and select any desired LUT in ImageJ's .lut format, such as Rainbow Smooth40 (included in Supplemental File 2 and on GitHub). Make sure the LUT has a dark color assigned to the 0 value.
- Set the display contrast by clicking on Image | Adjust | Brightness/Contrast. In the Brightness/Contrast window, click Set, and in the dialog window that appears, enter the desired values for the Minimum and Maximum displayed values. To maximize the color differences observed, set these to approximately the minimum and maximum obtained across all images. Set all the images in an experiment to the same contrast levels.
NOTE: Do not click Apply, because this will change the pixel values and prevent the generation of an accurate calibration bar in the next step. - Add a color calibration bar by clicking on Analyze | Tools | Calibration bar. In the dialog window that appears, check the Overlay option to remove the bar, if desired, by clicking on Image | Overlay | Remove Overlay.
- If desired, add a scale bar by clicking on Analyze | Tools | Scale Bar. In the dialog window that appears, set the desired size, location, and color for the bar. Check the Overlay option to maintain the color of the bar regardless of the LUT used.
- Generate an RGB image for publication by clicking on Image | Overlay | Flatten. Save the resulting image.
NOTE: This image is only for presentation or publication. The intensity values are altered, so it cannot be measured. The bars are also permanently burned on the image.
- To create an intensity modulated image:
- Click on Image | Duplicate to generate a copy of the image, then close the original.
- If the image is a Z stack, either select a single slice or generate a Z projection to view all mitochondria.
- Set the display contrast of the ratio image by clicking on Image | Adjust | Brightness/Contrast, and in the Brightness/Contrast window, click Set. In the dialog window that appears, enter the desired values for the Minimum and Maximum displayed values. To maximize the color differences observed, set these to approximately the minimum and maximum obtained across all images, and set all the images in an experiment to the same contrast levels. Click Apply to rescale the pixel values.
- To create a calibration bar, duplicate this enhanced image, and generate a bar by navigating to Analyze | Tools | Calibration Bar. Convert the image and overlay to RGB format by clicking on Image | Overlay | Flatten. If desired, paste this bar on the intensity-modulated RGB image obtained in step 5.6.2.12 for presentation or publication.
- Create a new image with the same width and height as the image by clicking on File | New | Image…. In the dialog window that opens, set the parameters as follows: Type: RGB; Fill with: Black; Width and Height: 9width and height of the image); Slices: 1.
- Convert the new stack to an HSB (hue, saturation, brightness) image stack by clicking on Image | Type | HSB Stack.
- Click on the contrast-adjusted ratio image by navigating to Edit | Select All, then Edit | Copy. Then, click on the HSB stack, select the first slice (Hue), and click on Edit | Paste to transfer the ratio values.
- Select slice 2 (Saturation) of the HSB stack. Set the foreground color to white by navigating to Edit | Options | Colors. Click on Edit | Select All, then Edit | Fill, and in the dialog window that opens, select No to fill only the current slice with white.
- Open the raw data image and split the channels by clicking on Image | Color | Split Channels.
- Generate the average of the two ratio channels using the Image Calculator and clicking on Process | Image Calculator. In the dialog window that appears, set the operation to Average, and set Image1 and Image2 to the numerator and denominator images, respectively. Check Create New Window.
- Click on the average image created above by navigating to Edit | Select All, then Edit | Copy. Then, click on the HSB stack, select the third slice (Value), and click on Edit | Paste to transfer the intensity values.
- Convert the HSB stack into the RGB color space by clicking on Image | Type | RGB Color. Save the resulting image.
- To produce an unmodulated colorized image:
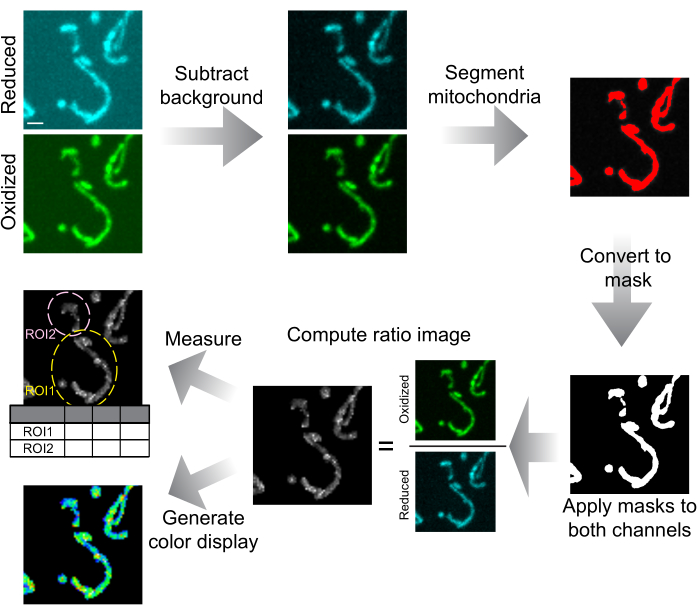
Figure 6: Schematic of image analysis and presentation. Spinning-disk confocal images are first background-subtracted. Mitochondria are segmented by thresholding and converted to masks for each slice. These masks are applied to both channels and the masked images are used to calculate the ratio image. ROIs are drawn to measure the mtHyPer7 ratio in cells or subcellular regions. Colorized ratio images can also be generated for data presentation. Scale bar = 1 µm. Abbreviation: ROI = region of interest. Please click here to view a larger version of this figure.
Representative Results
To confirm that mtHyPer7 is a suitable probe to assess mitochondrial H2O2, the localization of mtHyPer7 and its effect on yeast mitochondria and cell health were assessed. To assess the targeting of mtHyPer7, mitochondria were stained with the vital mitochondria-specific dye MitoTracker Red in control yeast cells and yeast that express mtHyPer7. Using MitoTracker Red staining, mitochondria were resolved as long tubular structures that aligned along the mother-bud axis and accumulated in the tip of the bud and the tip of the mother cell distal to the bud41. Mitochondrial morphology was similar in the control and mtHyPer7-expressing cells. In addition, mtHyPer7 co-localized with MitoTracker Red-stained mitochondria (Figure 3A). Thus, mtHyPer7 was efficiently and quantitatively targeted to mitochondria without affecting normal mitochondrial morphology or distribution.
Next, additional validation experiments were performed to assess the effect of mtHyPer7 on cellular fitness and sensitivity to oxidative stress in mitochondria. The growth rates of mtHyPer7-expressing cells in rich or synthetic glucose-based media (YPD or SC, respectively) are similar to those of untransformed wild-type cells (Figure 3B,D). To assess the possible effects on oxidative stress in mitochondria, control and mtHyPer7-expressing yeast were treated with low levels of paraquat, a redox-active small molecule that accumulates in mitochondria and results in elevated superoxide levels in the organelle24,27. If mtHyPer7 expression either induces oxidative stress in mitochondria or protects mitochondria from oxidative stress, then mtHyPer7-expressing cells should exhibit an increased or decreased sensitivity to paraquat treatment, respectively. Treatment with low levels of paraquat resulted in a decrease in yeast growth rates. Moreover, the growth rates of wild-type and mtHyPer7-expressing cells in the presence of paraquat were similar (Figure 3C,E). Therefore, expression of the biosensor did not create significant cellular stresses in budding yeast cells or alter the sensitivity of yeast to mitochondrial oxidative stress.
Given the evidence that mtHyPer7 is suitable for studying mitochondrial H2O2 in budding yeast, it was tested whether mtHyPer7 could detect mitochondrial H2O2 in mid-log phase wild-type yeast and changes in mitochondrial H2O2 induced by externally added H2O2. An H2O2 titration experiment was performed and the mean oxidized:reduced (O/R) mtHyPer7 ratio was measured in mitochondria.
The automated analysis scripts produced several output files.
The ratio image (ratio.tif): A Z stack consisting of the ratio of the background-corrected, thresholded 488 nm and 405 nm stacks. This stack can be viewed in Fiji without additional processing; however, enhancing the contrast and/or colorizing the image as described in protocol step 4.3 or 5.6 prior to viewing is recommended.
The mask image (mask.tif): A Z stack consisting of the thresholded mitochondrial areas used for analysis. This image must be used to evaluate the accuracy of the thresholding.
The ROIs used for analysis (ROIs.zip): When this file is opened in Fiji, along with the source image, the ROIs are superimposed on the image to cross-reference the results table and source image. Each ROI is renamed with a cell number and ROI number.
Measurement tables (NumResults.csv, DenomResults.csv, Results.csv) containing the area, mean, and integrated density of thresholded mitochondria for each slice in the numerator, denominator, and ratio images, respectively. In slices where mitochondria were absent or out of focus, NaN is recorded.
A log file (Log.txt) documenting the options used for background and noise correction, thresholding, and ratio calculation.
The O/R ratio of mtHyPer7 showed a dose-dependent response to H2O2 concentration, which reached a plateau at 1-2 mM externally added H2O2 (Figure 7). Surprisingly, the HyPer7 ratio was lower in the cells exposed to 0.1 mM H2O2 than in the control cells, although this difference was not statistically significant. One explanation for this phenomenon may be a hormesis response, in which exposure to a low level of stressor may induce stress responses, such as antioxidant mechanisms, which in turn lower the amount of ROS detectable by the probe. Higher levels of stressors, in contrast, may overwhelm the internal stress responses and result in a higher readout from HyPer7.
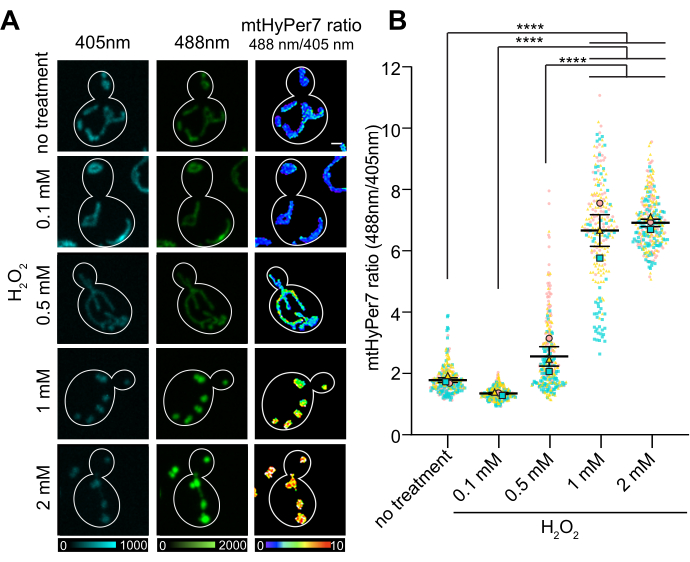
Figure 7: Response of mtHyPer7 to externally added H2O2. (A) Maximum projection of 405 nm and 488 nm excited images and average intensity projection of ratiometric images of mtHyPer7 in mid-log phase cells exposed to different concentrations of H2O2. Pseudocolor indicates the oxidized:reduced mtHyPer7 ratio (scale at bottom). Scale bar = 1 µm. The cell outline is shown in white; n > 100 cells per condition. (B) Quantification of the mtHyPer7 oxidized:reduced ratio in budding yeast cells treated with different concentrations of H2O2. The means of five independent trials are shown, with differently shaped and colored symbols for each trial. Mean ± SEM of the ratio of oxidized:reduced mtHyPer7: 1.794 ± 0.07627 (no treatment), 1.357 ± 0.03295 (0.1 mM), 2.571 ± 0.3186 (0.5 mM), 6.693 ± 0.5194 (1 mM), 7.017 ± 0.1197 (2 mM). p < 0.0001 (one-way ANOVA with Tukey's multiple comparisons test). p values are denoted as: ****p < 0.0001. Please click here to view a larger version of this figure.
Finally, previous studies revealed that fitter mitochondria, which are more reduced and contain less superoxide, are preferentially inherited by yeast daughter cells4, and that cytosolic catalases are transported to and activated in yeast daughter cells42,43,44,45. These studies document the asymmetric inheritance of mitochondria during yeast cell division and a role for this process in daughter cell fitness and lifespan, as well as mother-daughter age asymmetry. To test whether differences in H2O2 exist between mothers and buds, mtHyPer7 was measured in buds and mother cells. Differences in mitochondrial H2O2 were observed within yeast cells, and a subtle but statistically significant decrease in H2O2 biosensor readout was detected in mitochondria in the bud compared to those in the mother cell (Figure 8). These findings are consistent with previous findings that mitochondria in the bud are better protected from oxidative stress. They also provide documentation that mtHyPer7 can provide a quantitative readout for mitochondrial H2O2 with cellular and subcellular resolution in budding yeast.
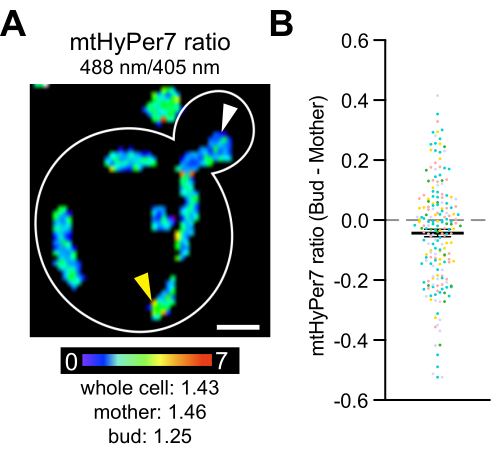
Figure 8: Mitochondrial H2O2 level is lower in the daughter cell. (A) Maximum projection of a ratiometric image of mtHyPer7 in a representative cell. Pseudocolor indicates the oxidized:reduced mtHyPer7 ratio (scale at bottom). Subcellular differences in the ratio are evident (arrowheads). Scale bar = 1 µm. Cell outline: white. (B) Difference between the mtHyPer7 ratio in the bud and mother cell. For each individual cell, the mother ratio value was subtracted from the bud ratio value and plotted as a point. n = 193 cells pooled from five independent experiments, shown with differently colored symbols for each experiment. Mean ± SEM of the difference between the mtHyPer7 ratio in the bud and mother cells: -0.04297 ± 0.01266. p = 0.0008 (paired t-test) Please click here to view a larger version of this figure.
Supplemental Table S1: Strains used in this study. List of yeast strains used. Please click here to download this File.
Supplemental Table S2: Plasmids used in this study. List of plasmids used. Please click here to download this File.
Supplemental Table S3: Primers used in this study. List of primers used. Please click here to download this File.
Supplemental File 1: Protocol for biosensor plasmid construction. Please click here to download this File.
Supplemental File 2: Scripts for automated analysis. For each script, sample input files and output files are supplied. Please click here to download this File.
Discussion
In this protocol, a method is described for using mtHyPer7 as a biosensor to assess mitochondrial H2O2 in living budding yeast cells. The biosensor is constructed using a CRISPR-based method and introduced into a conserved gene-free region in the yeast genome without the use of selectable markers. Compared to plasmid-borne biosensors, integrated ones are expressed in all cells and at consistent levels, providing more reliable quantification results. No selectable markers are used to generate mtHyPer7-expressing cells, which allows for use of a wider range of strain backgrounds and facilitates the genetic modification of biosensor-expressing cells. The mtHyPer7 protein is correctly targeted to mitochondria without noticeable effects on mitochondrial morphology, function, distribution, or cellular growth rates. mtHyPer7 shows a dose-dependent response to externally added H2O2. Moreover, mtHyPer7 is able to report on the heterogeneity of mitochondrial quality with subcellular resolution. Finally, using a spinning-disk confocal microscope as opposed to wide-field microscopy for imaging mitochondrial-targeted biosensors causes less photobleaching to fluorophores and generates high-resolution images for analyzing subcellular differences.
Limitations and alternative approaches
This method is not suitable for imaging cells for more than 10 min, as the cells will dry out under the coverslip. For longer-term imaging, it is better to use the agar pad method46 or to immobilize cells in a glass-bottom culture dish filled with SC medium.
The choice of biosensor should be guided by the concentration of the target under experimental conditions. If the sensitivity of HyPer7 is too high, a different HyPer version would be recommended, such as HyPer3 or HyPerRed47,48. However, it should be noted that other HyPer probes are more pH-sensitive. For higher sensitivity, the peroxiredoxin-based roGFP might be more appropriate (roGFP2Tsa2ΔCR)27.
The oxidation steady state of the H2O2 sensor is tied to both oxidation and reduction rates. The oxidation rate of biosensors is mainly caused by H2O2, but the reduction rate depends on the antioxidant reduction systems active in the cell and organelle. It has been shown that HyPer7 is predominantly reduced by the thioredoxin system in yeast cytosol, and its reduction is faster than that of roGFP2Tsa2ΔCR27. Therefore, the different reduction mechanisms and response dynamics of the probe should be taken into account when interpreting measurements of H2O2 biosensors. In particular, to infer H2O2 levels from the biosensor readout, it must be assumed that the reduction system maintains a constant capacity during the experiment. As an alternative to the scripts described here, a variety of other software has been made freely available for the analysis of redox sensors49.
Critical steps
With any biosensor, it is critical to demonstrate that the biosensor itself does not affect the process being measured. Therefore, comparing the growth and mitochondrial morphology of strains under each experimental condition is important. Here, mitochondrial morphology is assessed using MitoTracker Red, which stains mitochondria in a membrane potential-dependent manner. However, comparison of the mitochondria in untransformed and biosensor-transformed cells can be accomplished by staining with tetramethylrhodamine methyl ester (TMRM), an alternative membrane potential-sensing mitochondrial vital dye, or MitoTracker Green, which stains mitochondria independent of membrane potential. If deleterious effects are suspected, reducing the expression level or changing the integration site may help.
Validating the dose-response behavior of the probe and the signal-to-noise ratio of the imaging technique are also essential for collecting robust results. If the variability within a group exceeds the variability between groups, differences become difficult to detect. Intragroup variability may result from true population variation, or from noise in the detection process. The key steps to increase the signal:noise ratio are image acquisition (pixel value range and noise), background subtraction, and thresholding.
Noise effects can also be reduced during the calculation steps. The most straightforward approach is to calculate the weighted mean intensity from the ratio image measurements (Results.csv), where each pixel represents the local ratio between the excitation efficiencies. This produces a "pixelwise" ratio. However, if the image signal:noise ratio is low, more robust results can be obtained by calculating the weighted mean intensity for an ROI in both the numerator and the denominator channels, and then calculating the ratio between these two weighted means ("regionwise" ratio).
To select a thresholding method, the Fiji command Image | Adjust | Auto Threshold can be used to automatically try all built-in Fiji methods. To evaluate segmentation (thresholding), a saved mask is converted into a selection by clicking on Edit | Selection | Create Selection, added to the ROI Manager (by pressing T), and then activated on the raw image file. If mitochondria are not adequately detected, a different segmentation method should be attempted.
When comparing images, it is essential to acquire all the images with identical imaging conditions, as well as to display all the images with identical contrast enhancement.
Mitochondrial movement must be considered when optimizing imaging conditions. If the mitochondria move significantly between excitation at 405 and 488 nm, the ratio image will not be accurate. It is recommended to keep the exposure time <500 ms and to change excitation by the fastest method available (e.g. a trigger pulse or acousto-optic tunable filter). When capturing a Z stack, both excitations should be performed for each Z step before moving to the next Z step.
For display of the results, changes in hue (color) are more obvious to the human eye than changes in intensity. Therefore, the ratio value is converted to a color scale for easier visual interpretation. Colorized images can be unmodulated, where all the mitochondrial pixels appear at the same brightness, or intensity-modulated, where pixel intensity in the original image is used to set intensities in the colorized image.
Modification and troubleshooting
As an alternative to confirming mitochondrial function by challenge with paraquat, cells may be replica-plated or inoculated into fermentable and non-fermentable carbon sources.
For background subtraction, rolling ball subtraction (by navigating to Process | Subtract Background…) may also be used to remove illumination nonuniformity. It should be ensured that the presence of cells does not alter the background being subtracted (by checking the option to Create Background and examining the result).
In summary, the mtHyPer7 probe provides a consistent, minimally invasive method to relate the morphological and functional status of yeast mitochondria in living cells, and allows the study of an important cellular stressor and signaling molecule in a genetically tractable, easily accessible model system.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors thank Katherine Filpo Lopez for expert technical assistance. This work was supported by grants from the National Institutes of Health (NIH) (GM122589 and AG051047) to LP.
These studies used the Confocal and Specialized Microscopy Shared Resource of the Herbert Irving Comprehensive Cancer Center at Columbia University, funded in part through the NIH/NCI Cancer Center Support Grant P30CA013696.
Materials
| 100x/1.45 Plan Apo Lambda objective lens | Nikon | MRD01905 | |
| Adenine sulfate | Sigma-Aldrich | #A9126 | |
| Bacto Agar | BD Difco | #DF0145170 | |
| Bacto Peptone | BD Difco | #DF0118170 | |
| Bacto Tryptone | BD Difco | #DF211705 | |
| Bacto Yeast Extract | BD Difco | #DF0127179 | |
| BamHI | New England Biolabs | R0136S | |
| BglII | New England Biolabs | R0144S | |
| Carbenicilin | Sigma-Aldrich | C1389 | |
| Carl Zeiss Immersol Immersion Oil | Carl Zeiss | 444960 | |
| Dextrose (D-(+)-Glucose) | Sigma-Aldrich | #G8270 | |
| E. cloni 10G chemical competent cell | Bioserch Technologies | 60108 | |
| FIJI | NIH | Schindelin et al 2012 | |
| G418 (Geneticin) | Sigma-Aldrich | A1720 | |
| GFP emission filter | Chroma | 525/50 | |
| Gibson assembly | New England Biolabs | E2611 | |
| Graphpad Prism 7 | GraphPad | https://www.graphpad.com/scientific-software/prism/ | |
| H2O2 (stable) | Sigma-Aldrich | H1009 | |
| HO-pGPD-mito-roGFP-KanMX6-HO | Pon Lab | JYE057/EP41 | Liao et al 20201 |
| Incubator Shaker | New Brunswick Scientific | E24 | |
| KAPA HiFi PCR kit | Roche Sequencing and Life Science, Kapa Biosystems, Wilmington, MA | KK1006 | |
| L-arginine hydrochloride | Sigma-Aldrich | #A8094 | |
| laser | Agilent | 405 and 488 nm | |
| L-histidine hydrochloride | Sigma-Aldrich | #H5659 | |
| L-leucine | Sigma-Aldrich | #L8912 | |
| L-lysine hydrochloride | Sigma-Aldrich | #L8662 | |
| L-methionine | Sigma-Aldrich | #M9625 | |
| L-phenylalanine | Sigma-Aldrich | #P5482 | |
| L-tryptophan | Sigma-Aldrich | #T8941 | |
| L-tyrosine | Sigma-Aldrich | #T8566 | |
| mHyPer7 plasmid | This study | JYE116 | |
| Microscope coverslips | ThermoScientific | 3406 | #1.5 (170 µm thickness) |
| Microscope slides | ThermoScientific | 3050 | |
| MitoTracker Red CM-H2Xros | ThermoFisherScientific | M7513 | |
| NaCl | Sigma-Aldrich | S9888 | |
| NEBuilder HiFi Assembly Master Mix | New England Biolabs | E2621 | |
| Nikon Elements | Nikon | Microscope acquisition software | |
| Nikon Ti Eclipse inverted microscope | Nikon | ||
| Paraquat (Methyl viologen dichloride hydrate) | Sigma-Aldrich | Cat. #856177 | |
| RStudio | Posit.co | Free desktop version | |
| Spectrophotometer | Beckman | BU530 | |
| Stagetop incubator | Tokai Hit | INU | |
| Uracil | Sigma-Aldrich | #U1128 | |
| Yeast nitrogen base (YNB) containing ammonium sulfate without amino acids | BD Difco | #DF0919073 | |
| YN2_1_LT58_X2site | Addgene | 177705 | Pianale et al 2021 |
| Zyla 4.2 sCMOS camera | Andor |
References
- vander Bliek, A. M., Sedensky, M. M., Morgan, P. G. Cell biology of the mitochondrion. Genetics. 207 (3), 843-871 (2017).
- McBride, H. M., Neuspiel, M., Wasiak, S. Mitochondria: more than just a powerhouse. Current Biology. 16 (14), 551-560 (2006).
- Shi, R., Hou, W., Wang, Z. -. Q., Xu, X. Biogenesis of iron-sulfur clusters and their role in DNA metabolism. Frontiers in Cell and Developmental Biology. 9, 735678 (2021).
- McFaline-Figueroa, J. R., et al. Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age asymmetry in budding yeast. Aging Cell. 10 (5), 885-895 (2011).
- Higuchi-Sanabria, R., et al. Mitochondrial anchorage and fusion contribute to mitochondrial inheritance and quality control in the budding yeast Saccharomyces cerevisiae. Molecular Biology of the Cell. 27 (5), 776-787 (2016).
- Higuchi-Sanabria, R., et al. Role of asymmetric cell division in lifespan control in Saccharomyces cerevisiae. FEMS Yeast Research. 14 (8), 1133-1146 (2014).
- Lam, Y. T., Aung-Htut, M. T., Lim, Y. L., Yang, H., Dawes, I. W. Changes in reactive oxygen species begin early during replicative aging of Saccharomyces cerevisiae cells. Free Radical Biology & Medicine. 50 (8), 963-970 (2011).
- Laun, P., et al. Aged mother cells of Saccharomyces cerevisiae show markers of oxidative stress and apoptosis. Molecular Microbiology. 39 (5), 1166-1173 (2001).
- Doudican, N. A., Song, B., Shadel, G. S., Doetsch, P. W. Oxidative DNA damage causes mitochondrial genomic instability in Saccharomyces cerevisiae. Molecular and Cellular Biology. 25 (12), 5196-5204 (2005).
- Roca-Portoles, A., Tait, S. W. G. Mitochondrial quality control: from molecule to organelle. Cellular and Molecular Life Sciences. 78 (8), 3853-3866 (2021).
- Sies, H., Berndt, C., Jones, D. P. Oxidative stress. Annual Review of Biochemistry. 86, 715-748 (2017).
- Sies, H., Jones, D. P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nature Reviews Molecular Cell Biology. 21 (7), 363-383 (2020).
- Imlay, J. A., Fridovich, I. Assay of metabolic superoxide production in Escherichia coli. The Journal of Biological Chemistry. 266 (11), 6957-6965 (1991).
- Fridovich, I. Mitochondria: are they the seat of senescence. Aging Cell. 3 (1), 13-16 (2004).
- Quinlan, C. L., Perevoshchikova, I. V., Hey-Mogensen, M., Orr, A. L., Brand, M. D. Sites of reactive oxygen species generation by mitochondria oxidizing different substrates. Redox Biology. 1 (1), 304-312 (2013).
- Griendling, K. K., Minieri, C. A., Ollerenshaw, J. D., Alexander, R. W. Angiotensin II stimulates NADH and NADPH oxidase activity in cultured vascular smooth muscle cells. Circulation Research. 74 (6), 1141-1148 (1994).
- Griendling, K. K., Sorescu, D., Ushio-Fukai, M. NAD(P)H oxidase: role in cardiovascular biology and disease. Circulation Research. 86 (5), 494-501 (2000).
- Edmondson, D. E., Binda, C., Wang, J., Upadhyay, A. K., Mattevi, A. Molecular and mechanistic properties of the membrane-bound mitochondrial monoamine oxidases. Biochemistry. 48 (20), 4220-4230 (2009).
- Ramsay, R. R., Singer, T. P. The kinetic mechanisms of monoamine oxidases A and B. Biochemical Society Transactions. 19 (1), 219-223 (1991).
- Ramsay, R. R. Kinetic mechanism of monoamine oxidase A. Biochemistry. 30 (18), 4624-4629 (1991).
- Handy, D. E., Loscalzo, J. Redox regulation of mitochondrial function. Antioxidants & Redox Signaling. 16 (11), 1323-1367 (2012).
- Wood, Z. A., Schröder, E., Robin Harris, J., Poole, L. B. Structure, mechanism and regulation of peroxiredoxins. Trends in Biochemical Sciences. 28 (1), 32-40 (2003).
- Slade, L., et al. Examination of the superoxide/hydrogen peroxide forming and quenching potential of mouse liver mitochondria. Biochimica et Biophysica Acta. General Subjects. 1861 (8), 1960-1969 (2017).
- Pak, V. V., et al. Ultrasensitive genetically encoded indicator for hydrogen peroxide identifies roles for the oxidant in cell migration and mitochondrial function. Cell Metabolism. 31 (3), 642-653 (2020).
- Topell, S., Hennecke, J., Glockshuber, R. Circularly permuted variants of the green fluorescent protein. FEBS Letters. 457 (2), 283-289 (1999).
- Belousov, V. V., et al. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nature Methods. 3 (4), 281-286 (2006).
- Kritsiligkou, P., Shen, T. K., Dick, T. P. A comparison of Prx- and OxyR-based H2O2 probes expressed in S. cerevisiae. The Journal of Biological Chemistry. 297 (1), 100866 (2021).
- Baird, G. S., Zacharias, D. A., Tsien, R. Y. Circular permutation and receptor insertion within green fluorescent proteins. Proceedings of the National Academy of Sciences. 96 (20), 11241-11246 (1999).
- Abedi, M. R., Caponigro, G., Kamb, A. Green fluorescent protein as a scaffold for intracellular presentation of peptides. Nucleic Acids Research. 26 (2), 623-630 (1998).
- Onukwufor, J. O., et al. A reversible mitochondrial complex I thiol switch mediates hypoxic avoidance behavior in C. elegans. Nature Communications. 13 (1), 2403 (2022).
- Vega, M., et al. Antagonistic effects of mitochondrial matrix and intermembrane space proteases on yeast aging. BMC Biology. 20 (1), 160 (2022).
- Torello Pianale, L., Rugbjerg, P., Olsson, L. Real-time monitoring of the yeast intracellular state during bioprocesses with a toolbox of biosensors. Frontiers in Microbiology. 12, 802169 (2022).
- Imani, M., Mohajeri, N., Rastegar, M., Zarghami, N. Recent advances in FRET-based biosensors for biomedical applications. Analytical Biochemistry. 630, 114323 (2021).
- Zadran, S., et al. Fluorescence resonance energy transfer (FRET)-based biosensors: visualizing cellular dynamics and bioenergetics. Applied Microbiology and Biotechnology. 96 (4), 895-902 (2012).
- Gietz, R. D., Woods, R. A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods in Enzymology. 350, 87-96 (2002).
- Liao, P. -. C., Wolken, D. M. A., Serrano, E., Srivastava, P., Pon, L. A. Mitochondria-associated degradation pathway (MAD) function beyond the outer membrane. Cell Reports. 32 (2), 107902 (2020).
- Higuchi-Sanabria, R., Swayne, T. C., Boldogh, I. R., Pon, L. A. Live-cell imaging of mitochondria and the actin cytoskeleton in budding yeast. Methods in Molecular Biology. 1365, 25-62 (2016).
- Liao, P. -. C., Yang, E. J., Pon, L. A. Live-cell imaging of mitochondrial redox state in yeast cells. STAR Protocols. 1 (3), 100160 (2020).
- Schindelin, J., et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Collins, T. J. ImageJ for microscopy. BioTechniques. 43, 25-30 (2007).
- Chazotte, B. Labeling mitochondria with MitoTracker dyes. Cold Spring Harbor Protocols. 2011 (8), 990-992 (2011).
- Aguilaniu, H., Gustafsson, L., Rigoulet, M., Nyström, T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 299 (5613), 1751-1753 (2003).
- Erjavec, N., Larsson, L., Grantham, J., Nyström, T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes & Development. 21 (19), 2410-2421 (2007).
- Erjavec, N., Cvijovic, M., Klipp, E., Nyström, T. Selective benefits of damage partitioning in unicellular systems and its effects on aging. Proceedings of the National Academy of Sciences. 105 (48), 18764-18769 (2008).
- Erjavec, N., Nyström, T. Sir2p-dependent protein segregation gives rise to a superior reactive oxygen species management in the progeny of Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences. 104 (26), 10877-10881 (2007).
- Davidson, R., Liu, Y., Gerien, K. S., Wu, J. Q. Real-time visualization and quantification of contractile ring proteins in single living cells. Methods in Molecular Biology. 1369, 9-23 (2016).
- Bilan, D. S., et al. HyPer-3: a genetically encoded H2O2 probe with improved performance for ratiometric and fluorescence lifetime imaging. ACS Chemical Biology. 8 (3), 535-542 (2013).
- Ermakova, Y. G., et al. Red fluorescent genetically encoded indicator for intracellular hydrogen peroxide. Nature Communications. 5 (1), 5222 (2014).
- Fricker, M. D. Quantitative redox imaging software. Antioxidants & Redox Signaling. 24 (13), 752-762 (2016).
- Yang, E. J., Pernice, W. M., Pon, L. A. A role for cell polarity in lifespan and mitochondrial quality control in the budding yeast Saccharomyces cerevisiae. iSCIENCE. 25 (3), 103957 (2022).

