Bacterial Co-Incubation Assay: A Fluorescence Microscopy-Based Technique to Visualize Intraspecific Bacterial Competition at the Single-Cell Level
Abstract
Source: Smith, S., et al., Quantification of Interbacterial Competition using Single-Cell Fluorescence Imaging. J. Vis. Exp. (2021).
This video demonstrates a fluorescence microscopy-based method to study bacterial competition at the single-cell level. This method provides insights into the structure and function of bacterial communities.
Protocol
1. Coincubate bacterial strains
- Choose two bacterial strains for single-cell bacterial competition assays. Here, two strains of V. fischeri are used: a target strain (ES11424) and an inhibitor strain (MJ1125) that is known to kill the target strain using the type VI secretion system on chromosome II (T6SS2)1, which is a contact-dependent killing mechanism.
- Transform strains with stable plasmids encoding genes for different fluorescent proteins (e.g., GFP or RFP) to visually distinguish strain types on the microscope. Here, the inhibitor strain is tagged with a GFP-encoding plasmid (pVSV102), and the target strain is tagged with a dsRed-encoding plasmid (pVSV208).
- Start with mid-log cultures for both strains, measure and record the optical density at 600 nm (OD600) for all samples.
- Normalize each sample to an OD600 = 1.0, which corresponds to approximately 109 CFU/mL for V. fischeri, by diluting the culture with LBS medium.
- Mix the two competing strains together at a 1:1 ratio based on volume by adding 30 µL of each normalized strain to a labeled 1.5 mL tube. Vortex the mixed-strain culture for 1-2 s.
NOTE: In some cases, it may be appropriate to mix cocultures in different ratios. For example, when one strain grows much faster than the other, it may be necessary to start the slower-growing strain at a numerical advantage in order to observe the competition. Optimization may also be required if OD600 does not correspond to similar CFU/mL for both strains. - Repeat step 1.5 for each biological replicate and treatment. In the example shown here, this will result in a total of four mixed-strain tubes: two biological replicates with the wild-type inhibitor strain mixed with the target strain and two biological replicates with the type VI secretion system mutant strain mixed with the target strain.
- To ensure competing cells are sufficiently dense for contact-dependent killing in the coincubation on the agar pad, concentrate each mixed culture 3-fold by centrifuging the mixed culture in a standard 1.5 mL centrifuge tube for 1 min at 21,130 x g, discarding the supernatant, and resuspending each pellet in 20 µL LBS medium. Repeat for each sample.
NOTE: Some bacterial cells are sensitive to damage by centrifugation at high rcf; in such cases, the mixed culture can be centrifuged for 3 min at 4600 x g. Additionally, when quantifying contact-dependent competition, it is important to ensure sufficient cell density on the slide to observe killing. In this article, "crowded" treatments, where killing is observed, had approximately 10 cells/20 µm2.
2. Slide setup
- When using an upright microscope, place a ~5 mm2 agarose pad onto a standard 1 mm glass slide. Spot 2 µL of a mixed culture onto the agarose pad and place a #1.5 coverslip (25 mm2) over the spot. See Figure 1B.
- When using an inverted microscope, spot 2 µL of a mixed culture onto the #1.5 coverslip bottom of a 35 mm Petri dish and place a ~5 mm2 agarose pad over the coincubation spot. Place a 12 mm circular glass coverslip over the agarose pad. See Figure 1C for an example.
- Repeat steps 2.1 or 2.2, depending on the microscope setup used, for the remaining three mixed cultures, resulting in four slides or dishes to be imaged.
- Allow slides to sit on the benchtop for approximately 5 min before proceeding. This allows cells to settle on the agar pad and eliminate movement during the imaging process.
3. Fluorescence microscopy
- Begin by focusing on cells using white light (phase contrast or DIC) to minimize the effects of photo-bleaching. Based on the average size of a single bacterial cell, use a 60x or 100x oil objective.
- Adjust the exposure time and acquisition settings for each channel so that cells are visible in the appropriate channel with minimal background detection.
NOTE: It is appropriate to use different exposure times for different channels, but the same exposure time should be used across all biological replicates and treatments for a given channel. - For each sample, select at least five fields of view (FOV) and acquire images in each appropriate channel using the acquisition settings (See examples in Figure 2). Save the XY points from each FOV so that the same FOV can be imaged during the final time point. Imaging the same FOV at each time point is necessary to determine the proportion of area occupied by target or inhibitor cells during the analysis steps.
NOTE: In this example, the fluorescence of GFP is detected using a filter with an excitation wavelength of 467 – 498 nm and an emission filter of 513 – 556 nm and is false-colored green. The Fluorescence of dsRed is detected using a filter with an excitation wavelength of 542 – 582 nm and an emission filter of 603 – 678 nm and is false-colored magenta. - After 2 h, repeat the above step for each sample using the previously saved XY points (Figure 2).
NOTE: The timing of subsequent images may need to be optimized for organisms with different growth rates or competitive mechanisms.
Representative Results
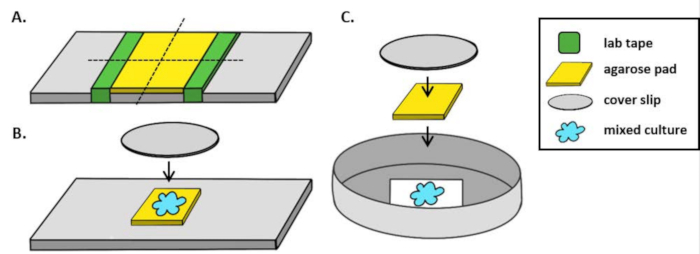
Figure 1: Agarose pad preparation and slide setup for coincubation assays. (A) Setup for making 2% agarose pads. Five layers of lab tape (green) are wrapped around a cover slip at two points approximately 20 mm apart. Next, warm 2% agarose in marine Phosphate-Buffered Saline (yellow) is pipetted between the pieces of tape and immediately covered with a 25 mm2 cover slip and allowed to solidify for at least 1 h at room temperature. Use a razor blade to cut the agarose pad into ~5 mm2 pieces and use tweezers to transfer the pad to a new slide for imaging. (B) When imaging on an upright microscope, place the 5 mm2 agarose pad directly onto the slide, followed the mixed culture (blue) and a 12 mm circular #1.5 coverslip. (C) When imaging on an inverted microscope, spot the mixed culture directly onto the #1.5 glass coverslip bottom of a 35 mm Petri dish and place an agarose pad on top of the culture followed by a second 12 mm circular coverslip to flatten the agarose pad.
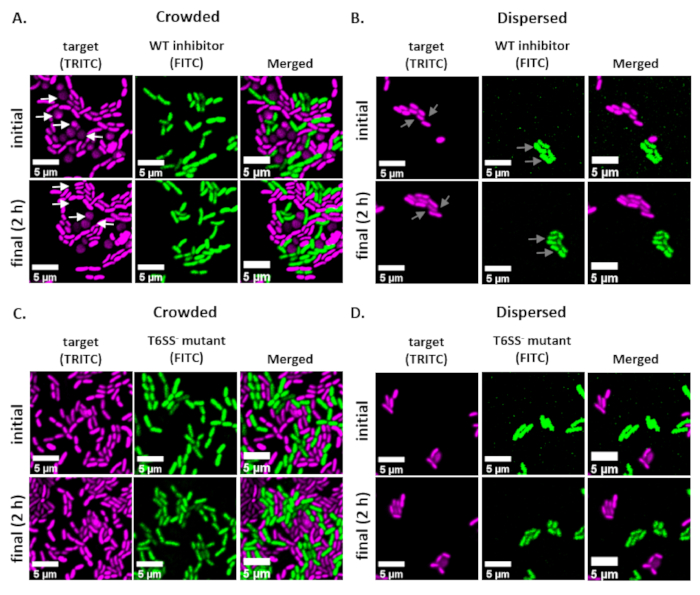
Figure 2: Time-lapse images of coincubation spots in either crowded or disperse conditions. (A) Representative images at initial and final time points where a mixed culture of target and wild-type inhibitor was concentrated 3x prior to spotting on the slide to force cell-cell contact between strains. White arrows in TRITC channel indicate examples of target cells that round or lyse throughout the course of the experiment. (B) Representative images where a mixed culture of target and wild-type inhibitor was spotted without concentrating so that cells are disperse and there is minimal cell-cell contact between strains. Gray arrows in FITC and TRITC channels indicate examples of cell division throughout the course of the experiment. (C) Representative images where mixed culture of target and T6SS– mutant was concentrated 3x prior to spotting on the slide to force cell-cell contact between strains. (D) Representative images where mixed culture of target and T6SS– mutant was spotted without concentrating so that cells are disperse and there is minimal cell-cell contact between strains. Scale bars = 5 µm and are consistent across all images; TRITC channel is false-colored magenta, FITC channel is false-colored green. Deconvolution was performed on all images; background was subtracted, and brightness/contrast adjusted uniformly across all images.
Disclosures
The authors have nothing to disclose.
Materials
| 1.5 mL Microcentrifuge tube | Fisher | 05-408-129 | |
| 10 uL single channel pipette | |||
| 1000 uL single channel pipette | |||
| 20 uL single channel pipette | |||
| 200 uL single channel pipette | |||
| Agarose | Fisher | BP165-25 | Low melting agarose |
| Cellvis 35 mm Dish | Fisher | NC0409658 | #1.5 cover glass bottom |
| Chloramphenicol | Sigma | C0378 | stock (20 mg/mL in Ethanol); final concentration in media (2 μg /mL LBS) |
| FIJI image analysis sofware | ImageJ | https://imagej.net/Fiji/Downloads | open-source software |
| Fisherbrand Cover Glasses: Circles | Fisher | 12-545-81P | #1.5 cover glass; 12 mm diameter |
| Kanamycin Sulfate | Fisher | BP906-5 | stock (100 mg/mL in water, filter sterilize); final concentration in media (1 μg/mL LBS) |
| Lens Cleaning Tissue Paper | Fisher | S24530 | |
| Petri Plates | Fisher | FB0875713 | sterile with lid |
| Razor Blades | Fisher | S65921 | |
| Semi-micro Cuvettes | VWR | 97000-586 | |
| Spectrophotometer | |||
| Thermo Scientific Gold Seal Plain Microscope Slides | Fisher | 12-518-100B | |
| Thermo Scientific Richard-Allan Scientific Cover Glass | Fisher | 22-050-235 | #1.5 cover glass, 25 mm2 |
| Type F Immersion Oil | Fisher | NC0297589 | |
| Upright or inverted fluorescence microscope with camera and imaging software | Images in this article were acquired on a Nikon TI-2 inverted fluorescent microscope outfitted with an ORCA-Fusion Digital CMOS camera using NIS-Elements software. | ||
| Vortex | |||
| Water bath | Used to keep agarose warm prior to pipetting | ||
| LBS media | |||
| 1M Tris Buffer (pH ~7.5) | 50 mL 1 M stock buffer (62 mL HCl, 938 mL DI water, 121 g Trizma Base) | ||
| Agar Technical | Fisher | DF0812-17-9 | 15 g (Add only for plates) |
| DI water | 950 mL | ||
| Sodium Chloride | Fisher | S640-3 | 20 g |
| Tryptone | Fisher | BP97265 | 10 g |
| Yeast Extract | Fisher | BP9727-2 | 5 g |
| mPBS (marine PBS) | Phosphate buffered saline with marine salts added; used for making agarose pad | ||
| 10X PBS | Fisher | ICN1960454 | |
| Instant Ocean Sea Salt | Instant Ocean | SS1-160P | Adjust concentration to appropriate salinity; 20 psu used here |
| Sterile Vacuum Filter Units | Fisher | SCGVU01RE | Used to filter-sterilize mPBS |
| Vacuum pump | Used to filter-sterilize mPBS |

